Abstract
In higher plants phloem and xylem are responsible for long-distance transport of water, nutrients, and signals that act systemically at short or long-distance to coordinate developmental processes. The formation of the plant vascular system is a complex process that integrates signaling events and gene regulation at transcriptional and posttranscriptional levels. Thanks to transcriptomic and proteomic analysis we start to better understand the mechanisms underlying the formation and the functioning of the vascular system. The role of the DNA-binding with one finger (Dof TFs), a group of plant-specific transcription factors, recently emerged as part of the transcriptional regulatory networks acting on the formation and functioning of the vascular tissues. More than half of the members of this TF family are expressed in the vascular system. In addition some of them have been proposed to be mobile proteins, suggesting a possible role in the control of short- or long-distance signaling as well. This review summarizes the current knowledge on Dof TFs family in Arabidopsis with a special focus on their role in vascular development and functioning.
Keywords: transcription factors, DNA-binding with one finger, vascular system, Arabidopsis
Introduction
The gradual colonization of the land through plant evolution has been possible thanks to the differentiation of complex vascular tissues that provide mechanical support and allow long-distance transport of water and nutrients. In land plants, vascular tissues are comprised of two conducting tissues the phloem and the xylem, and intervening cambium that can generate the different conducting cell types (Scarpella and Helariutta, 2010). Whereas the xylem, whose vessels are dead cells with highly lignified walls, ensures the transport of water, mineral salts, and some hormones, the phloem controls the transport and distribution of photosynthetic products from leaves to meristems and other sink organs such as fruits, tubers, and roots. The sieve elements (SE) are the phloem conducting cells and are associated with one or a few CCs. In general the meristematic mother cell of a CC-SE complex divides unequally in the longitudinal axis. One daughter cell develops into one or several metabolically hyperactive CCs with a dense cytoplasm and numerous mitochondria. The second one goes through a controlled disintegration process and differentiates into the SE, which is a highly specialized and unique developmental program (Juergensen et al., 2003). In the last 15 years, major insights have been obtained in the identification of the regulatory actors controlling the establishment of the vascular system as well as the fate of the vascular cell types (reviewed in Zhou et al., 2011). Many transcription factors including bHLH, HD-ZIP, ARF-GAP, AT-Hook, and Dof proteins families were shown to be involved, suggesting the existence of complex transcriptional regulatory networks in which transcription factors are likely to physically interact as shown recently by several bHLH and Dof TFs (Zhang et al., 1995; Kang and Singh, 2001; Skirycz et al., 2008). Some of these families of transcription factors are conserved among eukaryotes but the Dof TFs family, which is the focus of this review, is of particular interest for being specific to plants.
The first DNA-binding with one f inger (Dof) transcription factor has been identified in maize (ZmDof1) and was shown to be involved in light response and transcriptional regulation of genes involved in carbon metabolism (Yanagisawa and Sheen, 1998; Yanagisawa, 2001). Dof TFs are plant-specific and are characterized by a particular zinc finger domain, comprising a conserved region of 50 amino acids with a C2–C2 finger structure, associated to a basic region, that binds specifically to DNA sequences with a 5′-(A/T)AAAG-3′ core (Figure 1). A particular feature of these TFs is that the nucleus localization signal (NLS), which directs Dof proteins to the nucleus in Arabidopsis thaliana (Krebs et al., 2010), is an atypical bipartite NLS with a 17 amino acid long linker between its flanking basic regions (Figure 1). This bipartite NLS is highly conserved in plant Dof transcription factors (Krebs et al., 2010) and its feature allowed identifying Dof genes in a various set of plant species (for details, see below). Along plant evolution, Dof transcription factors might have originated from a common ancestor, likely represented by the single Chlamydomonas reinhardtii gene, and then expanded in the different taxonomic groups of vascular plants through recurrent duplication events (Moreno-Risueno et al., 2007). In Gymnosperms and lower plants (Selaginella moellendorffii and Physcomitrella patens) so far analyzed eight to nine Dof TF genes are found (Moreno-Risueno et al., 2007). In Angiosperms an average of 30 genes are found with 27 in Brachypodium distachyon (Hernando-Amado et al., 2012), 30 in rice (Oryza sativa) (Gaur et al., 2011), 36 Dof genes in Arabidopsis (reviewed in Yanagisawa and Schmidt, 1999; Moreno-Risueno et al., 2007), 37 in tomato (Solanum lycopersicon) (Cai et al., 2013), or 41 in poplar (Populus trichocarpa) (Yang and Tuskan, 2006), indicating additional recent duplications in higher plants (Moreno-Risueno et al., 2007). This diversification appears to be associated to the establishment and differentiation of the vascular system during the same period of time, which raises the question whether members of this plant-specific transcription factor family are involved in the formation of the vascular system or its functioning. Indeed besides their involvement in the regulation of different processes such as carbon assimilation (Yanagisawa, 2001; Tanaka et al., 2009), light signaling (Park et al., 2003; Ward et al., 2005; Gabriele et al., 2010), seed development or germination (Papi et al., 2000; Gualberti et al., 2002; Dong et al., 2007; Rueda-Romero et al., 2012), flowering (Sawa et al., 2007; Fornara et al., 2009), stomata functioning (Gardner et al., 2009; Negi et al., 2013), or response to phytohormones (Kang and Singh, 2001; Kang et al., 2003; Nakano et al., 2006) most of them are strikingly expressed in the vascular tissues suggesting a function in long-distance signaling (Gualberti et al., 2002; Papi et al., 2002; Ward et al., 2005; Fornara et al., 2009; Rueda-Romero et al., 2012). Recently several Dof TFs were shown to have a central role in the development of the vascular system (Konishi and Yanagisawa, 2007; Guo et al., 2009; Gardiner et al., 2010; Kim et al., 2010). Based on these observations it is possible to speculate that the plant-specific Dof TFs are likely to regulate directly or indirectly a wealth of processes associated to the development of the vascular system or to its role in long-distance signaling. In the present review we aim at summarizing the current knowledge about the Dof TF family highlighting their role in the control of vascular development and functioning.
Figure 1.
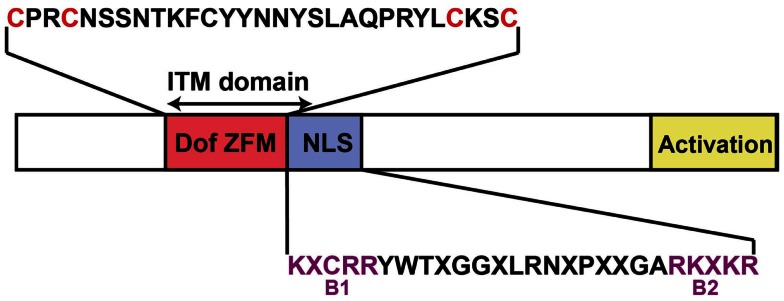
Schematic representation of an Arabidopsis Dof transcription factor structure. The Dof Zing Finger Motif (Dof ZFM), the nuclear localization signal (NLS), and the activation domain are indicated in pink, purple, and yellow respectively. The cysteine residues for putative coordination of zinc are shown in red letters in the Dof domain amino acid sequences. The consensus sequences of the bipartite NLS (B1 and B2 basic regions in purple letters) are based on Krebs et al. (2010). X represents any amino acids. The double-ended arrow indicated the intercellular trafficking motif (ITM) that confers the capacity to mediate selective intercellular trafficking (Chen et al., 2013). This motif includes the ZFM and the first motif of the bipartite NLS.
All Arabidopsis Dof TFs are Potentially Expressed in the Vascular Tissues
In Arabidopsis thaliana, out of the 36 Dof TFs, 20 have been found expressed in the vascular tissues (Table 1 and references there in). In silico studies also indicate the presence of cis-acting elements potentially driving specific expression in the vascular tissues in the promoter of these genes. In plants, mainly through 5′-deletion experiments, several positive cis-acting promoter elements have been identified to drive vascular-specific expression of reporter genes. The study of the rice tungro bacilliform virus (RTVB) present only in the phloem of infected plants allowed the identification of a virus promoter functional in plants (Bhattacharyya Pakrasi et al., 1993; Yin and Beachy, 1995). This promoter contains four domains allowing a strong phloem-specific expression (Yin et al., 1997a,b): a GATA motif which had already been shown to be implicated in light dependent phloem-specific gene expression in plants (Lam and Chua, 1989); an ASL box characterized by the presence of a specific GCA direct repeat sequence present only in phloem-specific promoters and missing in xylem-specific promoters; a Box II containing CCA/TGG repeat and a CCCC sequence that because of their presence in the promoter of plant genes expressed in vascular tissue might be important cis-elements for expression in vascular tissues. These three cis-elements, Box II, the ASL box, and the GATA motif were shown to act in combination (synergistically and/or additively) to confer phloem-specific expression (Yin et al., 1997a). Similarly the study of the coconut foliar decay virus (CFDV) led to the identification of a 13 bp motif sequence (Hehn and Rohde, 1998) which is highly conserved in several phloem-specific promoters as reported by Yoshida et al. (2002). Finally, the 26 bp RSE regulatory element was found to drive a vascular-specific expression of the GRP1 (Glycine-rich protein1) (Keller and Baumgartner, 1991). All these motifs act synergistically or additively and Yin et al. (1997a) have suggested that at least two of these motifs are required for phloem expression. More recently, Ruiz-Medrano et al. (2011) identified degenerate sequences containing CT/GA- and GT/GA-rich repeats within many of the promoters of 150 Arabidopsis genes homologous to members of the pumpkin phloem transcriptome. Interestingly the CT/GA motifs resemble cognate sites for the Dof family of transcription factors, which bind to the core sequence AAAG/CTTT (Higo et al., 1999; Yanagisawa and Schmidt, 1999).
Table 1.
Arabidopsis Dof genes phylogeny and nomenclature.
 |
Searching for these cis-elements in the 1000 bp upstream sequences of the Arabidopsis Dof TF family members we showed that at least two of these vascular-specific motifs were found in the promoter region of 31 Dof genes display (Table 2). Among the remaining five, the promoter of AtDof2.1 and AtDof1.8 genes contain three and five GATA motifs respectively and the promoter of AtDof4.4 contains two ASL boxes (Table 2). These observations suggest that the vast majority, if not all, of Arabidopsis Dof transcription factors are likely expressed in the vascular system (Table 2).
Table 2.
Synthesis of the predicted vascular-related regulatory cis-elements motifs in AtDof promoters.
| AtDof genes | Predicted Regulatory c/s-Elements1 |
Transcriptomic databases2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Box II motif | ASL box motif | GATA motif | 13 bp motif | RSE motif | Ph | PC | VB | |
| AtDofl.1 | 1 | – | 1 | – | – | |||
| AtDof1.2 | – | 2 | 4 | – | – | |||
| AtDof1.3 | – | 4 | 5 | – | – | |||
| AtDof1.4 | – | 1 | 6 | 1 | – | |||
| AtDof1.5 | – | 2 | 6 | – | – | |||
| AtDof1.6 | – | – | 1 | – | – | |||
| AtDofl.7 | – | – | 1 | – | – | |||
| AtDof1.8 | – | – | 5 | – | – | |||
| AtDofl.10 | – | – | 1 | 2 | – | |||
| AtDof2.1 | – | – | 3 | – | – | |||
| AtDof2.2 | – | 3 | 5 | – | – | |||
| AtDof2.3 | – | 2 | 4 | – | – | |||
| AtDof2.4 | – | 1 | 5 | – | – | |||
| AtDof2.5 | – | 2 | 3 | – | – | |||
| AtDof3.1 | – | – | 1 | – | – | |||
| AtDof3.2 | – | 1 | 2 | – | – | |||
| AtDof3.3 | – | 1 | 2 | 1 | – | |||
| AtDof3.4 | – | 1 | 2 | – | – | |||
| AtDof3.5 | – | 2 | 4 | 1 | – | |||
| AtDof3.6 | – | 2 | 3 | – | – | |||
| AtDof3.7 | – | – | 4 | – | – | |||
| AtDof4.1 | – | – | 4 | 1 | – | |||
| AtDof4.2 | – | 1 | 2 | – | – | |||
| AtDof4.3 | – | 1 | 1 | – | – | |||
| AtDof4.4 | – | 2 | – | – | – | |||
| AtDof4.5 | – | 2 | 2 | – | – | |||
| AtDof4.6 | – | 1 | 2 | 1 | – | |||
| AtDof4.7 | – | 1 | 5 | – | – | |||
| AtDof5.1 | – | 1 | 3 | – | – | |||
| AtDof5.2 | – | – | 3 | 1 | – | |||
| AtDof5.3 | – | 1 | 1 | 2 | – | |||
| AtDof5.4 | – | 1 | 3 | – | – | |||
| AtDof5.5 | – | 4 | 2 | – | – | |||
| AtDof5.6 | – | 1 | 2 | 1 | – | |||
| AtDof5.7 | – | – | 2 | – | 1 | |||
| AtDof5.8 | – | 2 | 4 | – | – | |||
1Box II motif: TGGNCCCCN (Yin et al., 1997a,b); ASL box: GCA N(20) GCA (Yin et al., 1997a); GATA motif: WNMNGATA (Gilmartin et al., 1990; Yin et al., 1997a); 13bp motif: ATAAGXAXXXXGA (Yoshida et al., 2002); RSE motif: GXXXXXXACTTTCXTXT (Hatton et al., 1995; Hauffe et al., 1993; Keller and Baumgartner, 1991).
2AtDof genes found in the transcriptomic databases related to the vascular system of Arabidopsis (Birnbaum et al., 2003; Zhao et al., 2005: Mustroph et al., 2009; Gandotra et al., 2013), Poplar (Schrader et al., 2004), and Celery (Vilaine et al., 2003).
Grey color indicates the tissue in which the gene has been found to be expressed.
Ph, phloem; PC, procambium; VB, vascular bundle.
The mining of RNA profiling datasets at the resolution of the vascular tissue (phloem, cambium, xylem) and vascular cell type (CC, SE) (Hertzberg et al., 2001; Birnbaum et al., 2003; Vilaine et al., 2003; Schrader et al., 2004; Zhao et al., 2005; Lee et al., 2006; Brady et al., 2007; Ruiz-Medrano et al., 2011; Gandotra et al., 2013) further indicate an expression of most Dof genes in the vascular system. Indeed, the expression of Dof TF genes could be found, either in the xylem, or the phloem or both (Table 2). AtDof1.1, AtDof2.2, AtDof3.2, AtDof3.6, AtDof3.7, AtDof5.3, and AtDof5.5 were identified in the global phloem cells transcript databases established in Arabidopsis (Birnbaum et al., 2003; Zhao et al., 2005). In addition poplar orthologs of AtDof1.6, AtDof3.4, AtDof4.6 were found in the transcriptomic analysis of the wood forming region (Hertzberg et al., 2001; Schrader et al., 2004) and the celery orthologs of AtDof1.8 and AtDof4.6 were expressed in the phloem of celery (Vilaine et al., 2003), suggesting that AtDof1.6 and AtDof1.8 which promoter contains one and five GATA motifs respectively could also be expressed in the vascular region. Besides these genes and the ones already experimentally described as expressed as well in the vascular system (Table 1), the recent transcriptome analysis of microdissected provascular/procambial cells or complete vascular bundle by Gandotra et al. (2013) allowed identifying new AtDof genes (AtDof1.3, AtDof1.4, AtDof1.7, AtDof1.8, AtDof2.2, AtDof3.1, AtDof4.5, and AtDof4.7) potentially expressed in vascular tissue at different step of its development.
In conclusion it is likely that almost all the members of the Arabidopsis Dof TFs family are at least expressed in the vascular system, being so far one of the TF family displaying such a strong tissue-specificity.
AtDof Transcription Factors are Potential Mobile Transcription Factors
Coordinated growth and development of multi-cellular organisms require mechanisms that allow for extensive cell-to-cell communication which can be mediated by signaling molecules directly transported from one cell to the other. Sessile nature of plant growth also necessitated the evolution of mechanisms that rapidly transmit signaling molecules in response to environmental changes or pathogen attacks. These mobile signals in plants can be proteins, RNAs, small RNAs, and other small molecules. They are transmitted through the vascular system in a long-distance (between organs), or through plasmodesmata in a short distance (between cell types) (Lucas et al., 2009; Dinant and Suárez-López, 2012).
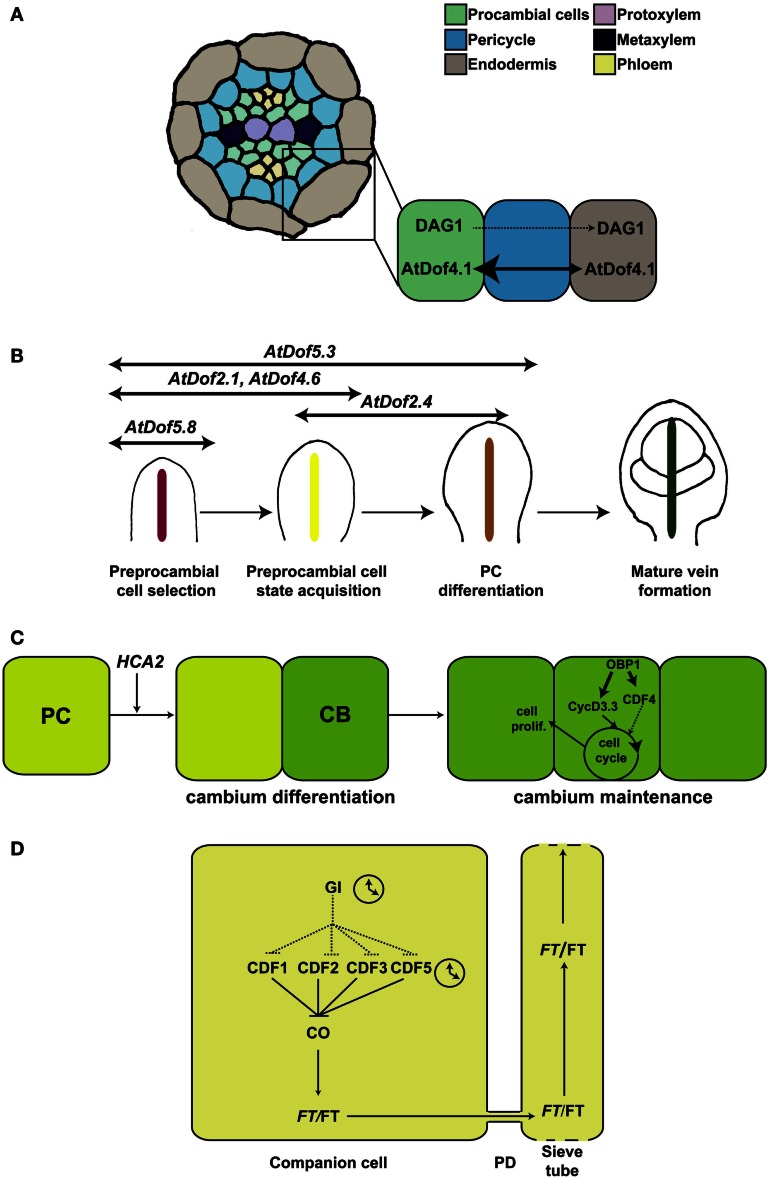
By comparing the translational and transcriptional patterns of several TFs, including Dofs, expressed in a cell-type specific manner in Arabidopsis root, Lee et al. (2006) found that the translational pattern was broader or different than the transcriptional pattern for two Dof TFs, AtDof3.7/DAG1 and AtDof4.1, suggesting that these TFs could move out from the cell/tissue they were expressed. In the case of AtDof4.1 the transcriptional expression domain was contained in the translational one indicating that the expansion in the expression pattern must occur after the synthesis of the mRNA (from pericycle to endodermis, Figure 2A). Thus it is likely that it occurred via cell-to-cell protein movement, as shown for the CAPRICE protein (Kurata et al., 2005). For AtDof3.7/DAG1, because the transcriptional AtDof3.7prom:GFP pattern (in the stele, Figure 2A) was shown expressed in a different tissue compared the one described in the published root mRNA expression map (Birnbaum et al., 2003) the protein might not be expressed in its native tissue. Nevertheless, the expansion of the pattern is unlikely to be because of mRNA movement since the mRNA and the translational expression pattern were also different (transcriptional and translation fusions expressed in the stele and endodermis cell layer respectively; Figure 2A) (Lee et al., 2006).
Figure 2.
The Dof TFs are involved in different steps of plant development. (A) Non-cell-autonomous function of DAG1 and AtDof4.1 in the root. Schematic representation of an Arabidopsis root showing the different cell types. The black box focuses on stele, pericycle, and endodermis in which the cell-to-cell movement of DAG1 (Lee et al., 2006) (dashed line) and AtDof4.1 (Chen et al., 2013) (plain line) occur. The arrow and its size indicate the direction and the intensity of the protein movement respectively. (B) Pattern of expression of several Dof TFs at early stage of formation of the leaf vascular tissues. The PIN1 expression domain defines the preprocambial cell selection zones (purple). Then the preprocambial cell state acquisition is controlled by the expression of ATBH-8 and SHR transcription factors (yellow). The preprocambial cells will give rise gradually to the procambium (PC) cells (brown) that will differentiate into phloem and xylem tissues and form the mature vein (green). The involvement of the AtDof s gene in these processes is essentially based on the colocalization of their expression patterns and those of the already known regulators PIN1 and ATHB-8. (C) Hypothetic role of Dof TFs for cell cycle during cambium formation/maintenance. The transcription factor AtDof5.6/HCA2 has been suggested to be involved in the transition between the procambium (PC) and the cambium (CB), and the AtDof3.6/OBP3 is supposed to control cambium maintenance through the control of the cell cycle. (D) Schematic representation for the role of CFD1, 2, 3, and 5 on the GI/CO/FT signaling pathways. In the phloem companion cell some of the steps of the photoperiodic flowering pathway are taking place. The core of this pathway is composed of GIGANTEA (GI), CONSTANS (CO), and FLOWERING LOCUS T (FT). In the sieve tube, the arrow indicates the movement of FT/FT as well as the sap flux direction. The clock symbol next to GI and CDF genes indicates that their expression is under the control of the circadian clock. PD, plasmodesmata.
In addition, cell-type specific expression maps from Arabidopsis root and cell-type specific translatome data were produced by several groups (Birnbaum et al., 2003; Nawy et al., 2005; Lee et al., 2006; Brady et al., 2007; Mustroph et al., 2009) and represent an extremely valuable source of information for predicting putative mobile transcription factors. By analyzing these data sets Lee and Zhou (2012) further proposed that seven Dof TFs (AtDof1.1, AtDof2.1, AtDof2.2, AtDof2.4, AtDof3.2/DOF6, AtDof3.6/OBP3, AtDof3.7/DAG1) are potential mobile transcription factor RNAs.
A direct experimental proof of the intercellular movement of a non-cell-autonomous Dof TFs has recently been brought by Chen et al. (2013) who showed that AtDof4.1/IDT1 (INTERCELLULAR TRAFFICKING DOF 1, IDT1) moves between the root stele and the endodermis in a plasmodesmata-selective pathway. This movement seems to be possible in either ways but more efficient from the endodermis to the stelar tissues (Figure 2A) (Chen et al., 2013). Moreover, the authors demonstrated that IDT1 movement requires an intercellular trafficking motif (ITM) comprising the Zinc Finger motif and the nuclear localization signal (NLS (Figure 1) (Chen et al., 2013). By looking for the existence of an ITM sequence in the different members of the Dof TF family, Chen et al. (2013) also showed that in contrast to AtDof5.4 and AtDof2.2, AtDof3.7/DAG1, and AtDof4.5 were also able to move from cell to cell.
In conclusion, some of the Dof TFs are able to move from cell to cell, however, although several Dof genes have been found expressed in the phloem, there is so far no evidence for long-distance transport since neither RNAs nor Dof proteins have been found in transcriptome and proteome datasets of phloem sap (Giavalisco et al., 2006; Deeken et al., 2008; Kehr and Buhtz, 2008).
AtDof Transcription Factors are Involved in the Control of Vascular Development
Control of the procambium formation during leaf development
Vascular development is initiated by the procambium formation, which is followed by the differentiation of the procambium into phloem and xylem. It is now well established that in the leaf primordia the procambium formation begins with the specification of undifferentiated ground meristem cells, by still unknown positioning information, that follows redirection of polar auxin flow to distinct convergent points by the auxin efflux carrier AtPIN1 (Scarpella et al., 2006). The ATHB-8 gene [a Class III homeodomain-leucine zipper (HD-ZIP III)], a differentiation-promoting transcription factor of the vascular system (Baima et al., 1995), follows the expression of AtPIN1 and the consequent restricted routes for auxin transport that define the sites of procambium formation (Scarpella et al., 2006), defining ATHB-8 as one of the earliest marker for the acquisition of procambial cell identity. Recently, several Dof transcription factors were shown to be expressed during the very early steps of procambium formation. Konishi and Yanagisawa (2007) showed that the GUS expression driven by the AtDof5.8 promoter was similar to that of PIN1 at very early stages of vascular development either in embryos, young leaf primordia, roots, or flower buds. These observations suggest that AtDof5.8 might play a role in the de novo formation of procambium from ground meristem cells and in the very early processes of vasculature formation (Konishi and Yanagisawa, 2007) (Figure 2B). The same authors showed that the timing of expression of AtDof2.4, in contrast to AtDof5.8, was quite similar to that of the ATHB-8 gene during procambium formation (Baima et al., 1995; Scarpella et al., 2004) suggesting that AtDof2.4 might play a role in the processes following the primary formation of procambium in various organs (Figure 2B). AtDof2.1, AtDof4.6, and AtDof5.3 are expressed in the root vascular system (Birnbaum et al., 2003) but recently, Gardiner et al. (2010) showed that during leaf development they are also expressed in overlapping subepidermal domains with a comparable dynamics. Expression of AtDof2.1 and AtDof4.6 was sustained at all stages of vein formation, while that of AtDof5.3 stopped during procambium differentiation. The expression domains of AtDof2.1, AtDof4.6, and AtDof5.3 overlapped with that of ATHB-8 expression, suggesting that these three Dof TFs are expressed at preprocambial stages (Gardiner et al., 2010). AtDof2.1, AtDof4.6, and AtDof5.3 expression was always initiated in wide domains that became laterally confined over time while ATHB-8 expression domains are always narrower and comprised in the Dof expression domain. In addition the fact that discrete AtDof2.1, AtDof4.6, and AtDof5.3 expression domains do not show ATHB-8 expression may suggest that the expression of AtDof2.1, AtDof4.6, and AtDof5.3 is initiated prior to the acquisition of the ATHB-8 preprocambial cell state (Figure 2B) (Gardiner et al., 2010).
AtDof Genes Control Vascular and Interfascicular Cambium Formation and Activity
The vascular cambium is the lateral meristem source of secondary xylem and secondary phloem. The fascicular cambium, initiated from the procambium, is present between the xylem and the phloem of a vascular bundle. Additionally an interfascicular cambium can also appear during secondary growth, giving rise to interfascicular fibers in the floral stem in species like Arabidopsis. By studying the Arabidopsis gain-of-function mutant high cambial activity2 (hca2), which shows a precocious formation of the interfascicular cambium and its subsequent cell division, Guo et al. (2009) showed that the phenotype was due to the overexpression of AtDof5.6 gene. Repression of AtDof5.6/HCA2 activity led to the disruption of interfascicular cambium formation and development in inflorescence stems. HCA2 promotes interfascicular cambium formation at a very early stage of inflorescence stem development. In addition Guo et al. (2009) showed that AtDof5.6/HCA2 is preferentially expressed in the vasculature of all the organs, but more particularly in the cambium, the phloem, and the interfascicular parenchyma cells of inflorescence stems. In the hca2 mutant high cambial activity and high phloem proliferation activity were detected not only in inflorescence stems, but also in petioles and leaf major veins suggesting that, in addition to interfascicular cambium formation/development, AtDof5.6/HCA2 may also be involved in other developmental aspects during vascular tissue development, for instance, (pro)cambium initiation and activity (Figure 2C).
Additionally two Dof TFs (AtDof3.4/OBP1 and AtDof2.3) are suspected to be involved in the control of the maintenance of the cambium and/or cambial activity by controlling the cell cycle (Skirycz et al., 2008) (Figure 2C). AtDof3.4/OBP1 is highly expressed in tissue with high cell proliferation activity (cell cultures, calli, developing embryo, apical, and root meristems, fascicular procambium in the stem), and while looking for direct targets of AtDof3.4/OBP1 several core cell cycle genes and transcription factors were identified (Skirycz et al., 2008). OBP1 modulates cell cycle activity by affecting the expression of CDKA regulators, S-phase specific transcription factors and components of the replication machinery, and is therefore important for cell cycle onset. OBP1 most likely operates through a direct regulation of CYCD3.3 and AtDof2.3 gene expression since it was shown to interact physically with their respective promoter of AtDof2.3 and CYCD3.3 (Skirycz et al., 2008) (Figure 2C).
AtDof TFs are Involved in the Regulation of Long-Distance Signaling
AtDof TFs regulate the photoperiodic flowering response
In order to adapt to the seasonal changes, plants synchronize their developmental program to be able to flower during the longest days of the year. The molecular genetics studies have allowed identifying actors involved in this photoperiodic flowering pathway. The circadian-clock controlling flowering signaling pathway takes place in the phloem CC, and recruits GIGANTEA (GI), CONSTANS (CO), and FLOWERING LOCUS T (FT) genes (Kobayashi and Weigel, 2007; Turck et al., 2008). The clock protein CO, which is expressed in the CC, induces FT transcription, and the FT protein moves long-distance through the phloem sap to reach the meristem where it triggers flowering (Corbesier et al., 2007). Additional actors in this pathway were identified by screening a library of Arabidopsis transcription factors systematically expressed in CC (Imlau et al., 1999; Fornara et al., 2009). These transcription factors, that all belong to the subfamily A of Dof TFs family (Moreno-Risueno et al., 2007), were named CYCLING DOF FACTOR (CDF), suggesting that this clade would almost be exclusively involved in flowering response (Table 1). Five Dof TFs genes (AtDof5.5/CDF1, AtDof5.2/CDF2, AtDof3.3/CDF3, AtDof2.3/CDF4, and AtDof1.10/CDF5) have been shown to be expressed in the vasculature throughout the plant and were shown to negatively regulate the transcription of CO (Imaizumi et al., 2005; Fornara et al., 2009) (Figure 2D). Although these Dof TFs were not shown to move in the phloem sap, they indirectly act on long-distance signaling by regulating the expression of CO.
AtDof TFs contribute to the regulation of light signaling
Yanagisawa and Sheen (1998) reported for the first time the role of a Dof TF in light signaling. They showed that the mRNA level of the maize gene ZmDof1 and its ability to bind DNA were regulated by light. In Arabidopsis, four Dof TFs have been suggested to be involved in photomorphogenesis processes like germination and hypocotyl growth (Papi et al., 2000, 2002; Gualberti et al., 2002; Park et al., 2003; Ward et al., 2005). During seed germination, light is required to convert the inactive Pr form of phytochrome into the active Pfr that will trigger the germination. In this context, the knockout mutant in AtDof3.7/DAG1 requires substantially less phytochrome signaling than the wild type to germinate (Papi et al., 2002). Later on, a null mutant in a gene closely related to DAG1 and named AtDof2.5/DAG2, has been characterized by Gualberti et al. (2002). They showed that DAG2 substantially reduced the capacity of seeds to germinate in the absence of light, while DAG1 enhanced the dark germination (Gualberti et al., 2002). These two Dof TFs have therefore opposite effects during seed germination in response to light and are acting as repressor and activator respectively (Papi et al., 2000, 2002; Gualberti et al., 2002). Interestingly both these transcription factors are expressed specifically in the vascular system but neither in the seed or in the embryo as one could expect (Gualberti et al., 2002; Papi et al., 2002) suggesting that they indirectly control long-distance light related signaling pathways involved in embryo and seed development.
Two other Dof TFs, AtDof1.5/COG1 and AtDof3.6/OBP3, have been shown to impact the seedling morphogenesis and especially the hypocotyl elongation which is also a light signaling related process. Park et al. (2003) showed that the activation tagged mutant cog1-D, in which AtDof1.5/COG1 gene is overexpressed, displays a longer hypocotyl prominently under red and far-red lights. They suggested that COG1 is a negative regulator in the phyA- and phyB-signaling pathways (Park et al., 2003). On the contrary, OBP3 gene, which accumulates in the vascular tissues during the light period, is suggested to act as a positive regulator of the phyB-signaling pathway (Ward et al., 2005). In addition, Ward et al. (2005) showed that OBP3 negatively regulate the cryptochrome-mediated seedling photomorphogenesis.
AtDof TFs potentially control the phloem sugar transport
In Arabidopsis, a plant species that mainly transports sucrose for long-distance allocation of carbohydrates, the transition from sink-leaf to source-leaf is paralleled by the expression of AtSUC2 that encodes a companion-cell (CC) specific H+-sucrose symporter (Truernit and Sauer, 1995; Stadler and Sauer, 1996). In 2008, an elegant 5′-deletion experiment combined with an in silico analysis of the AtSUC2 promoter, allowed identifying several cis-regulatory sequences in the minimal promoter sequence required to drive AtSUC2 vascular expression, including three Dof-binding motifs and one motif recognized by HD-ZIP transcription factor (Schneidereit et al., 2008). Only the simultaneous deletion of at least one Dof-binding motif and the HD-ZIP binding site resulted in the loss of AtSUC2 promoter activity, suggesting that a Dof TF in coordination with an HD-ZIP regulates the expression of the SUC2 gene. So far no Dof TFs involved in AtSUC2 transcriptional regulation have been identified in Arabidopsis. However, it has been clearly established that the ZmDof1 and ZmDof2 from maize control the expression of genes involved in carbon metabolism (Yanagisawa, 2001). Their two orthologs in Arabidopsis AtDof1.4 and AtDof3.5 could thus be proposed to regulate AtSUC2 expression, which would give new insights in the fine-tuning of the sink-source transition.
Toward an Integrated Transcriptional Network for the Control of Vascular Development and/or Functioning
Structurally, like for other zinc fingers, the Dof domain is known to be a bi-functional domain that mediates not only DNA-binding but also protein–protein interactions (Yanagisawa, 2002). When looking at cis-elements present in Dof TFs promoters, one can observe that each of the promoter displays at least one cis-element related to other transcription factor (bHLH, MADS boxes, ARF, Homeobox, MYB, bZIP, WRKY) (for more details, see the AGRIS interface, http://arabidopsis.med.ohio-state.edu/AtcisDB/). Altogether these data suggest that interactions between Dof TFs and TFs belonging to other families are theoretically possible. Experimentally, OBP1 was shown to directly interact with the promoter of a bZIP transcription factor but also with the promoter of a member of the same family AtDof2.3 (Zhang et al., 1995; Kang and Singh, 2001; Skirycz et al., 2008). Moreover Kang et al. (2003) showed that two bHLH members (bHLH038 and bHLH039) are targets of the vascular expressed OBP3 transcription factor. Interestingly, bHLH039 was also found to be highly expressed in phloem tissue during hypocotyl secondary growth in Arabidopsis (Zhao et al., 2005). In the regulation of leaf axial patterning, a direct binding of AtDof5.1 on the promoter of the HD-ZIP, REVOLUTA have also been demonstrated (Kim et al., 2010). Besides these interactions at the transcriptional level, protein–protein interactions between the Dof TFs, CDF1, CDF2, and CDF3 and the F-box FKF1 have also been observed in the frame of the photoperiodic flowering response (Imaizumi et al., 2005). These few examples illustrate the fact that Dof TFs and TFs of other families (bHLH, bZIP, HD-ZIP, ARF, F-box) have the potential to interact together at the transcriptional as well as at the protein level and therefore be part of transcriptional networks involved in different developmental processes such as vascular development. Such transcriptional network, involving a bHLH complex, has been recently shown to control the establishment of the embryonic vascular tissue (De Rybel et al., 2013). This discovery opens a new level of complexity in the molecular mechanisms underlying the vascular development. Similar complexes involving Dof TFs could also be speculated. Indeed besides TARGET OF MONOPTEROS (TMO5/bHLH135), a Dof TF (AtDof5.3/TMO6) has been also identified as a potential indirect target of MP transcription factor (Schlereth et al., 2010) and has been described as one of the transcription factors participating in Arabidopsis vein formation (Gardiner et al., 2010). TMO6 could therefore be a good candidate to explore the involvement of Dof TFs in the regulation of the early steps of procambium formation. Altogether, TMO6 as well as the other Dof TFs mentioned in this review could constitute the starting point to study the potential implication of the Dof TFs in the transcriptional networks underlying vascular development.
Conclusion and Future Prospects
In the context of the vascular system development and functioning, which integrates a large variety of internal or external stimuli, regulators are needed to fine-tune the responses to changes in the environment. In this review, we present evidences that the Dof TFs family is likely to participate in such fine-tuning of the responses to the different stimuli in the vascular system. By modulating a variety of transcription factors, by protein-DNA and/or protein–protein interactions, the Dof TFs could therefore act at the cross-talk of various developmental pathways directly or indirectly linked to the vascular development and/or functioning. One of the next challenges, besides the functional characterization of the still undescribed Dofs TFs, will therefore be identifying the actors involved in an integrated regulatory network. Combination of the high-throughput data collection at the resolution of the vascular cell types and developmental stages as well as the use of the next generation sequencing technology will certainly allow identifying genes network regulating the fundamental features of the vascular development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank S. Dinant for stimulating discussions and critical reading of the manuscript.
References
- Baima S., Nobili F., Sessa G., Lucchetti S., Ruberti I., Morelli G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya Pakrasi M., Peng J., Elmer J. S., Laco G., Shen P., Kaniewska M. B., et al. (1993). Specificity of a promoter from the rice tungro bacilliform virus for expression in phloem tissues. Plant J. 4, 71–79 10.1046/j.1365-313X.1993.04010071.x [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D. E., Wang J. Y., Jung J. W., Lambert G. M., Galbraith D. W., et al. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960 10.1126/science.1090022 [DOI] [PubMed] [Google Scholar]
- Brady S. M., Orlando D. A., Lee J.-Y., Wang J. Y., Koch J., Dinneny J. R., et al. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 10.1126/science.1146265 [DOI] [PubMed] [Google Scholar]
- Cai X., Zhang Y., Zhang C., Zhang T., Hu T., Ye J., et al. (2013). Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 10.1111/jipb.12043 [DOI] [PubMed] [Google Scholar]
- Chen H., Ahmad M., Rim Y., Lucas W. J., Kim J.-Y. (2013). Evolutionary and molecular analysis of Dof transcription factors identified a conserved motif for intercellular protein trafficking. New Phytol. 198, 1250–1260 10.1111/nph.12223 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- De Rybel B., Mãller B., Yoshida S., Grabowicz I., Barbier De Reuille P., Boeren S., et al. (2013). A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G., Hedrich R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem based transcripts hidden in complex datasets of microarray experiments. Plant J. 55, 746–759 10.1111/j.1365-313X.2008.03555.x [DOI] [PubMed] [Google Scholar]
- Dinant S., Suárez-López P. (2012). “Multitude of long-distance signal molecules acting via phloem,” in Biocommunication of Plants, eds Witzany G., Baluska F. (New York: Springer; ), 89–121 [Google Scholar]
- Dong G., Ni Z., Yao Y., Nie X., Sun Q. (2007). Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol. Biol. 63, 73–84 10.1007/s11103-006-9073-3 [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K., Gissot L., Sauerbrunn N., Rühl M., Jarillo J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86 10.1016/j.devcel.2009.06.015 [DOI] [PubMed] [Google Scholar]
- Gabriele S., Rizza A., Martone J., Circelli P., Costantino P., Vittorioso P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61, 312–323 10.1111/j.1365-313X.2009.04055.x [DOI] [PubMed] [Google Scholar]
- Gandotra N., Coughlan S. J., Nelson T. (2013). The Arabidopsis leaf provascular cell transcriptome is enriched in genes with roles in vein patterning. Plant J. 74, 48–58 10.1111/tpj.12100 [DOI] [PubMed] [Google Scholar]
- Gardiner J., Sherr I., Scarpella E. (2010). Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int. J. Dev. Biol. 54, 1389. 10.1387/ijdb.093006jg [DOI] [PubMed] [Google Scholar]
- Gardner M. J., Baker A. J., Assie J.-M., Poethig R. S., Haseloff J. P., Webb A. A. (2009). GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J. Exp. Bot. 60, 213–226 10.1093/jxb/ern292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V. S., Singh U., Kumar A. (2011). Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol. Biol. Rep. 38, 2827–2848 10.1007/s11033-010-0429-z [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Kapitza K., Kolasa A., Buthz A., Kehr J. (2006). Towards the proteome of Brassica napus phloem sap. Proteomics 6, 896–909 10.1002/pmic.200500155 [DOI] [PubMed] [Google Scholar]
- Gilmartin P. M., Sarokin L., Memelink J., Chua N.-H. (1990). Molecular light switches for plant genes. Plant Cell 2, 369. 10.1105/tpc.2.5.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti G., Papi M., Bellucci L., Ricci I., Bouchez D., Camilleri C., et al. (2002). Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14, 1253–1263 10.1105/tpc.010491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Qin G., Gu H., Qu L.-J. (2009). Dof5. 6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21, 3518–3534 10.1105/tpc.108.064139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton D., Sablowski R., Yung M. H., Smith C., Schuch W., Bevan M. (1995). Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 7, 859–876 10.1046/j.1365-313X.1995.07060859.x [DOI] [PubMed] [Google Scholar]
- Hauffe K. D., Lee S. P., Subramaniam R., Douglas C. J. (1993). Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL-1 expression in transgenic tobacco. Plant J. 4, 235–253 10.1046/j.1365-313X.1993.04020235.x [DOI] [PubMed] [Google Scholar]
- Hehn A., Rohde W. (1998). Characterization of cis-acting elements affecting strength and phloem specificity of the coconut foliar decay virus promoter. J. Gen. Virol. 79, 1495–1499 [DOI] [PubMed] [Google Scholar]
- Hernando-Amado S., González-Calle V., Carbonero P., Barrero-Sicilia C. (2012). The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 12:202 10.1186/1471-2229-12-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M., Aspeborg H., Schrader J., Andersson A., Erlandsson R., Blomqvist K., et al. (2001). A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. U.S.A. 98, 14732–14737 10.1073/pnas.261293398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300 10.1093/nar/27.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong-Feng Z., Yu-Qin Z., Wei W., Hao-Wei C., Qing-Xin S., Yun-Feng L., et al. (2013). The transcription factor AtDOF4. 2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem. J. 449, 373–388 10.1042/BJ20110060 [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A., Kay S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Sci. Signal. 309, 293. [DOI] [PubMed] [Google Scholar]
- Imlau A., Truernit E., Sauer N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11, 309–322 10.2307/3870862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergensen K., Scholz-Starke J., Sauer N., Hess P., Van Bel A. J., Grundler F. M. (2003). The companion cell-specific Arabidopsis disaccharide carrier AtSUC2 is expressed in nematode-induced syncytia. Plant Physiol. 131, 61–69 10.1104/pp.008037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.-G., Foley R. C., Oñate-Sánchez L., Lin C., Singh K. B. (2003). Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 35, 362–372 10.1046/j.1365-313X.2003.01812.x [DOI] [PubMed] [Google Scholar]
- Kang H.-G., Singh K. B. (2001). Characterization of salicylic acid-responsive. Plant J. 21, 329–339 10.1046/j.1365-313x.2000.00678.x [DOI] [PubMed] [Google Scholar]
- Kehr J., Buhtz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59, 85–92 10.1093/jxb/erm176 [DOI] [PubMed] [Google Scholar]
- Keller B., Baumgartner C. (1991). Vascular-specific expression of the bean GRP 1.8 gene is negatively regulated. Plant Cell 3, 1051–1061 10.1105/tpc.3.10.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-S., Kim S. J., Abbasi N., Bressan R. A., Yun D.-J., Yoo S.-D., et al. (2010). The DOF transcription factor Dof5. 1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. Plant J. 64, 524–535 10.1111/j.1365-313X.2010.04346.x [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D. (2007). Move on up, it’s time for change-mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21, 2371–2384 10.1101/gad.1589007 [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2007). Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol. Biochem. 45, 623–629 10.1016/j.plaphy.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Krebs J., Mueller-Roeber B., Ruzicic S. (2010). A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. J. Plant Physiol. 167, 583–586 10.1016/j.jplph.2009.11.016 [DOI] [PubMed] [Google Scholar]
- Kurata T., Ishida T., Kawabata-Awai C., Noguchi M., Hattori S., Sano R., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132, 5387–5398 10.1242/dev.02139 [DOI] [PubMed] [Google Scholar]
- Lam E., Chua N.-H. (1989). ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1, 1147–1156 10.1105/tpc.1.12.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Colinas J., Wang J. Y., Mace D., Ohler U., Benfey P. N. (2006). Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 103, 6055–6060 10.1073/pnas.0510607103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Zhou J. (2012). “Function and identification of mobile transcription factors,” in Advance in Plant Biology; Short and Long Distance Signaling, eds Kragler F., Hulskamp M. (New York: Springer; ), 61–86 [Google Scholar]
- Lucas W. J., Ham B.-K., Kim J.-Y. (2009). Plasmodesmata–bridging the gap between neighboring plant cells. Trends Cell Biol. 19, 495–503 10.1016/j.tcb.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno Á., Martínez M., Vicente-Carbajosa J., Carbonero P. (2007). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics 277, 379–390 10.1007/s00438-006-0186-9 [DOI] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M. E., Jang C. J. H., Holtan H. E., Repetti P. P., Galbraith D. W., et al. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18843–18848 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Ohtsuki N., Tsujimoto Y., Fujimura T., Shinshi H. (2006). Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J. Plant Res. 119, 407–413 10.1007/s10265-006-0287-x [DOI] [PubMed] [Google Scholar]
- Nawy T., Lee J.-Y., Colinas J., Wang J. Y., Thongrod S. C., Malamy J. E., et al. (2005). Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17, 1908–1925 10.1105/tpc.105.031724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J., Moriwaki K., Konishi M., Yokoyama R., Nakano T., Kusumi K., et al. (2013). A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 23, 479–484 10.1016/j.cub.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M., Sabatini S., Altamura M. M., Hennig L., Schafer E., Costantino P., et al. (2002). Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 128, 411–417 10.1104/pp.010488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M., Sabatini S., Bouchez D., Camilleri C., Costantino P., Vittorioso P. (2000). Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 14, 28–33 [PMC free article] [PubMed] [Google Scholar]
- Park D. H., Lim P. O., Kim J. S., Cho D. S., Hong S. H., Nam H. G. (2003). The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 34, 161–171 10.1046/j.1365-313X.2003.01710.x [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P., Barrero-Sicilia C., Gómez-Cadenas A., Carbonero P., Oñate-Sánchez L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949 10.1093/jxb/err388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cázares B., Ham B.-K., Li G., Lucas W. J. (2011). Vascular expression in Arabidopsis is predicted by the frequency of CT/GA-rich repeats in gene promoters. Plant J. 67, 130–144 10.1111/j.1365-313X.2011.04581.x [DOI] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Sci. Signal. 318, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E., Francis P., Berleth T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131, 3445–3455 10.1242/dev.01182 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Helariutta Y. (2010). Chapter eight-vascular pattern formation in plants. Curr. Top. Dev. Biol. 91, 221–265 10.1016/S0070-2153(10)91008-9 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027 10.1101/gad.1402406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E. H., et al. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–917 10.1038/nature08836 [DOI] [PubMed] [Google Scholar]
- Schneidereit A., Imlau A., Sauer N. (2008). Conserved cis-regulatory elements for DNA-binding-with-one-finger and homeo-domain-leucine-zipper transcription factors regulate companion cell-specific expression of the Arabidopsis thaliana SUCROSE TRANSPORTER 2 gene. Planta 228, 651–662 10.1007/s00425-008-0767-4 [DOI] [PubMed] [Google Scholar]
- Schrader J., Nilsson J., Mellerowicz E. J., Berglund A., Nilsson P., Hertzberg M., et al. (2004). A high-resolution transcript profile across the wood forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16, 2278–2292 10.1105/tpc.104.024190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Radziejwoski A., Busch W., Hannah M. A., Czeszejko J., Kwasniewski M. A., et al. (2008). The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 56, 779–792 10.1111/j.1365-313X.2008.03641.x [DOI] [PubMed] [Google Scholar]
- Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47, 10–24 10.1111/j.1365-313X.2006.02767.x [DOI] [PubMed] [Google Scholar]
- Stadler R., Sauer N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109, 299–306 [Google Scholar]
- Tanaka M., Takahata Y., Nakayama H., Nakatani M., Tahara M. (2009). Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 230, 737–746 10.1007/s00425-009-0979-2 [DOI] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of B-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196, 564–570 10.1007/BF00203657 [DOI] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 10.1146/annurev.arplant.59.032607.092755 [DOI] [PubMed] [Google Scholar]
- Vilaine F., Palauqui J. C., Amselem J., Kusiak C., Lemoine R., Dinant S. (2003). Towards deciphering phloem: a transcriptome analysis of the phloem of Apium graveolens. Plant J. 36, 67–81 10.1046/j.1365-313X.2003.01855.x [DOI] [PubMed] [Google Scholar]
- Ward J. M., Cufr C. A., Denzel M. A., Neff M. M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17, 475–485 10.1105/tpc.104.027722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. (2001). Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J. 21, 281–288 10.1046/j.1365-313x.2000.00685.x [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560 10.1016/S1360-1385(02)02362-2 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Schmidt R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214 10.1046/j.1365-313X.1999.00363.x [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Sheen J. (1998). Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10, 75–89 10.2307/3870630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Tuskan G. A. (2006). Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142, 820–830 10.1104/pp.106.083642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Beachy R. N. (1995). The regulatory regions of the rice tungro bacilliform virus promoter and interacting nuclear factors in rice (Oryza sativa L.). Plant J. 7, 969–980 10.1046/j.1365-313X.1995.07060969.x [DOI] [PubMed] [Google Scholar]
- Yin Y., Chen L., Beachy R. (1997a). Promoter elements required for phloem-specific gene expression from the RTBV promoter in rice. Plant J. 12, 1179–1188 10.1046/j.1365-313X.1997.12051179.x [DOI] [PubMed] [Google Scholar]
- Yin Y., Zhu Q., Dai S., Lamb C., Beachy R. N. (1997b). RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 16, 5247–5259 10.1093/emboj/16.17.5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Mohri T., Nishiguchi M., Tazaki K. (2002). Robinia pseudoacacia inner-bark lectin promoter expresses GUS also predominantly in phloem of transgenic tobacco. J. Plant Physiol. 159, 757–764 10.1078/0176-1617-0814 [DOI] [Google Scholar]
- Zhang B., Chen W., Foley R. C., Büttner M., Singh K. B. (1995). Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 7, 2241–2252 10.1105/tpc.7.12.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Craig J. C., Petzold H. E., Dickerman A. W., Beers E. P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138, 803–818 10.1104/pp.105.060202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Sebastian J., Lee J.-Y. (2011). Signaling and gene regulatory programs in plant vascular stem cells. Genesis 49, 885–904 10.1002/dvg.20795 [DOI] [PubMed] [Google Scholar]