Abstract
Small cell lung cancer (SCLC) is a subtype of lung cancer with poor prognosis. Expression array analysis of 23 SCLC cases and 42 normal tissues revealed that EZH2 and other PRC2 members were highly expressed in SCLC. ChIP-seq for H3K27me3 suggested that genes with H3K27me3(+) in SCLC were extended not only to PRC2-target genes in ES cells but also to other target genes such as cellular adhesion-related genes. These H3K27me3(+) genes in SCLC were repressed significantly, and introduction of the most repressed gene JUB into SCLC cell line lead to growth inhibition. Shorter overall survival of clinical SCLC cases correlated to repression of JUB alone, or a set of four genes including H3K27me3(+) genes. Treatment with EZH2 inhibitors, DZNep and GSK126, resulted in growth repression of SCLC cell lines. High PRC2 expression was suggested to contribute to gene repression in SCLC, and may play a role in genesis of SCLC.
Lung cancer is the most leading cause of cancer-related deaths in the world, accounting for about 1.4 million deaths each year1. Small cell lung cancer (SCLC) is a histologic subtype of lung cancer with a distinct clinical and biological feature, and represents approximately 15% of all the lung cancer cases2,3. SCLC is strongly related to cigarette smoking, and approximately 90% of cases are reported to be smokers4,5. The prognosis is poor mainly due to early dissemination and rapid growth6,7. While SCLC shows high response to initial therapy, most cases relapse and become refractory to treatment. The 5-year survival is as poor as 15–25% for cases at limited stage, and <1% for cases at extensive stage6,8,9.
Frequent mutation of TP53 and RB1 is reported in SCLC10. Sutherland et al. reported that cell type-restricted inactivation of Trp53 and Rb1 in mouse lung neuroendocrine cells and alveolar type 2 cells induced formation of lung tumors with extrapulmonary metastasis resembling SCLC11. Whole genome sequencing of SCLC cell line NCI-H209 revealed >20,000 somatic substitutions including 134 of those in coding exons, and rearrangement of CHD712. Rudin et al. analyzed exome, transcriptome and copy number aberration of clinical SCLC samples, normal tissues, SCLC cell lines and normal cell lines, and identified 22 significantly mutated cancer-associated genes in SCLC, including TP53 and RB1. Furthermore they found SOX2 amplification and RLF-MYCL1 fusion13. Peifer et al. also performed integrative analysis of exome/genome sequencing, transcriptome, and copy number aberrations in 29 SCLC. They reported TP53 and RB1 inactivation, mutations in PTEN, SLIT2, and EPHA2, and recurrent mutation of histone modifiers e.g. CREBBP, EP300, and MLL14.
Polycomb Repressive Complexes (PRCs) are known to modify epigenetic status and repress their target genes to establish and maintain the cell fates15,16. EZH2 is a catalytic component of PRC2 and functions as a histone methyltransferase for H3 lysine 27 residue (H3K27), and its overexpression in cancer was reported first in prostate cancer, followed by breast cancer. EZH2 overexpression correlates to cancer progression and poorer prognosis17,18. On the other hand, SWI/SNF antagonizes PRCs and activates PRC target genes and contributes to cell differentiation. Mutation in proteins in SWI/SNF complex leads to loss of antagonistic function to polycomb complex protein19. Those mutations or high expression of EZH2 may play a role in maintenance of cell stemness and thus carcinogenesis20. As for lung cancer, 3-Deazaneplanocin A (DZNep), an inhibitor of PRC2, inhibits growth of non-small cell lung cancer cells compared to non-cancerous bronchial epithelial and fibroblast cells21. Furthermore, immunohistochemical analysis suggested that elevated EZH2 expression is associated with non-small cell lung cancer progression and metastasis22.
Involvement of PRC2 in SCLC, however, has not been elucidated. Here we report high expression levels of EZH2 and other PRC2 components in SCLC. Genes with H3K27me3(+) in SCLC cell lines but H3K27me3(−) in normal small airway epithelial cell (SAEC), (i) did not overlap with PRC2-target genes in ES cells, (ii) showed lower expression levels not only in SCLC cell lines but also in clinical SCLC samples, and (iii) showed significant enrichment of GO-terms e.g. immune response, cell adhesion, and plasma membrane. While JUB is the most repressed gene with such GO-terms and with H3K27me3 mark in all the three SCLC cell lines, JUB introduction lead to growth inhibition. Shorter overall survival of clinical SCLC cases correlated to repression of JUB alone, or a set of four genes including PRC target genes and marker genes of classic type SCLC23,24,25. It is suggested that high expression of PRC2 contributes to gene repression in SCLC, and the gene repression may play a role in genesis of SCLC.
Results
Microarray expression analysis in SCLC and normal tissues
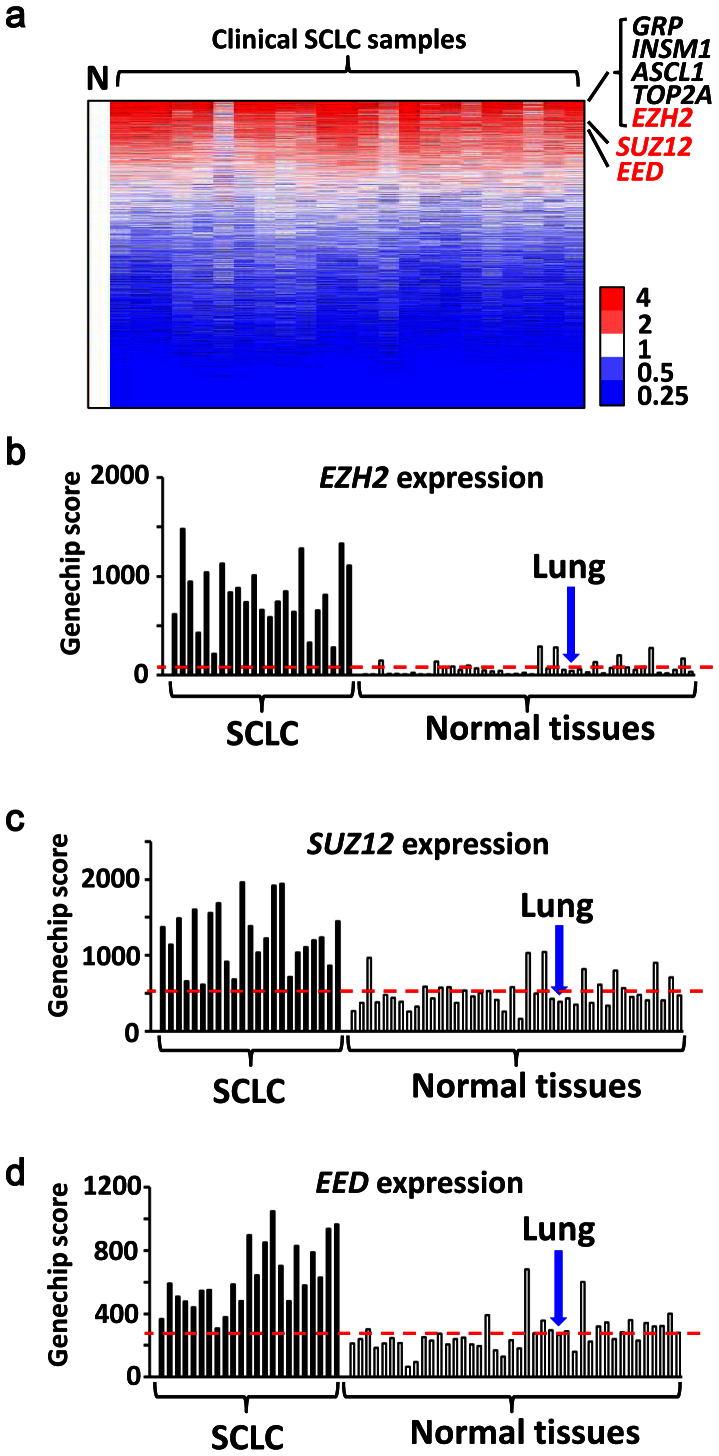
Gene expressions in 23 clinical SCLC samples and 42 normal tissues including the normal lung were analyzed on genome-wide scale using microarray, and average expression levels of SCLC were compared to those of normal tissues. Among 11,037 genes with Genechip score >200 in at least one sample, 71 genes showed higher expression in SCLC by >10-fold compared to normal tissues (Supplementary Table S1). The most highly expressed genes in SCLC samples included GRP, INSM1 and ASCL1, which were reported to be highly expressed in classic type SCLC with neuroendocrine features23,24,25, TOP2A, which encodes a DNA topoisomerase active in fast-growing tumors and is a target of chemotherapy in SCLC26, and EZH2, a member of PRC2 and a methyltransferase for histone H3 lysine K27 (Fig. 1). Other PRC2 members, SUZ12 and EED, were also highly expressed in SCLC compared to the normal tissues. The expression levels of PRC2 members in SCLC were also significantly higher than squamous cell carcinoma and adenocarcinoma of the lung (Supplementary Fig. S1).
Figure 1. Expression levels of PRC2 components in SCLC.
Gene expression levels in 23 clinical SCLC samples were analyzed using expression arrays, and compared to 42 normal tissues including the lung. (a) Heatmap of gene expression in SCLC. Expression levels were shown by the ratio to the average expression level of all the normal samples (N), and genes were sorted by the descending order of average expression levels in SCLC samples. Ezh2 was one of the most highly expressed genes. Expression levels of EZH2 (b) and other PRC2 members, SUZ12 (c) and EED (d), in SCLC samples were significantly higher than normal samples (P = 3 × 10−10, 7 × 10−9, 1 × 10−8, respectively, t-test). Red bar, the average expression level of the normal samples. Expression levels were shown by Genechip Score.
Mapping of histone modification
We performed ChIP-seq for H3K27me3 mark in three SCLC cell lines and in SAEC, and obtained 9280, 9352, 15704 and 7589 peaks in Lu130, H209, DMS53, and SAEC, respectively (Fig. 2a). We then prepared ChIP samples by additionally performed ChIP experiments, and performed quantitative ChIP-PCR to validate ChIP-seq results (Fig. 2b). To confirm overlapping of H3K27me3(+) regions and PRC2-target regions, we also performed ChIP-seq for SUZ12 in a SCLC cell line Lu130. SUZ12(+) regions were well-overlapped to H3K27me3(+) regions as reported27,28 (Supplementary Fig. S2).
Figure 2. Identification of genes modified with H3K27me3 in SCLC cell lines.
(a) ChIP-seq of H3K27me3 in SAEC and SCLC cell lines. Genes not modified with H3K27me3 in either SAEC or SCLC cell lines (GAPDH), modified both in SAEC and SCLC cell lines (ADAM33), and not modified in SAEC but in SCLC cell lines (ITGB4), were representatively shown. Brown bars, regions for ChIP-PCR. (b) The results of ChIP-seq in GAPDH, ADAM33 and ITGB4 were validated by real-time PCR using ChIP samples that were obtained in additionally performed ChIP experiments. (c) Expression of H3K27me3(+) genes was significantly lower than H3K27me3(−) genes in each sample (*P < 10−20, t-test). (d) PRC2-target genes in ES cells28 occupied 3.5% among all the genes (left). Among genes with H3K27me3 in SAEC only, in SAEC and 1 SCLC cell line, and in SAEC and 2–3 SCLC cell lines (center), PRC2-target genes in ES cells occupied 14.3%, 23.8%, and 27.0%, respectively, indicating significant overlap (*P < 10−30, P < 10−30, P < 10−30, Fisher's exact test.). Genes with H3K27me3 in 1 SCLC cell line or 2–3 SCLC cell lines but not in SAEC (right), however, were not significantly overlapped with PRC2-target genes in ES cells, 3.2% (P = 0.66) or 3.8% (P = 0.65), respectively.
Genes with and without H3K27me3 around promoter region (within ± 2kb from transcription start site) were regarded as H3K27me3(+) and H3K27me3(−) genes, respectively. When expression levels of H3K27me3(+) genes were compared to H3K27me3(−) genes within each sample, H3K27me3(+) genes showed significant repression: 0.11-fold in SAEC, 0.27-fold in Lu130, 0.08-fold in H209, and 0.11-fold in DMS53 (P < 1 × 10−20, t-test), confirming that ChIP-seq analysis for H3K27me3 was properly performed (Fig. 2c).
These H3K27me3(+) genes were compared with reported PRC2-target genes in ES cells28. Genes with H3K27me3 in SAEC only or genes with H3K27me3 in both SAEC and SCLC cell lines, showed significant overlap with PRC2-target genes in ES cells. Genes with H3K27me3 in SCLC cell lines but not in SAEC, however, were not significantly overlapped with PRC2-target genes in ES cells (Fig. 2d).
Genes repressed with H3K27me3 in SCLC
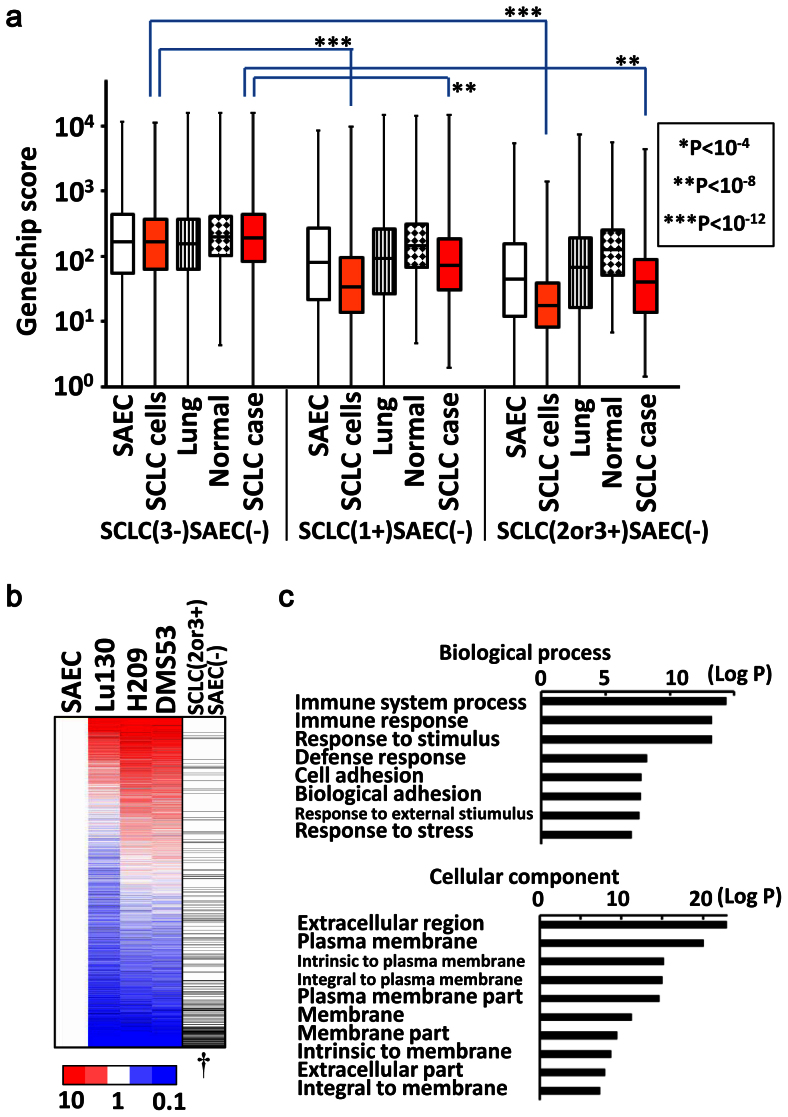
The 8,654 genes showing H3K27me3(−) in 3 SCLC cell lines and also H3K27me3(−) in SAEC (SCLC(3-)SAEC(−) genes), showed similar expression levels among SAEC, 3 SCLC cell lines, the normal lung, other normal tissues, and clinical SCLC cases (Fig. 3a). But the 644 genes showing H3K27me3(+) in 1 of 3 SCLC cell lines but H3K27me3(−) in SAEC (SCLC(1+)SAEC(−) genes), showed lower expression levels in SCLC cell lines as well as clinical SCLC cases compared to the SCLC(3-)SAEC(−) genes. The 343 genes showing H3K27me3(+) in 2 or 3 SCLC cell lines but H3K27me3(−) in SAEC (SCLC(2or3+)SAEC(−) genes), showed further reduced expression levels in SCLC cell lines as well as clinical SCLC cases compared to the SCLC(3-)SAEC(−) genes. These data indicated that genes with H3K27me3(+) in SCLC cell lines were significantly repressed not only in the analyzed SCLC cell lines, but also in clinical SCLC samples (Fig. 3a).
Figure 3. Repression of genes with H3K27me3 in SCLC.
(a) Gene expression in clinical samples. As for SCLC(3-)SAEC(−) genes showing H3K27me3(−) in all the three SCLC cell lines and H3K27me3(−) in SAEC (left), gene expression levels were similar among SAEC, SCLC cell lines, the lung, the normal other tissues, and clinical SCLC samples. As for SCLC(1+)SAEC(−) genes showing H3K27me3(+) in one SCLC cell line but H3K27me3(−) in SAEC (middle), expression levels were significantly lower in SCLC cell lines and clinical SCLC samples when compared between SCLC(3-)SAEC(−) and SCLC(1+)SAEC(−) categories. As for SCLC(2or3+)SAEC(−) genes showing H3K27me3(+) in two or three SCLC cell lines but H3K27me3(−) in SAEC (right), expression levels were even lower in SCLC cell lines and clinical SCLC samples when compared between SCLC(3-)SAEC(−) and SCLC(2or3+)SAEC(−) categories. (b) Significant repression of H3K27me3(+) genes in SCLC. Gene expression levels were shown by the ratio to those of SAEC, and all the genes were sorted by the descending order of average expression levels in SCLC cell lines. Genes in SCLC(2or3+)SAEC(−) category (black bar in the most right column) showed significant enrichment downward (†P < 10−15, Kolmogorov-Smirnov test), indicating that H3K27me3 modification at promoter regions correlated to gene repression. (c) GO-terms with significant enrichment. Immune response (P = 5.1 × 10−14), cell adhesion (P = 1.7 × 10−8), extracellular region (P = 1.3 × 10−23), plasma membrane (P = 8.1 × 10−21), and terms similar to these, showed significant enrichment in SCLC(2or3+)SAEC(−) genes.
When genes were sorted by fold expression level between SAEC and mean of 3 SCLC cell lines in descending order, the SCLC (2or3+)SAEC(−) genes were significantly enriched downward (P < 1 × 10−15, Kolmogorov-Smirnov test), i.e. were significantly repressed in SCLC cell lines (Fig. 3b). The 343 SCLC(2or3+) SAEC(−) genes showed significant enrichment of GO-terms e.g. immune response (P = 5.1 × 10−14), cell adhesion (P = 1.7 × 10−8) and other similar terms in Biological Process, which were different from GO-terms of PRC2-target genes in ES cells related to differentiation and development. Extracellular region (P = 1.3 × 10−23), plasma membrane (P = 8.1 × 10−21) and other similar GO-terms were enriched in Cellular Component (Fig. 3c).
Introduction of repressed genes in SCLC cell line
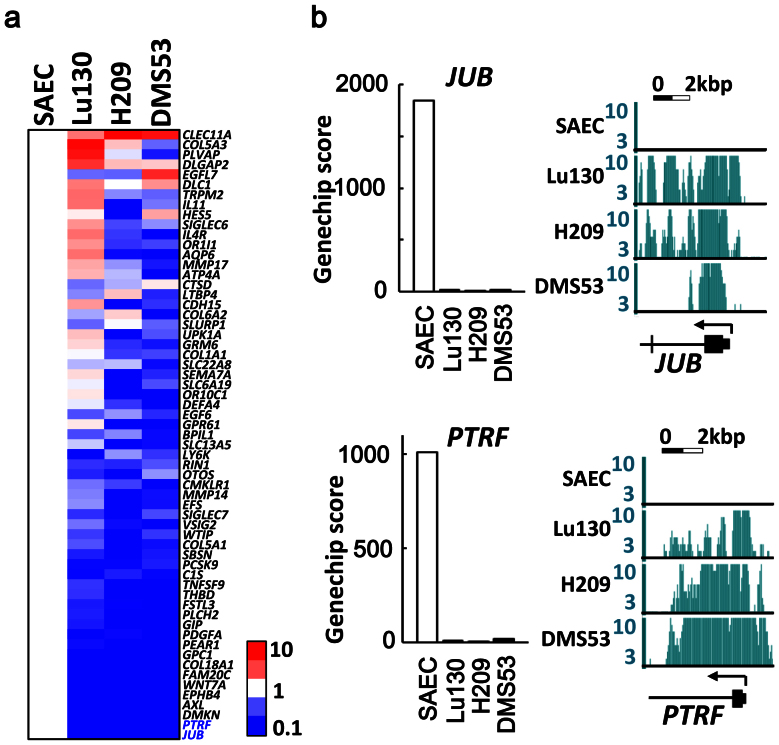
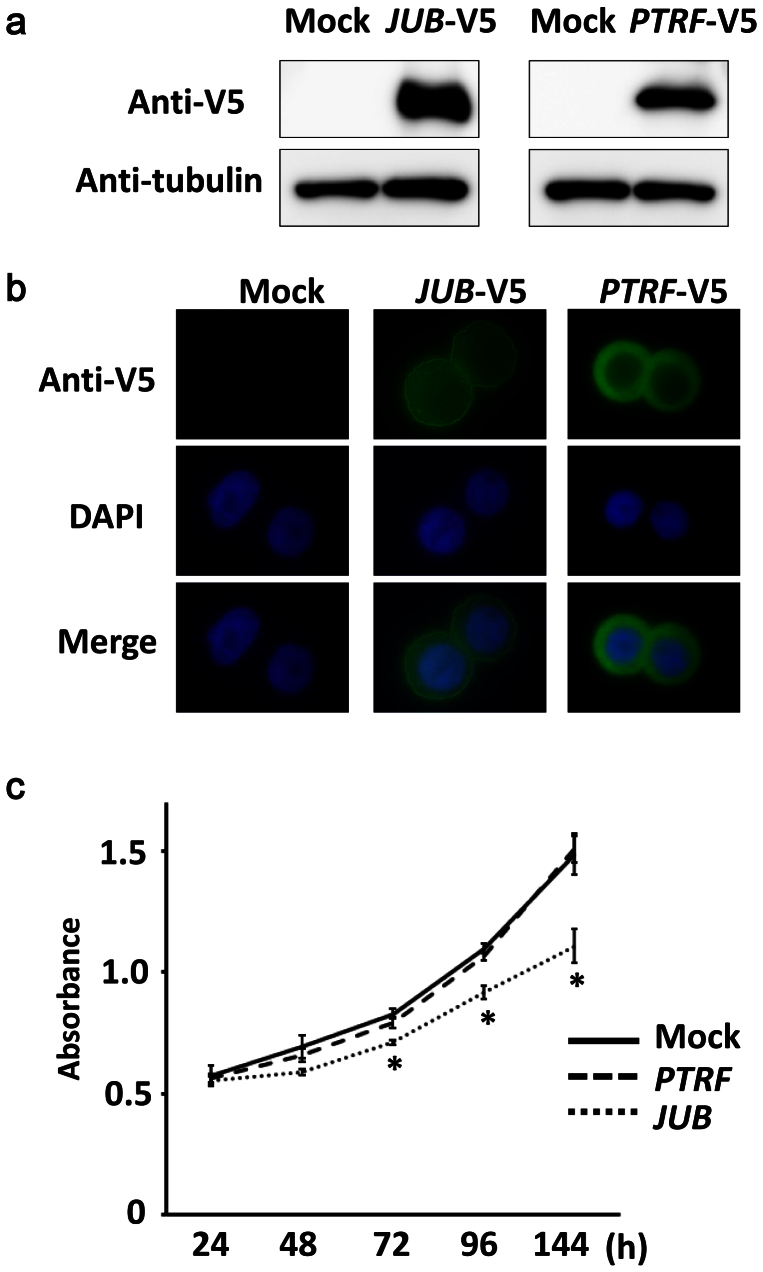
Genes possessing H3K27me3 mark in all the three SCLC cell lines, and relating to the four GO-terms of immune response, cell adhesion, extracellular region, or plasma membrane, were sorted by fold expression level between SAEC and mean of 3 SCLC cell lines in descending order (Fig. 4a). The most repressed genes were JUB and PTRF, which showed decreased expression to 0.01-fold in SCLC cell lines compared to SAEC, and H3K27me3 mark specifically in SCLC cell lines (Fig. 4b). These two genes were introduced in an SCLC cell line DMS53 using lentiviral system, and the protein expressions were confirmed by western blotting (Fig. 5a and Supplementary Fig. S3). JUB was distributed to the cellular membrane, while PTRF was localized in the cytoplasm (Fig. 5b). Introduction of JUB inhibited cellular growth, whereas introduction of PTRF did not affect cellular growth (Fig. 5c), suggesting that the most repressed gene related to cell adhesion, JUB, might function as a growth-suppressor in SCLC cells and its repression by PRC2 might contribute to genesis of SCLC.
Figure 4. Genes most repressed with H3K27me3 in SCLC.
(a) Genes that showed H3K27me3 in all the three SCLC cell lines, and that were included in the four GO-terms of Immune response, cell adhesion, extracellular region, and plasma membrane. They were sorted by the descending order of average expression levels in SCLC cell lines, and the most repressed genes were JUB and PTRF. (b) SCLC cell lines showed low expression levels and H3K27me3 modifications of JUB and PTRF.
Figure 5. Introduction of JUB and PTRF in SCLC cell line.
JUB or PTRF cDNA with V5-tag was introduced in SCLC cell line DMS53 by lentivirus infection. (a) Expression was confirmed by western blotting. Blots were cropped in the figure, but the gels were run under the same experimental conditions, and full-length blots are presented in Supplementary Fig. S3. (b) JUB and PTRF expression by immunofluorescence. Introduced JUB was mainly observed at plasma membrane, and introduced PTRF was mainly observed in the cytoplasm. (c) Growth curve analyzed by WST-8 assay. PTRF-introduced cells showed similar growth to mock cells infected with empty lentivirus, whereas JUB-introduced cells showed suppressed growth significantly (*P < 0.05, t-test). Bars, standard deviation in triplicated analysis.
JUB repression correlated to shorter survival
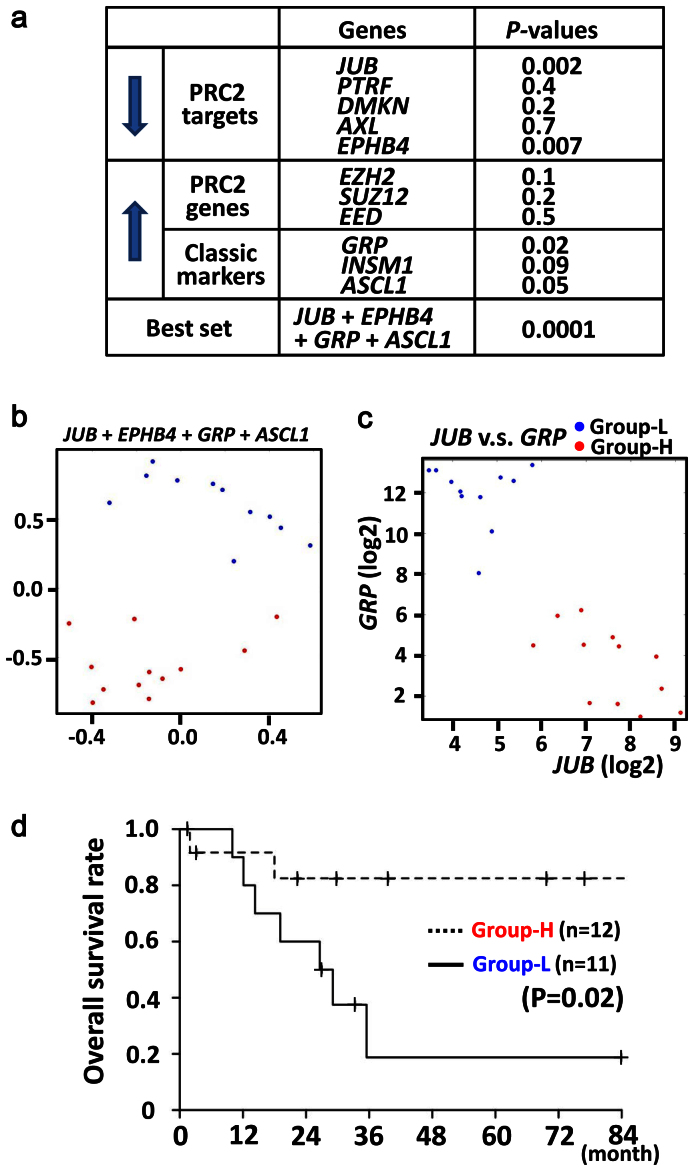
Since gene repression by PRC2 was suggested to contribute to genesis of SCLC, we wanted to analyze whether the repression correlated to poorer prognosis of clinical SCLC or not. We therefore examined the dependency of overall survival time on JUB and other highly repressed PRC-target genes (PTRF, DMKN, AXL, EPHB4) (Fig. 4a) as well as highly expressed PRC2 complex genes (EZH2, SUZ12, EED) and also highly expressed classic type marker genes (GRP, INSM1, ASCL1) (Fig. 1a), using the Cox proportional-hazards regression. Repression of JUB or EPHB4 showed strong correlation to shorter survival (P = 0.002 or P = 0.007, respectively), while high expression of classic type marker, GRP, also correlated to shorter survival (P = 0.02) (Fig. 6a). The most relevant predictor variables were analyzed by the AIC based criteria, the stepAIC function of R software, and a set of four genes, JUB, EPHB4, GRP and ASCL1, was selected as the most relevant one (P = 0.0001) among all the possible combination of one through 11 genes. The K-means sample clustering with the four genes using Orange software29 showed that the optimal cluster size was two, and the two groups of samples were shown by the multi dimensional scaling plot (Fig. 6b). The one group (namely Group-L) could be simply characterized with low JUB and high GRP expression, and the other one (namely Group-H), opposite (Fig. 6c). To analyze whether the classification into the two clusters reflect distinct prognosis, Kaplan-Meyer survival analysis was also performed. Group-L showed shorter overall survival than Group-H (P = 0.02, log-rank test) (Fig. 6d).
Figure 6. Lower expression of JUB and poorer prognosis.
(a) For five of repressed PRC-target genes (downward arrow) and six of highly expressed genes (upward arrow), the dependency of overall survival time of SCLC cases on each gene was analyzed using the Cox proportional-hazards regression, e.g. P = 0.002 for JUB. Among all the possible combination of one through 11 genes, the most relevant predictor variables were the set of the four genes, JUB, EPHB4, GRP and ASCL1 (bottom, P = 0.0001). (b) The K-means sample clustering using the four genes showed that the optimal cluster size of SCLC cases was two, and the two groups of samples (blue and red spots) were shown by the multi dimensional scaling plot. (c) The one group (blue) could be simply characterized with low JUB and high GRP expression, and thus called Group-L. The other one (red) showed high JUB and low GRP expression, and thus called Group-H. (d) Group-L showed shorter recurrence-free survival than Group-H (P = 0.02, log-rank test).
To confirm the robustness of the set of four genes to classify SCLC, we analyzed reported RNA-seq data of SCLC14. The optimal cluster size was again two, revealed by the K-means sample clustering with the four genes using Orange29, and the two groups of samples were shown by the multi dimensional scaling plot (Supplementary Fig. S4a). Again, the Group-L could be simply characterized with low JUB and high GRP expressions, and the Group-H, opposite (Supplementary Fig. S4b).
Treatment of SCLC cell lines with EZH2 inhibitors
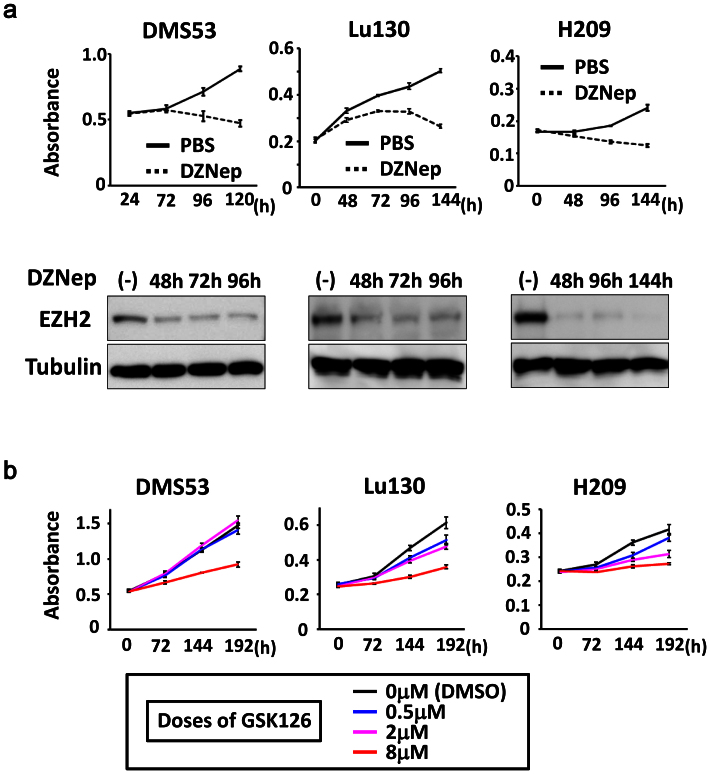
To get insight into clinical application of EZH2 inhibitors on SCLC, we next analyzed effect of an EZH2 inhibitor, DZNep30,31, on the three SCLC cell lines in vitro. When SCLC cell lines were treated with 5 μM DZNep for 5–6 days, western blot analysis showed decrease of EZH2 protein level (Fig. 7a and Supplementary Fig. S5), which is consistent with the previous report32. WST-8 assay showed repression of cellular growth in the three SCLC cell lines, suggesting that EZH2 inhibitors might be effective for therapy on SCLC.
Figure 7. Growth inhibition of SCLC cell lines by treatment with EZH2 inhibitors.
(a) SCLC cell lines, Lu130, H209, and DMS53, were treated with 5 μM DZNep, and growth curve was analyzed by WST-8 assay. Inhibition of cellular growth by DZNep treatment was observed in all the three cell lines, compared to vehicle (PBS) treatment. Protein levels of EZH2 were confirmed to be decreased by DZNep treatment, by western blot analysis. Blots were cropped in the figure, but the gels were run under the same conditions, except that 7% gel was used for EZH2 and 12% gel was used for α-tubulin in analyses of Lu130 and H209. Full-length blots are presented in Supplementary Fig. S5. (b) SCLC cell lines were treated with 0.5, 2, and 8 μM GSK126, and growth curve was analyzed by WST-8 assay. Inhibition of cellular growth by GSK126 treatment was observed at 8 μM in all the three cell lines, while Lu130 and H209 were more sensitive to GSK126, even at lower doses.
SCLC cell lines were then treated with a selective EZH2 inhibitor, GSK126, at different doses of 0.5, 2, and 8 μM (Fig. 7b). All the three cell lines showed substantial inhibition of cellular growth at high dose (8 μM). Although DMS53 was not sensitive to lower doses (0.5 μM and 2 μM), growth inhibition was observed in part at lower doses in Lu130 and H209.
Discussion
We here report high expression levels of EZH2 and other PRC2 components in SCLC. Genes targeted by PRC2 in SCLC cell lines were shown to be repressed in SCLC cell lines and clinical SCLC samples. While JUB is the most repressed gene with H3K27me3 mark in all the three SCLC cell lines, JUB introduction lead to growth inhibition of SCLC cells. Shorter overall survival of clinical SCLC cases significantly correlated to lower expression of JUB alone (P = 0.002), or a set of PRC2 target genes (JUB, EPHB4) and classic type marker genes (GRP, ASCL1) (P = 0.0001). Treatment of three SCLC cell lines with EZH2 inhibitors, DZNep and GSK126, induced growth inhibition. These data suggested that high expression of PRC2 contributed to gene repression in SCLC, and the repression may play a role in worse prognosis of SCLC.
PRC2 complex has a histone methyltransferase activity and represses gene expression by methylation of H3K27 residue. PRC2-target genes in ES cells28 show enrichment of genes related to development and differentiation, and repression of those genes in ES cells or adult tissue stem cells plays a role in maintenance of stemness33. In this study, genes with H3K27me3 in SAEC only and genes with H3K27me3 both in SAEC and SCLC cells were significantly overlapped with PRC2-target genes in ES cells, and showed enrichment of GO-terms such as development and differentiation, similar to PRC2-target genes in ES cells. Genes with H3K27me3 in SCLC cells but not in SAEC, however, were not overlapped with PRC2-target genes in ES cells (Fig. 2d). It might be suggested that H3K27me3 marks in SCLC might be extended to not only PRC2-target genes in ES cells but also other target genes such as cellular adhesion-related genes, due to highly expressed PRC2 components.
It was reported that high expression levels of EZH2 in prostate cancer and breast cancer correlated to metastasis and invasion, and thus poorer prognosis17,18. Inhibition of PRC2 by DZNep treatment resulted in growth inhibition of cancer cell lines with high expression of EZH2, compared to normal cell lines. DAB21P and p21CDKN1A were reported to be PRC2 target genes and play a role in these cancers34,35,36. In SCLC, high expression of EZH2 may extend PRC2-target genes including JUB, and its repression in SCLC correlated to poorer prognosis. DZNep treatment of SCLC cell lines also resulted in growth inhibition, while DZNep is known as PRC2 inhibitor30,31.
Recently, EZH2-specific inhibitors were reported to be developed and effective to diffuse large B-cell lymphoma cell lines37,38. Although GSK126 induced loss of H3K27me3 independent of EZH2 mutation status, six of seven lymphoma cell lines sensitive to GSK126 treatment were EZH2-mutation(+), while only two of 11 lymphoma cell lines less sensitive to GSK126 were EZH2-mutation(+). It was suggested that lymphoma cell lines with mutant EZH2 tend to be dependent on EZH2 activity on cell growth37. In our study, two of three SCLC cell lines showed growth inhibition in part at lower doses of GSK126. It might be suggested that elevated EZH2 in SCLC could induce the dependence of cellular growth on EZH2 activity, like the reported seven sensitive lymphoma cell lines (six EZH2-mutation(+) and one EZH2-mutation(−) cells), but that co-occurring alterations could perhaps weaken the dependency on EZH2 in some SCLC cells. PRC2-target medicine may become an important therapeutic strategy against cancer including SCLC, and application of EZH2-specific inhibitors should be further analyzed for SCLC.
While introduction of JUB showed growth suppression, introduction of PTRF had no effect. PTRF as well as CAV1 is known as structural component of caveolae, 50–100 nm flask-shaped invaginations of plasma membrane involved in numerous signal transductions39. Whereas CAV1 was reported as a tumor suppressor40,41 and PTRF was required for premature senescence induction through interaction with CAV142, CAV1 was also strongly repressed in the three SCLC cell lines and clinical SCLC samples (Supplementary Fig. 6). Introduction of PTRF together with CAV1 might be necessary to rule out tumor-suppressive function of PTRF in SCLC. Beside JUB and PTRF, genes with H3K27me3 in SCLC but not in SAEC included EPHB4, WNT7A, COL18A1, and THBD (Fig. 4). EPHB4 was reported to be a tumor suppressor gene silenced by DNA hypermethylation in acute lymphoblastic leukemia, and transduction of EPHB4 resulted in down-regulation of phosphorylated Akt43. WNT7A was reported to be decreased in non-small cell lung cancer (NSCLC), and its transfection into NSCLC cell lines reversed malignant phenotype44. COL18A1 is cleaved to be Endostatin, an endogenous inhibitor of angiogenesis, and systemic administration of recombinant Endostatin inhibited growth of transplanted cancers45. THBD was reported to be a tumor suppressor gene silenced by DNA hypermethylation in cancers46,47, and its suppression correlated to tumor invasion and poor prognosis48,49. Repression of these genes with H3K27me3 modification may also contribute to genesis of SCLC.
A cell adhesion-related protein JUB harbors LIM domain and is recruited to cadherin-dependent cell-cell adhesive complexes to form stable cell-cell junction50. JUB was also reported to bind to PRMT5 and repress SNAIL target genes in the nucleus51, and accumulation of JUB in the nucleus of P19 embryonal cells resulted in growth inhibition and spontaneous endodermal differentiation52. In this study, introduction of JUB in an SCLC cell DMS53 resulted in growth repression, but the protein was distributed mainly to the cellular membrane, not in the nucleus. It is yet to be elucidated whether the protein distributed to cell-cell adhesive complexes functions as growth suppressor.
Classic type of SCLC with neuroendocrine feature is characterized with expression of GRP as well as L-dopa-decarboxylase, neuron specific enolase, and creatine kinase-BB isoenzyme53. ASCL1 is essential for development of neuroendocrine cells and also highly expressed in classic SCLC, and knock-down of ASCL1 inhibited colony formation in soft agar and induced apoptosis in SCLC54,55. Whereas JUB repression alone significantly correlated to shorter survival (P = 0.002), SCLC was classified into two groups with distinct prognosis using a set of PRC2 target genes (JUB, EPHB4) and classic type marker genes (GRP, ASCL1), and the group with poorer outcome could be simply characterized with low expression of JUB and high expression of GRP (Fig. 6c). Further study is necessary whether these expressions are independent phenomena or features of classic type SCLC include gene repression with H3K27me3.
In summary, PRC2 components were highly expressed in SCLC and contributed to gene repression, and the repression may play a role in genesis of SCLC.
Methods
Clinical samples and cell lines
Primary SCLC samples were obtained from patients undergoing pulmonary resection at Department of Surgery, Cancer Institute Hospital, with written informed consents. These samples were immediately frozen with liquid nitrogen and stored at −80°C. The frozen materials were microscopically examined by two independent pathologists, and were dissected to enrich cancer cells when necessary. Three SCLC cell lines with variable characteristics were prepared: a monolayer cell line DMS53 with wild type RB and missense mutation of p53, a floating cell line Lu130 with wild type RB and missense mutation of p53, and another floating cell line H209 with missense mutation of RB and splice site mutation of p53 (Supplementary Table S2). H209 and DMS53 were purchased from ATCC (Manassas, VA), and Lu130 was from Riken BioResource Center Cell Bank (Tsukuba, Japan). SAEC was obtained from Lonza (Basel, Switzerland). Lu130 and H209 were cultured in RPMI1640 containing penicillin and streptomycin, supplemented with 10% fetal bovine serum at 37°C in 5% CO2. DMS53 was cultured in Dulbecco's modified Eagle's medium (DMEM) containing penicillin and streptomycin, supplemented with 10% fetal bovine serum at 37°C in 5% CO2. SAEC was cultured according to supplier's instruction using SAGM BulletKit and Reagent Pack (Lonza). This study was certified by Ethics Committee in The University of Tokyo and in the Cancer Institute.
RNA samples
Total RNA of 23 SCLC samples, three SCLC cell lines and SAEC was extracted using Trizol (Invitrogen). RNA samples of normal tissues to include variety of organs e.g. central nervous, respiratory, gastrointestinal, endocrine, immunological, skeletal, reproductive, urinary and circulatory systems, were purchased as follows: whole brain, cerebellum, adrenal gland, salivary gland, trachea, bone marrow, testis from Clontech (Mountain View, CA); cereberal cortex, pons, hippocampus, diencephalon, thalamus, tongue, esophagus, gallbladder, tonsil, artery, vein, adipose, lymph node, seminal vesicle from Biochain (Newark, CA); thymus, ovary, skeletal muscle, heart, small intestine, colon, pancreas, kidney, bladder, spleen, prostate from Ambion (Carlsbad, CA); skin, thyroid, breast, and uterus from Stratagene (Santa Clara, CA). The quality of RNA was controlled using Bioanalyzer 2100 (Agilent Technology, CA) and high quality RNA with RNA intensity number (RIN) ≥7.0 was used for array analysis.
Expression array analysis
For genome-wide transcription analysis, Affymetrix GeneChip Human Genome U133 plus 2.0 oligonucleotide arrays (Fremont, CA) was used. Data were collected and analyzed by GeneChip Scanner 3000 (Affymetrix). The GeneChip data were analyzed using the Affymetrix GeneChip Operating Software v1.3 by MAS5 algorithms, to obtain signal value (Genechip score) for each probeset. For global normalization, the average signal in an array was made equal to 100. Expression array data is available at GEO datasets (GSE43346). Expression levels of squamous cell carcinoma and adenocarcinoma of the lung analyzed also using U133 plus 2.0 oligonucleotide arrays were collected from GEO datasets (GSE2199), and global normalization was done by the same manner.
Gene ontology analysis
Gene annotation enrichment analysis was done for Gene Ontology (biological process and cellular component) using the Functional Annotation tool at DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/).
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% formaldehyde for 10 min at room temperature and were prepared for ChIP. ChIP using H3K27me3 (07-142, Upstate, rabbit polyclonal), or Suz12 (D39F6, Cell Signaling, rabbit monoclonal) antibody was performed as we previously reported56. Briefly, cells were crosslinked with 1% formalin for 10 min, and crosslinked cell lysates underwent fragmentation by sonication and incubated with antibodies bound to protein A-sepharose beads (50% slurry) overnight at 4°C. The beads were washed several times and eluted with elution buffer (1% SDS, 0.1 M NaHCO3). The eluates were treated with 1.5 μg of pronase 2 hr at 42°C then incubated at 65°C over night to reverse the crosslinks. The ChIP'ed DNA was purified by phenol/chroloform treatment and precipitated with LiCl and 70% ethanol.
ChIP-sequencing (Chip-seq)
Sample preparation for ChIP-seq was performed according to the manufacturer's instructions (Illumina), and sequencing was performed using Illumina Genome Analyzer IIx as we previously reported56. Briefly, size fractionated DNA was extracted and a single adenosine was added using Klenow exo– (3′ and 5′ exo minus; Illumina). Illumina adaptors were then added and DNA was subjected to 20 cycles of PCR according to manufacturer's instructions. We then purified DNA and performed cluster generation and 36 cycles of sequencing on the Illumina cluster station and 1G Analyzer following the manufacturer's instructions. 36-bp single end reads were mapped to the NCBI Build #36 (UCSC hg18) reference human genome, using the Illumina pipeline software version 1.4. The numbers of uniquely mapped reads for SAEC, Lu130, H209 and DMS53 were 19,527,119 (SAEC), 7,543,376 (Lu130), 18,418,930 (H209) and 21,354,066 (DMS53) for H3K27me3, and 20,025,034 (Lu130) for Suz12. Distribution of immunoprecipitated DNA fragments was analyzed using Model-based Analysis for Chip-Seq (MACS)57, with a window of 600 bp. Regions with p < 10−30 and p < 10−10 were regarded as H3K27me3(+) and SUZ12(+) regions, respectively. ChIP-seq data is available at GEO datasets (GSE43346).
Quantitative real-time ChIP-PCR
ChIP samples were amplified by real-time PCR using SYBR Green and iCycler Thermal Cycler (Bio-Rad Laboratories) and quantified by drawing the standard curve using 20, 2, 0.2, and 0.02 ng/μL sonicated genomic DNA of SAEC. The quantity of ChIP'ed DNA was shown as a ratio to Input DNA (Input %). The PCR primers and conditions are shown in Supplementary Table S3.
Lentiviral vectors
To introduce JUB and PTRF in SCLC cell, we constructed lentiviral vectors for JUB and PTRF by cloning full-length cDNAs by reverse-transcription PCR products from SAEC, into a CMV promoter driven expression vector pLentiV5 (Invitrogen, Carlsbad, CA) that contains puromycin resistance gene, and check the sequences. Lentivirus was prepared by transfection of pLentiV5 vector together with pMD2 and psPAX2 vectors in 293T cells. DMS53 was infected with the prepared viruses, and selected using puromycin at 2 ug/mL for two days.
Immunoblot analysis
Aliquots of protein were subjected to SDS/PAGE followed by immunoblot analysis using antibodies against EZH2 (#39103, Active Motif), V5 (R960-25, Invitrogen), and α-Tubulin (DM1A, Sigma) as primary antibodies, and against rabbit IgG (sc-2004, Santa Cruz, 1:5,000 dilution) and mouse IgG (sc-2005, Santa Cruz) as secondary antibodies. The antibodies used were well characterized previously by us and others56. Proteins were transferred to nitrocellulose and the resulting immunoblots were visualized using Amersham ECL Plus (GE Healthcare) and LAS-3000 (Fujifilm, Japan), and processed using MultiGauge software (Fujifilm).
Cellular immunofluorescence
Cells were washed with PBS three times and fixed with 4% paraformaldehyde for 5 min. After washed with PBS three times, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min. Introduced proteins were detected using antibodies against V5 (R960-25, Invitrogen) as primary antibody, and green-fluorescent Alexa Fluor 488 dye-labeled anti-mouse antibody (A11029, Invitrogen) as secondary antibody. Photographs were taken with Biozero BZ-8100 (KEYENCE, Osaka, Japan).
Growth curve
JUB- and PTRF-introduced DMS53 cells were seeded at density of 1 × 103 cells/well in 96-well plate, and cellular growth was analyzed using WST-8 kit (Doijndo, Japan) at 12, 36, 60, and 84 h. Cellular growth of Lu130, H209, and DMS53 with treatment by DZNep or GSK126 (Active Biochem, NJ) was also analyzed using WST-8 kit. DZNep was dissolved in PBS at 5 mM, and cells were cultured at the final concentration of 5 μM. GSK126 was dissolved in DMSO at 10 mM, and cells were cultured at 0.5, 2, and 8 μM.
Statistical analysis
Statistical analyses were performed using t-test and Fisher's exact test. K-means sample clustering was performed by Orange29 (http://orange.biolab.si/citation/). Kaplan-Meier survival analysis was performed by JMP 7 (http://www.jmp.com/) and P-value was calculated by log-rank test. Survival analysis by Cox proportional-hazards regression was performed by R software (http://www.R-project.org/). In overall survival analysis, the end of follow up period was 84 months from the primary surgery and the mean follow up time of the cases was 64 months. Death as a result of SCLC was the primary end point and deaths by other causes were censored.
Author Contributions
T.S. performed experiments, generated data and figures and co-wrote the manuscript. A.K. designed study, supervised the study, performed experiments, generated data and figures and co-wrote the manuscript. S.T. analysed data and generated figures. T.I. performed experiments. S.Y. analysed data. T.F. analysed data. R.Y. analysed data. Y.T. performed experiments. T.N. supervised the study. V.E.M. performed experiments. Y.I. collected clinical samples and information. M.I. supervised data. H.A. designed study and supervised the study.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Kaori Shiina, Hiroko Meguro and Kyoko Fujinaka for their technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the NFAT project from the New Energy and Industrial Technology Development Organization (NEDO), by JST Core Research for Evolutional Science and Technology (CREST) program, and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Ferlay J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127 (12), 2893–2917 (2011). [DOI] [PubMed] [Google Scholar]
- Read W. L., Page N. C., Tierney R. M., Piccirillo J. F. & Govindan R. The epidemiology of bronchioloalveolar carcinoma over the past two decades: analysis of the SEER database. Lung Cancer 45 (2), 137–142 (2004). [DOI] [PubMed] [Google Scholar]
- Devesa S. S., Bray F., Vizcaino A. P. & Parkin D. M. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer 117 (2), 294–299 (2005). [DOI] [PubMed] [Google Scholar]
- Brownson R. C., Chang J. C. & Davis J. R. Gender and histologic type variations in smoking-related risk of lung cancer. Epidemiology 3 (1), 61–64 (1992). [DOI] [PubMed] [Google Scholar]
- Otterson G., Lin A. & Kay F. Genetic etiology of lung cancer. Oncology (Williston Park) 6 (9), 97–104, 107; discussion 108, 111–102 (1992). [PubMed] [Google Scholar]
- Sher T., Dy G. K. & Adjei A. A. Small cell lung cancer. Mayo Clin Proc 83 (3), 355–367 (2008). [DOI] [PubMed] [Google Scholar]
- Gustafsson B. I., Kidd M., Chan A., Malfertheiner M. V. & Modlin I. M. Bronchopulmonary neuroendocrine tumors. Cancer 113 (1), 5–21 (2008). [DOI] [PubMed] [Google Scholar]
- Albain K. S., Crowley J. J. & Livingston R. B. Long-term survival and toxicity in small cell lung cancer. Expanded Southwest Oncology Group experience. Chest 99 (6), 1425–1432 (1991). [DOI] [PubMed] [Google Scholar]
- Lassen U. et al. Long-term survival in small-cell lung cancer: posttreatment characteristics in patients surviving 5 to 18+ years--an analysis of 1,714 consecutive patients. J Clin Oncol 13 (5), 1215–1220 (1995). [DOI] [PubMed] [Google Scholar]
- Sekido Y., Fong K. M. & Minna J. D. Molecular genetics of lung cancer. Annu Rev Med 54, 73–87 (2003). [DOI] [PubMed] [Google Scholar]
- Sutherland K. D. et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19 (6), 754–764 (2011). [DOI] [PubMed] [Google Scholar]
- Pleasance E. D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463 (7278), 184–190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin C. M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44 (10), 1111–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M. et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44 (10), 1104–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A. P. & Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9 (11), 773–784 (2009). [DOI] [PubMed] [Google Scholar]
- Margueron R. & Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 469 (7330), 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419 (6907), 624–629 (2002). [DOI] [PubMed] [Google Scholar]
- Kleer C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A 100 (20), 11606–11611 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A. A. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 10 (10), 669–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. G. et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18 (4), 316–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi J. et al. Epigenetic therapy with 3-deazaneplanocin A, an inhibitor of the histone methyltransferase EZH2, inhibits growth of non-small cell lung cancer cells. Lung Cancer 78 (2), 138–143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huqun et al. Enhancer of zeste homolog 2 is a novel prognostic biomarker in nonsmall cell lung cancer. Cancer 118 (6), 1599–1606 (2012). [DOI] [PubMed] [Google Scholar]
- Miyake Y., Kodama T. & Yamaguchi K. Pro-gastrin-releasing peptide(31–98) is a specific tumor marker in patients with small cell lung carcinoma. Cancer Res 54 (8), 2136–2140 (1994). [PubMed] [Google Scholar]
- Bhattacharjee A. et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 98 (24), 13790–13795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M. E. et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 98 (24), 13784–13789 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhatriwala H., Jafri N. & Salgia R. A review of topoisomerase inhibition in lung cancer. Cancer Biol Ther 5 (12), 1600–1607 (2006). [DOI] [PubMed] [Google Scholar]
- Kirmizis A. et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 18 (13), 1592–1605 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125 (2), 301–313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curk T. et al. Microarray data mining with visual programming. Bioinformatics 21 (3), 396–398 (2005). [DOI] [PubMed] [Google Scholar]
- Tan J. et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 21 (9), 1050–1063 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase A. & Cross N. C. Aberrations of EZH2 in cancer. Clin Cancer Res 17 (9), 2613–2618 (2011). [DOI] [PubMed] [Google Scholar]
- Zoabi M., Sadeh R., de Bie P., Marquez V. E. & Ciechanover A. PRAJA1 is a ubiquitin ligase for the polycomb repressive complex 2 proteins. Biochem Biophys Res Commun 408 (3), 393–398 (2011). [DOI] [PubMed] [Google Scholar]
- Juan A. H. et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev 25 (8), 789–794 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Tu S. W. & Hsieh J. T. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem 280 (23), 22437–22444 (2005). [DOI] [PubMed] [Google Scholar]
- Min J. et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med 16 (3), 286–294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T. et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res 9 (4), 418–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492 (7427), 108–112 (2012). [DOI] [PubMed] [Google Scholar]
- Knutson S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 8 (11), 890–896 (2012). [DOI] [PubMed] [Google Scholar]
- Hill M. M. et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132 (1), 113–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozza F. et al. Absence of caveolin-1 sensitizes mouse skin to carcinogen-induced epidermal hyperplasia and tumor formation. Am J Pathol 162 (6), 2029–2039 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. M. et al. Loss of caveolin-1 gene expression accelerates the development of dysplastic mammary lesions in tumor-prone transgenic mice. Mol Biol Cell 14 (3), 1027–1042 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volonte D. & Galbiati F. Polymerase I and transcript release factor (PTRF)/cavin-1 is a novel regulator of stress-induced premature senescence. J Biol Chem 286 (33), 28657–28661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S. Q. et al. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood 115 (12), 2412–2419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn R. A. et al. Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J Biol Chem 280 (20), 19625–19634 (2005). [DOI] [PubMed] [Google Scholar]
- O'Reilly M. S. et al. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell 88 (2), 277–285 (1997). [DOI] [PubMed] [Google Scholar]
- Kaneda A., Kaminishi M., Yanagihara K., Sugimura T. & Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res 62 (22), 6645–6650 (2002). [PubMed] [Google Scholar]
- Yagi K. et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res 16 (1), 21–33 (2010). [DOI] [PubMed] [Google Scholar]
- Suehiro T. et al. Thrombomodulin inhibits intrahepatic spread in human hepatocellular carcinoma. Hepatology 21 (5), 1285–1290 (1995). [PubMed] [Google Scholar]
- Liu P. L. et al. Decreased expression of thrombomodulin is correlated with tumor cell invasiveness and poor prognosis in nonsmall cell lung cancer. Mol Carcinog 49 (10), 874–881 (2010). [DOI] [PubMed] [Google Scholar]
- Marie H. et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem 278 (2), 1220–1228 (2003). [DOI] [PubMed] [Google Scholar]
- Hou Z. et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol 28 (10), 3198–3207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanungo J., Pratt S. J., Marie H. & Longmore G. D. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol Biol Cell 11 (10), 3299–3313 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney D. N. et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res 45 (6), 2913–2923 (1985). [PubMed] [Google Scholar]
- Sriuranpong V. et al. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol Cell Biol 22 (9), 3129–3139 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T. et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res 69 (3), 845–854 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda A. et al. Activation of Bmp2-Smad1 signal and its regulation by coordinated alteration of H3K27 trimethylation in Ras-induced senescence. PLoS Genet 7 (11), e1002359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9 (9), R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information