Summary
Mouse and human dendritic cells (DCs) are composed of functionally specialized subsets, but precise interspecies correlation is currently incomplete. Here, we showed that murine lung and gut lamina propria CD11b+ DC populations were comprised of two subsets: FLT3- and IRF4-dependent CD24+CD64− DCs and contaminating CSF-1R-dependent CD24−CD64+ macrophages. Functionally, loss of CD24+CD11b+ DCs abrogated CD4+ T cell-mediated interleukin-17 (IL-17) production in steady state and after Aspergillus fumigatus challenge. Human CD1c+ DCs, the equivalent of murine CD24+CD11b+ DCs, also expressed IRF4, secreted IL-23, and promoted T helper 17 cell responses. Our data revealed heterogeneity in the mouse CD11b+ DC compartment and identifed mucosal tissues IRF4-expressing DCs specialized in instructing IL-17 responses in both mouse and human. The demonstration of mouse and human DC subsets specialized in driving IL-17 responses highlights the conservation of key immune functions across species and will facilitate the translation of mouse in vivo findings to advance DC-based clinical therapies.
Highlights
-
•
Mucosal CD11b+ DCs consist of CD24+CD64− DCs and CD24−CD64+ macrophages
-
•
Mucosal CD24+CD11b+ DCs are IRF4-dependent
-
•
IRF4-dependent CD24+CD11b+ DCs secrete IL-23α and control mucosal IL-17 responses
-
•
Human CD1c+CD11b+ DCs are functional homologs of murine CD24+CD11b+ DCs
Introduction
Dendritic cells (DCs) initiate and regulate immune responses against pathogens and maintain tolerance to self-antigens (Banchereau et al., 2000; Steinman et al., 2003) and as such are potential targets of immunotherapeutic interventions (Palucka et al., 2010). DCs are heterogeneous and can be subdivided by anatomical location, origin, and function. DCs are found in lymphoid and nonlymphoid tissues (NLT). In mice, lymphoid tissue (LT) DCs are categorized into two groups: plasmacytoid DCs (pDCs) and classical DCs, and the latter include CD8+CD4–CD11b– (CD8+ DC), CD8–CD4+CD11b+ (CD4+CD11b+ DC), and CD8–CD4–CD11b+ DC populations (Shortman and Naik, 2007). In peripheral NLTs, DCs that populate the outer epidermal layer of stratified epithelia are termed Langerhans cells, whereas DCs in connective tissues are called interstitial DCs. The interstitial DC populations are distinguished by mutually exclusive surface expression of the integrins CD103 and CD11b in steady state (Ginhoux et al., 2009). In the gut lamina propria (LP), an additional subset expressing both CD103 and CD11b has been described (Annacker et al., 2005; Bogunovic et al., 2009).
Recent studies on DC differentiation and function have highlighted many parallels between LT and NLT DC populations, in particular between CD8+ and CD103+ DCs (for review, see Hashimoto et al., 2011). The NLT CD103+ DCs and CD8+ LT resident DCs constitute an independent DC lineage; both reliant upon the Fms-like tyrosine kinase 3 ligand receptor (FLT3) and the transcription factors (TFs) IRF8, ID2, and BATF3. Both populations are endowed with superior ability to cross-present viral, tumor, and self-antigens. We recently showed that a functionally-equivalent cross-presenting DC lineage is conserved in human peripheral tissues identifying the CD141hi DC subset with substantial cross-presenting capacities (Haniffa et al., 2012), similar to blood CD141+ DCs (Bachem et al., 2010; Crozat et al., 2010; Poulin et al., 2010). By using interspecies transcriptomic analysis, we demonstrated homology between human CD141+ DC lineage with mouse NLT CD103+ and CD8+ splenic DCs (Haniffa et al., 2012), as previously suggested for blood CD141+ DCs (Crozat et al., 2010).
In contrast, the relationships between NLT and LT CD11b+ DC populations and their unique immune specializations remain unclear. Several TFs controlling splenic CD11b+ DC development have been identified and include IRF2, IRF4, IKAROS, RELB, and the members of the Notch pathway, NOTCH2 and RBP/J (for review, see Hashimoto et al., 2011). Recent data have revealed that the gut LP CD103+CD11b+ DC also rely on NOTCH2 (Lewis et al., 2011), but the dependency of other NLT CD11b+ DCs on these TFs remains to be defined. In addition, NLT and LT CD11b+ DCs appear to be heterogeneous in both origin and differentiation. NLT CD11b+ DCs can be derived from both bone marrow (BM) DC specific progenitors and monocytes in steady state (Ginhoux et al., 2009) and are partially dependent on both FLT3 and the colony-stimulating factor 1 receptor (CSF-1R) (Ginhoux et al., 2009). This is corroborated by the demonstration of heterogeneity within murine splenic CD8−CD11b+ DCs, which are composed of endothelial cell-selective adhesion molecule (ESAM)+CD4+ and ESAM−CD4− fractions, the latter suggested as being monocyte-derived (Lewis et al., 2011).
Our recent analysis to define homologies between mouse and human DCs also predicted the existence of monocyte and/or macrophage cells within NLT CD11b+ DC population (Haniffa et al., 2012). Human peripheral tissue DCs are composed of three main subsets in the steady state: CD141hi DCs, equivalent to the CD8/CD103 murine lineage; CD14+ DCs, related to the monocyte and/or macrophage lineage; and CD1c+ DCs (Haniffa et al., 2012). Human CD1c+ DCs were positively associated with murine splenic CD4+ DCs but not tissue CD11b+ DCs. Surprisingly, human CD14+ DCs were positively associated with murine monocytes and NLT CD11b+ DCs, suggesting contamination by monocyte-macrophage cells within the NLT CD11b+ DC fraction.
We therefore hypothesized the existence of a specialized DC subset within the NLT CD11b+ fraction with defined TF dependence and function that is conserved between mouse and human. Here, we identify FLT3-dependent CD11b+CD24+CD64− bona fide DCs and CSF-1R-dependent CD11b+CD24−CD64+ macrophages in steady state within MHC class II (MHCII)+CD11c+CD11b+ cells in the lung and small intestine (SI). Mucosal CD24+CD11b+ DCs were dependent on IRF4 and secreted the T helper 17 (Th17) cell prone cytokine interleukin-23 (IL-23), instructing Th17 cell differentiation in steady state and after Aspergillus fumigatus (A. fumigatus) challenge. We also showed that human CD1c+ DCs express IRF4, secrete IL-23, and are the functional homologs of murine tissue CD11b+CD24+CD64− DCs.
Results
Lung CD11b+ DCs Are Heterogeneous
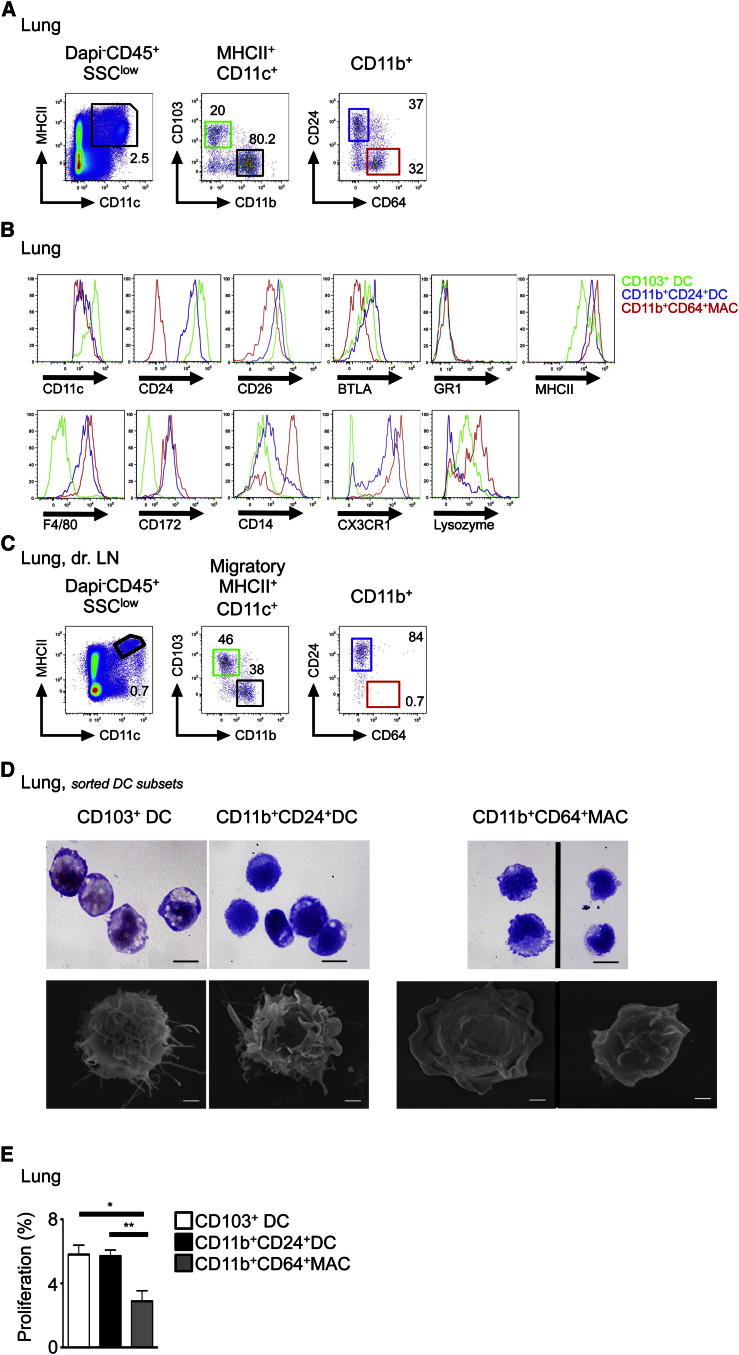
We first sought to identify specific markers that could discriminate bona fide DCs from monocyte-macrophage cells within tissue CD11b+MHCII+CD11c+ fraction. Lung SSCloMHCII+CD11c+CD11b+ fraction contained two distinct populations distinguished by the expression of the sialoglycoprotein CD24 (heat-stable protein) and CD64 (FcγR1) (Figure 1A; see also Figure S1A available online). CD24+CD64−CD11b+ cells were characterized by their expression of the DC markers CD11c, BTLA, and CD26 (Miller et al., 2012), while CD24-CD64+CD11b+ cells were characterized by their expression of the monocyte/macrophage markers CD14, the fractalkine receptor CX3CR1, and lysozyme (Figure 1B). Migratory CD11b+ DCs in the lung draining lymph nodes (LLN) expressed high amounts of CD24 and no CD64, suggesting that only CD11b+CD24+CD64− cells migrate to LLN (Figure 1C). Morphologically, CD24+CD64−CD11b+ cells exhibited fine dendrites and large cytoplasms characteristic of DCs, whereas CD24−CD64+CD11b+ cells were a mixture of larger cells with vacuoles and membrane ruffles alongside small monocyte-like cells with smooth membranes (Figure 1D).
Figure 1.
Lung CD11b+ DCs Are Heterogeneous
(A) Flow cytometry of mouse lung cell suspension. Gating strategy to identify CD103+ DCs (green gate), CD11b+CD24+ DCs (blue gate), and CD11b+CD64+ MACs (red gate) is shown.
(B and C) Histograms show relative expression of the indicated markers on lung DCs and MACs (B). Flow cytometry of mouse LLN cell suspension. Gating strategy as described in (A) was used in (C). Representative data from n > 5 shown for (A)–(C).
(D) Morphology of purified lung DCs and MACs visualized by GIEMSA staining and SEM.
(E) Percentage proliferation of lung DCs and MACs indicated by fluorescence levels in Fucci mice (mean fluorescent+ cells ± SEM, pooled results of three experiments, n = 6). See also Figure S1.
Tissue DCs proliferate in situ in the steady state (4%–6% in S-G2-M phase), whereas macrophages (MACs) are more quiescent (1%–2% in S-G2-M phase) (Ginhoux et al., 2009; Liu et al., 2009). By using Fucci-492 mice where cells in S, G2, and M phases of the cell cycle are fluorescent (Sakaue-Sawano et al., 2008), we observed that 5%–7% of lung CD103+ DCs and CD11b+CD24+ cells were in active cell cycle compared to 3% of CD11b+CD24− cells (Figure 1E). These results demonstrate the presence of two distinct populations in the steady state within the MHCII+CD11c+CD11b+ fraction; (1) a CD24+CD64−CD11b+ cell expressing DC-related antigens and higher proliferative status and (2) a CD24−CD64+CD11b+ cell expressing macrophage markers with low cell-cycle activity.
Lung CD11b+CD24+CD64− Cells Are Bona Fide DCs, whereas CD11b+CD24−CD64+ Cells Are Macrophages
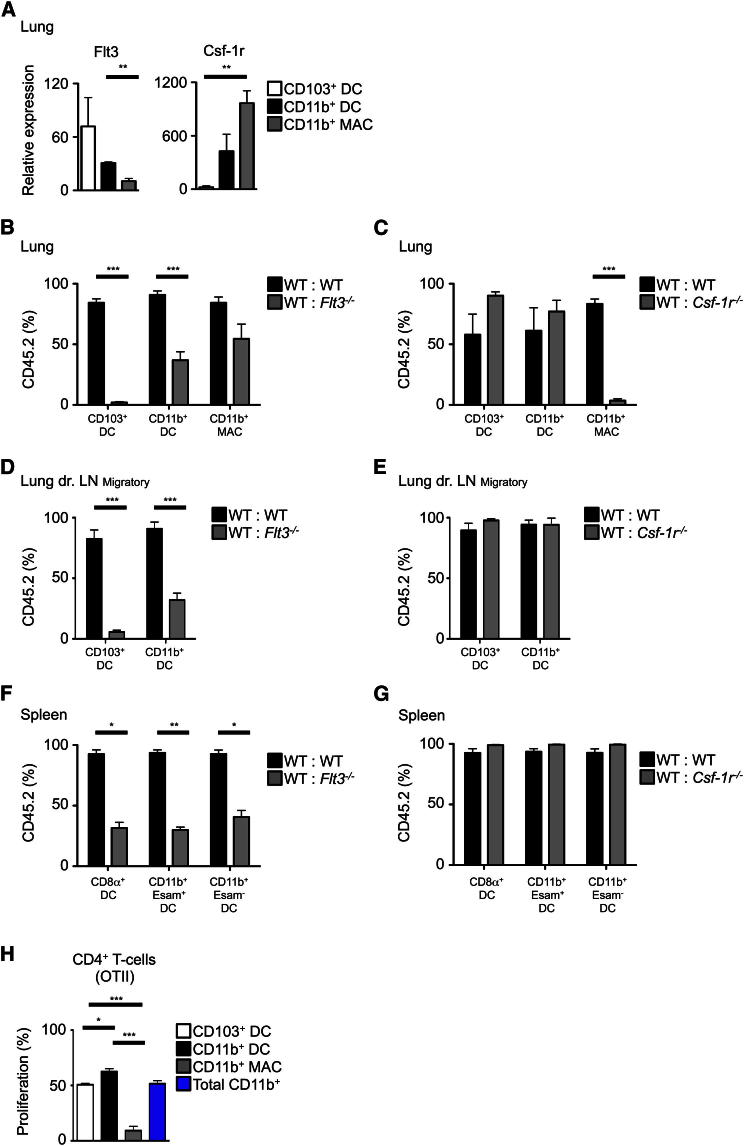
We hypothesized that CD24+CD64−CD11b+ cells represent bona fide lung DCs (named thereafter CD11b+ DCs), whereas CD24−CD64+CD11b+ cells correspond to interstitial macrophages (named thereafter CD11b+ MACs), known to express high amounts of MHCII (Bedoret et al., 2009). We investigated the dependency of CD11b+ DCs and CD11b+ MACs on the DC differentiation cytokine receptor FLT3 and the monocyte-macrophage differentiation cytokine receptor CSF-1R. Flt3 messenger RNA (mRNA) expression was highest on purified CD103+ DCs followed by CD11b+ DCs and at an extremely low level on CD11b+ MACs (Figure 2A; see Figure S1A for sorting strategy). In contrast, CD11b+ MACs had the highest expression of Csf-1r. We then tested their requirement for functional expression of FLT3 and CSF-1R by using mixed BM chimeras (Figures 2B–2G; Figure S2). Consistent with our previous report (Ginhoux et al., 2009), the ability of Flt3−/− BM progenitors to generate lung and LLN CD103+ DCs was dramatically reduced (Figures 2B and 2D). Flt3−/− BM progenitors were also less efficient at generating lung and LLN CD11b+ DCs (Figure 2B and 2D) at a similar level to splenic DC subsets, regardless of ESAM expression within CD11b+ DCs (Figure 2F). In contrast, Flt3−/− BM progenitors fully reconstituted the CD11b+ MAC population, suggesting its independence of FLT3 signaling. Csf-1r−/− progenitors efficiently generated CD103+ and CD11b+ DC subsets in the lung and LLN (Figures 2C and 2E), but not CD11b+ MACs (Figure 2C). Furthermore, Csf-1r deficiency did not affect any splenic DC subset (Figure 2G). We also tested the ex vivo ability of these different lung DC and MAC populations to present intratracheally delivered ovalbumin (OVA) to OTII-Rag1−/− CD4+ T cells recognizing the I-Ab restricted OVA323–339 peptide (Barnden et al., 1998). As expected, CD11b+ DCs induced greatest OTII proliferation, whereas the CD11b+ MACs were poor stimulators of OTII cells (Figure 2H).
Figure 2.
Lung CD11b+CD24+CD64− Cells Are Bona Fide DCs, whereas CD11b+CD24−CD64+ Cells Are Macrophages
(A) Relative expression of Flt3 and Csf-1r mRNA by lung DCs and MACs (n = 3, mean ± SEM).
(B–G) Percentage of CD45.2 and CD45.1 DCs and MACs in the indicated tissues from mixed BM chimeric mice (CD45.2 WT: CD45.1 WT; CD45.2 Flt3−/−: CD45.1 WT (B, D, F); CD45.2 Csf1r−/−: CD45.1 WT (C, E, G).
(H) Bar graph shows mean ± SEM of the indicated cell populations (pooled results of four experiments, n = 12). Percentage proliferation of OTII cells stimulated by OVA-loaded lung DCs and MACs (n = 3, mean ± SEM). See also Figure S2.
Lung CD11b+ DCs Are Dependent on IRF4
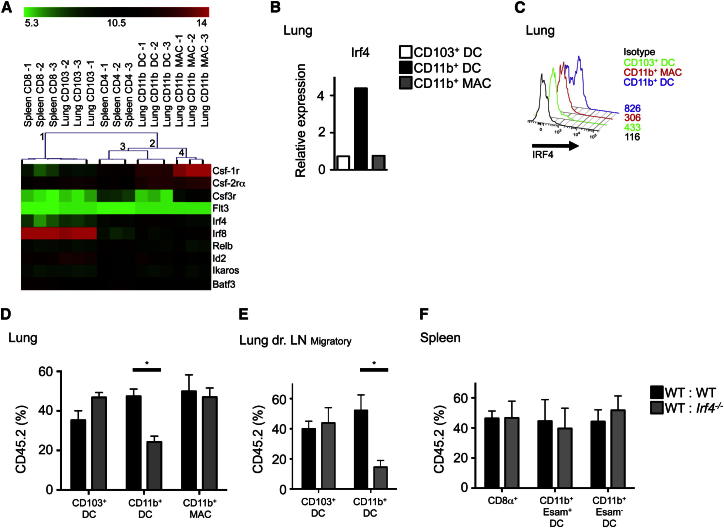
We next analyzed the expression of cytokine receptors and TFs known to be important for DC and macrophage differentiation by CD11b+ DCs and CD11b+ MACs (Table S1). By using microarray analysis, we identified genes that were differentially expressed by splenic CD8+ and lung CD103+ DCs compared to splenic CD11b+CD4+ DCs. Hierarchical clustering based on the differentially expressed genes showed that lung CD11b+ DCs clustered with splenic CD4+ DCs, whereas CD11b+ MACs were distantly related (Figure 3A), suggesting a similar developmental program between lung CD11b+ DCs and splenic CD11b+CD4+ DCs.
Figure 3.
Lung CD11b+ DCs Are IRF4 Dependent
(A) Hierarchical clustering of murine spleen and lung DC/MAC populations based on differential expression of selected genes (SAM algorithm, delta = 2.6).
(B) Irf4 mRNA expression by lung DCs and MACs (n = 3, mean).
(C) Relative expression of IRF4 by flow cytometry on lung CD11b+ DCs and MACs (n = 3).
(D–F) Percentage of CD45.2 and CD45.1 DCs and MACs in the indicated tissues from indicated mixed BM chimeric mice. Bar graphs show mean ± SEM of the indicated cell populations (pooled results of four experiments, n = 10, mean ± SEM). See also Figure S2 and Table S1.
Microarray data showed highest expression of Irf4 on lung CD11b+ DCs (Figure 3A). Q-PCR (Figure 3B) and flow cytometric (Figure 3C) analyses confirmed that lung CD11b+ DCs expressed higher amounts of IRF4 than either lung CD103+ DCs or CD11b+ MACs. Because IRF4 is known to regulate splenic CD4+ DC differentiation (Suzuki et al., 2004; Tamura et al., 2005), we tested whether lung CD11b+ DC differentiation was affected by IRF4 deficiency by using mixed BM chimeras. Lung CD11b+ DCs were adversely affected by IRF4 deficiency but CD103+ DCs and CD11b+ MACs were unaffected (Figure 3D). This defect was more dramatic in the LLN (Figure 3E). However, we did not detect any defects in the splenic CD11b+ DC population (Figure 3F), regardless of ESAM expression that correlates with CD4+ DCs (Lewis et al., 2011).
Small Intestinal LP CD24+CD103+CD11b+ DCs Are Also Dependent on IRF4
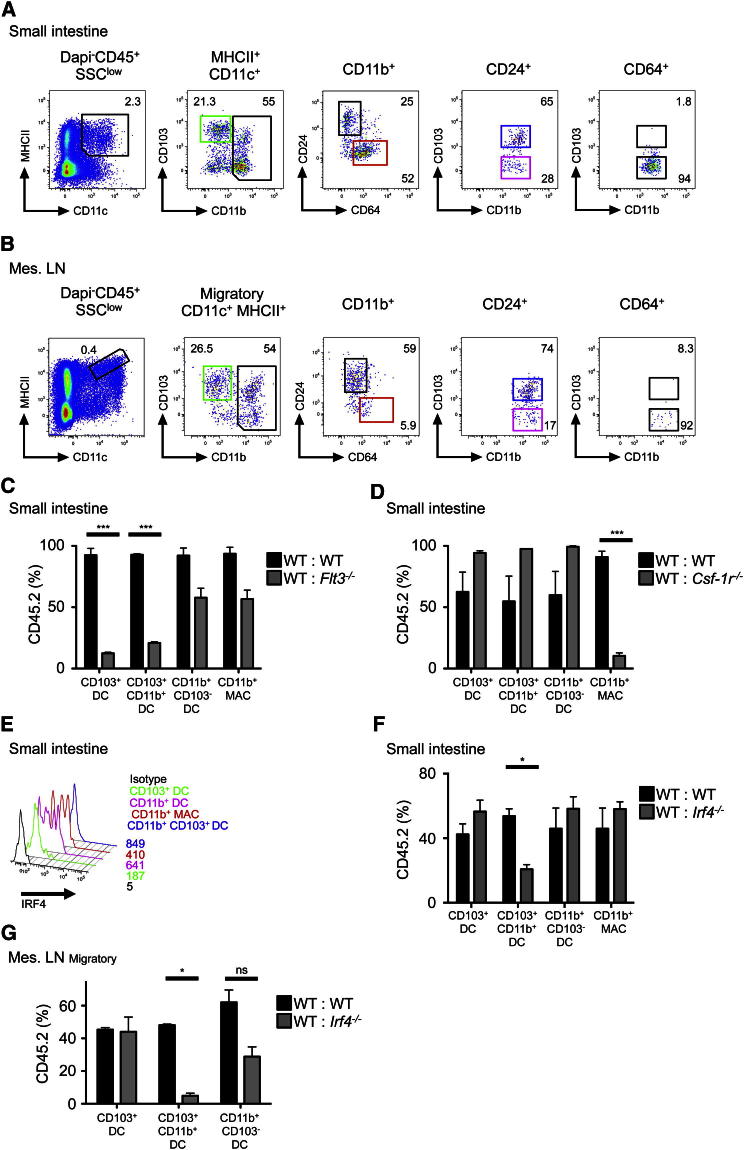
We extended our analysis to other NLTs (dermis, kidney, liver, and gut) to test whether similar CD11b+CD24+CD64− DCs were present in these organs. Only gut LP CD11b+ DCs in the small and large intestine segregated clearly into CD24+CD64− and CD24−CD64+ populations (Figure 4A; Figures S1B and S3A), whereas CD24 and CD64 were not fully discriminatory in the dermis, kidney, or liver (data not shown). Furthermore, there was no correlation between IRF4 and CD24 expression in these organs detectable by flow cytometry (data not shown).
Figure 4.
Gut Lamina Propria CD24+CD103+CD11b+ DCs Are Dependent on IRF4
(A) Flow cytometry of gut LP to identify CD103+ DCs, CD103+CD11b+ DCs, CD103-CD11b+ DCs, and CD11b+ MACs (n = 5).
(B) Flow cytometry of MLN to identify DCs and MACs as described in (A), (n = 5).
(C, D, F, and G) Percentage of CD45.2 and CD45.1 DCs and MACs in the indicated tissues from indicated BM chimeric mice.
(E) Pooled results from four experiments, n = 10 (mean ± SEM). IRF4 expression on SI-LP DCs and MACs by flow cytometry (n = 3). See also Figure S3.
The small intestinal LP (SI-LP) CD11b+ DC fraction contained a CD24+CD64− DC population (CD11b+ DCs) and CD24−CD64+ macrophages (CD11b+ MACs). However, unlike lung CD11b+ DCs, the SI-LP CD11b+ DCs were mostly CD103+ (CD11b+CD103+ DCs), with a minor CD103− fraction (CD11b+CD103− DCs) (Figure 4A), as previously described (Annacker et al., 2005; Bogunovic et al., 2009; Cerovic et al., 2012). CD11c expression, but not CD26 or BTLA, discriminated CD11b+ DCs (regardless of CD103 expression) from CD11b+ MACs (Figure S3B). In contrast, SI-LP CD11b+ MACs expressed higher amounts of the monocyte-macrophage markers CD14, CX3CR1, and lysozyme (Figure S3B). Similar to LLN, the CD11b+ fraction of migratory DCs (CD11cloMHCIIhi) in the mesenteric lymph nodes (MLN) expressed CD24 and not CD64, suggesting that only the CD11b+CD24+CD64− fraction migrates to MLN (Figure 4B), as previously reported (Bogunovic et al., 2009; Schulz et al., 2009). Mixed BM chimera experiments showed that both LP CD11b+CD103+ DCs and CD24+CD103+ DCs were FLT3 dependent and CSF-1R independent, whereas CD11b+ MACs were CSF-1R dependent (Figures 4C and 4D). LP CD11b+CD103− DCs were CSF-1R and FLT3 independent because their chimerism was similar to blood granulocytes used as engraftment control (Figure S2).
We also tested whether SI-LP CD11b+ DCs (regardless of CD103 expression) were dependent on IRF4. SI-LP CD11b+CD103− DCs expressed lower levels of IRF4 (Figure 4E) and were independent of IRF4, as were CD11b−CD103+ DC and CD11b+ MAC populations (Figures 4F and 4G). In contrast, LP CD11b+CD103+ DCs expressed the highest level of IRF4 (Figure 4E) and were partially dependent on IRF4 expression (Figure 4F). Their relationship with LP CD11b+CD103− DCs remains to be investigated. Furthermore, correlating with the lung data, migratory MLN CD11b+CD103+ DCs were also entirely dependent on IRF4 (Figure 4G). Analogous CD24+CD11b+ DCs, CD64+CD11b+ MACs, or an IRF4-dependent population was undetectable in the dermis, kidney, and liver (data not shown).
Lung CD11b+ DCs and Small Intestinal LP CD11b+CD103+ DCs Are Absent in Mice Lacking IRF4 in the DC Lineage
We next established whether the lung and SI-LP IRF4-dependent DC subsets shared functional specializations. We crossed the DC-specific Cre recombinase mouse (Itgax-cre) (Caton et al., 2007) with a mouse wherein the Irf4 gene (exons 1 and 2) is flanked by loxP sites and a GFP construct is placed in the reverse orientation, upstream of the Irf4 promoter region (Irf4fl/fl) (Klein et al., 2006). In these mice (Itgax-cre Irf4fl/fl), expression of the Cre recombinase leads to excision of both exons, preventing transcription of the Irf4 gene only in CD11c+ cells and concomitantly enabling GFP expression in recombined CD11c+ cells, allowing the tracking of Irf4-deficient cells (Klein et al., 2006). As expected, CD11c+MHCII+ DCs were GFP+ in all tissues tested (Figures S4A–S4C). Other populations expressing GFP were Gr1lo monocytes (80%), CD3+ T cells (25%), and B220+CD19+ B cells (15%) (Figure S4D). Mice exhibiting >25% GFP+ lymphocytes were excluded from our studies.
Absence of IRF4 expression in DCs led to a specific depletion of CD24+CD11b+ DCs in the lung and LLN and CD24+ CD11b+CD103+ DCs in the gut LP and MLN (Figures S4E–S4J). Other DC and MAC populations were unaffected, despite expressing similar levels of GFP (Figure S4). Similar to the data obtained from mixed BM chimeras, we did not detect any defects in DC populations in the dermis or liver but observed a slight decrease in the kidney CD11b+CD24+ DC fraction (Figures S4K–S4O).
We next investigated how IRF4 regulates mucosal tissue CD11b+ DC homeostasis. To examine whether IRF4 controlled CD11b+ DC homeostasis at the progenitor level, we quantified CD11c+ DC progenitors including pre-DCs (Liu et al., 2009) and CCR9− pre-pDCs (Schlitzer et al., 2012) in Itgax-cre Irf4fl/fl versus control littermate (wild-type, WT) BM. Both DC progenitors were GFP+ as a mark of IRF4 excision in Itgax-cre Irf4fl/fl (Figure S5A). We found no differences in their relative numbers (Figure S5B). We next tested whether IRF4 was playing a prosurvival role in CD11b+ DC homeostasis, a considerably difficult experiment to perform as very few CD11b+ DCs remained in the Itgax-cre Irf4fl/fl mice. As such, only the lung yielded enough cells for analysis. Because cells undergoing apoptosis show enhanced mitochondrial fission and fragmentation (Martinou and Youle, 2011), we performed a mitotracker assay that assessed mitochondrial network integrity in lung CD11b+ DCs lacking IRF4. The remaining lung CD11b+ DCs in Itgax-cre Irf4fl/fl mice showed significantly enhanced mitochondrial fragmentation, as observed during apoptosis, compared to WT mice (Figures S5C and S5D). These data suggest that IRF4 has a prosurvival role and its absence in mucosal tissue CD11b+ DCs leads to apoptosis, likely explaining the depletion of mucosal tissue CD11b+ DCs in Itgax-cre Irf4fl/fl mice.
IRF4-Dependent CD24+CD11b+ DCs Control Th17 Responses in Steady State
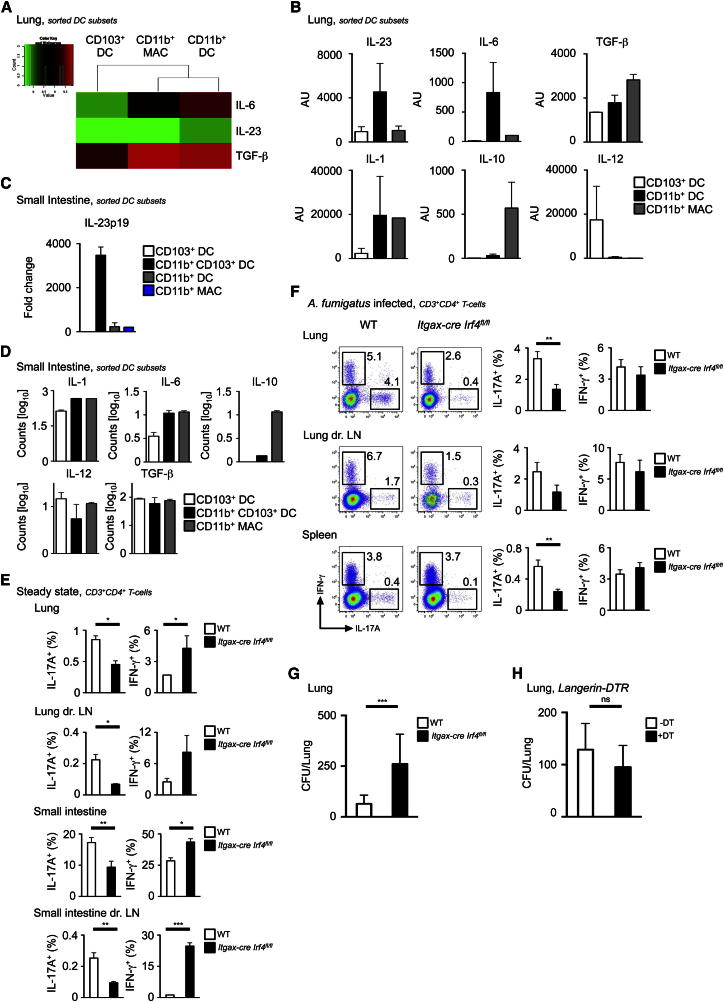
The loss of LP CD11b+CD103+ DCs, which correspond to our IRF4-dependent CD24+CD64−CD11b+CD103+ DC subset, is associated with reduced IL-17-producing CD4+ T cells in the intestine (Lewis et al., 2011). We also observed that CD11b+ DCs expressed Th17 cell-polarizing molecules, such as IL-23p19, IL-6, and TGF-β in our microarray data (Figure 5A). We quantified IL-23p19 mRNA expression from unstimulated sorted tissue DC and MAC populations (for sorting strategy, see Figure S1) and confirmed that lung CD11b+ DCs and SI-LP CD11b+CD103+ DCs homeostatically expressed IL-23p19, IL-6, TGF-β, and IL-1, but not IL-10 and only minute amounts of IL-12 mRNA (Figures 5B–5D), underlining the functional homology between these two subsets. We quantified the proportion of CD3+CD4+ T cells expressing interferon-γ (IFN-γ) or IL-17A in the lung and SI-LP of WT compared to Itgax-cre Irf4fl/fl by intracellular cytokine staining. As expected (Lewis et al., 2011), in Itgax-cre Irf4fl/fl mucosal tissues, the proportion of Th17 cells was reduced in steady state compared to WT, whereas Th1 cells were concomitantly increased (Figure 5E; Figure S5E).
Figure 5.
CD11b+CD24+ DCs in Lung and SI Control Th17 Responses
(A–D) Expression of indicated genes in lung DCs and MACs by microarray (A), by Q-PCR in lung (B), and Q-PCR and nanostring in SI-LP (C and D) DCs and MACs (mean ± SEM, n = 3).
(E) Percentage of IL-17A and IFN-γ expressing CD3+CD4+ T cells in steady state from lung, lymph node, SI-LP, and MLN of WT (white bars) and Itgax-cre Irf4fl/fl (black bars) mice (mean ± SEM, pooled results of two experiments, n = 10). (F) Same as (E) after A. fumigatus challenge.
(F) Representative dot plot shown left and bar graphs show composite data (mean ± SEM, pooled results of four experiments, n = 20).
(G) CFUs from homogenized lung tissue of either WT (white bars) or Itgax-cre Irf4fl/fl (black bars) mice 7 days after intranasal infection with A. fumigatus (mean ± SEM, pooled results of four experiments, n = 20).
(H) Same than (F) with Langerin-DTR mice (white bars) or Langerin-DTR mice injected with DT (black bars) (mean ± SEM, pooled results of two experiments, n = 10). See also Figures S4 and S5.
IRF4-Dependent CD24+CD11b+ Lung DCs Control Th17 Cell Responses
Next, we assessed the functional consequences of the absence of lung CD11b+ DCs in an infection model dependent on Th17 cell responses such as the fungal pathogen A. fumigatus, a prevalent environmental fungus that causes potentially lethal infections in immunosuppressed individuals (Hohl and Feldmesser, 2007; Romani, 2004).
We first infected WT and Itgax-cre Irf4fl/fl mice with a sublethal dose of live A. fumigatus conidia and monitored IFN-γ and IL-17A production by CD3+CD4+ T cells on day 7 of infection. Itgax-cre Irf4fl/fl mice exhibited decreased effector Th17 T cell responses compared to WT in the lung (Figure 5F). However, to determine whether this decreased Th17 cell response was specifically due to the absence of lung CD11b+ DCs, we performed a similar experiment in a mouse model (langerin-DTR) in which the CD11b−CD103+ DCs can be selectively ablated upon Diphtheria toxin (DT) administration (GeurtsvanKessel et al., 2008). There was no reduction in Th17 cell responses in CD103+ DC-depleted compared to CD103+ DC-sufficient mice (Figure S5F). Fungal burdens were assessed by quantitative culture of fungal colony-forming units (CFU) from homogenized infected lungs of WT, Itgax-cre Irf4fl/fl, and langerin-DTR mice (Figures 5G and 5H). As expected from the altered Th17 T cell response in Itgax-cre Irf4fl/fl, the fungal burdens were significantly higher in these mice (Figure 5G), although without associated mortality. In contrast, in line with the unchanged Th17 T cell response in DT treated langerin-DTR, the fungal burdens were similar to control levels (Figure 5H). These data identify an IRF4-dependent CD11b+CD24+CD64− DC population in mucosal tissues with functional specialization to instruct IL-17 responses during steady state and infection.
Transcriptomic Alignment of Mouse IRF4-Dependent CD11b+ DCs with Human CD1c+ DCs
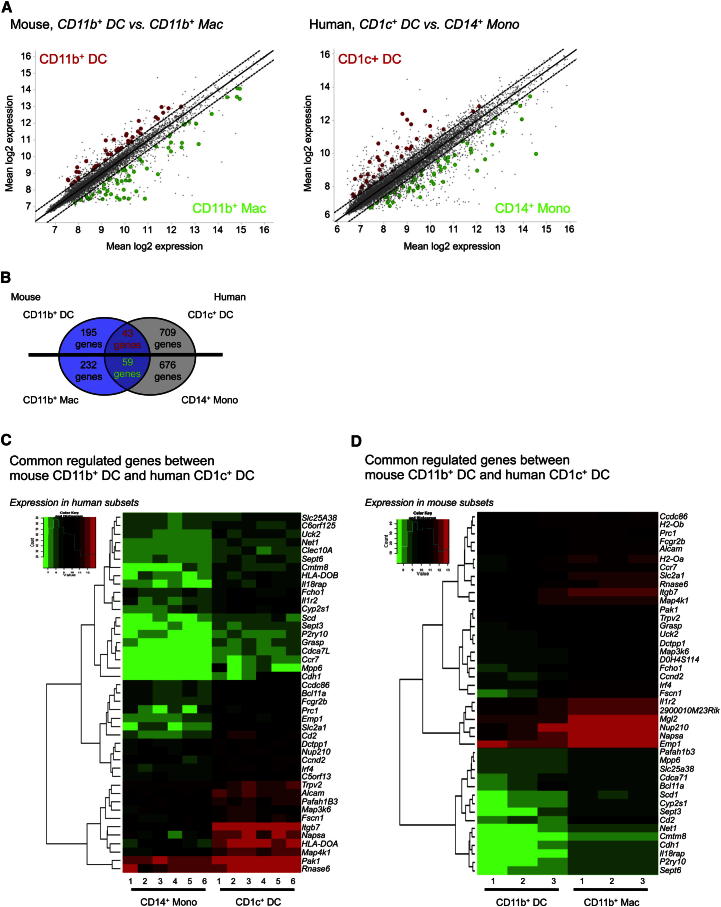
To address the putative relationship between mouse CD11b+ DCs and human CD1c+ DCs, we first identified the differentially expressed genes (DEG) between mouse lung CD11b+ DCs and CD11b+ MACs and then identified their human orthologs as described in the Experimental Procedures section. Given the logistical difficulties of obtaining sufficient material to perform microarray analysis on human lung DCs, we used microarray data of equivalent cells in human blood. We previously showed that CD14+ monocytes were positively associated with interstitial CD14+ DCs (Haniffa et al., 2012) and also cluster with interstitial MACs (data not shown). We predicted that DEG between CD11b+ DCs and CD11b+ MACs would correspond to DEG between human blood CD1c+ DCs and CD14+ monocytes. Therefore, we identified the DEG between human blood CD1c+ DCs and CD14+ monocytes (Figure 6A; Tables S2 and S3) and compared them between species. Forty-three genes were commonly differentially expressed in both mouse CD11b+ DCs and blood CD1c+ DCs compared to CD11b+ MACs and CD14+ monocytes, respectively (p = 2.32e−18) (Figure 6B) and include Irf4, Ccr7, and the cell-cycle genes Cdh1 and Cdca71 (Figures 6C and 6D). Fifty-nine genes were commonly differentially expressed in both mouse CD11b+ MACs and CD14+ monocytes compared to CD11b+ DCs and blood CD1c+ DCs respectively (p = 4.46e−33) (Figure 6B). The reverse approach identified only 14 genes commonly differentially expressed in both mouse CD11b+ DCs and blood CD14+ monocytes compared to CD11b+ MACs and CD1c+ DCs, respectively (p = 0.046) (data not shown). The higher enrichment of common DEG between mouse CD11b+ DCs and human CD1c+ DCs compared to CD11b+ DCs and CD14+ monocytes implies homology between human CD1c+ DCs and murine CD11b+ DCs.
Figure 6.
Transcriptomic Alignment of Mouse IRF4-Dependent CD11b+ DCs with Human CD1c+ DCs
(A) Scatterplot of mouse CD11b+ DCs versus CD11b+ MACs and human CD1c+ DCs and CD14+ monocytes. Common DEG for mouse CD11b+ DCs and human CD1c+ DCs are highlighted in red, whereas common DEG for mouse CD11b+ MACs and CD14+ monocytes are highlighted in green. All genes depicted are regulated < 1.5-fold.
(B) Analysis strategy depicting the number of DEG, which are shared and not shared between mouse CD11b+ DCs, mouse CD11b+ MACs, human CD1c+ DCs, and human CD14+ monocytes.
(C) Heatmap showing expression in CD14+ monocytes and CD1c+ DCs of common DEG identified between mouse CD11b+ DCs and human CD1c+ DCs.
(D) Same than (C) with mouse CD11b+ DCs and CD11b+ MACs. See also Figure S7 and Tables S2 and S3.
Human IRF4-Expressing CD1c+ DCs Induce IL-17 T Helper Responses following A. fumigatus Challenge
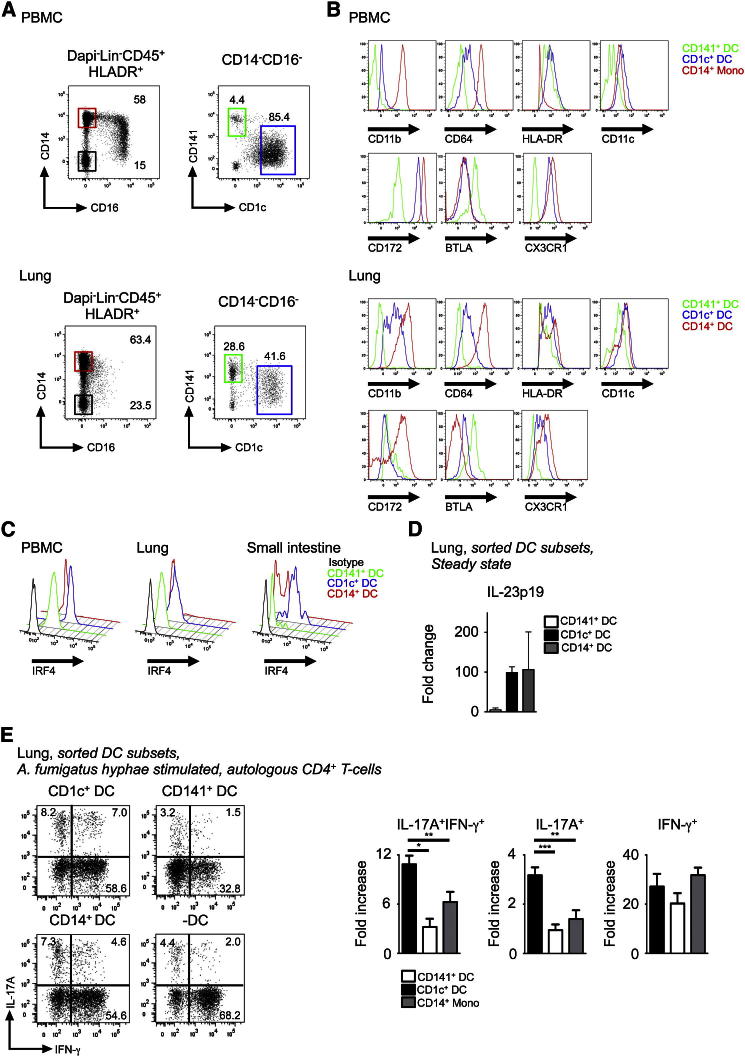
Phenotypic analysis of human blood and lung DC subsets for DC and macrophage antigens showed similar expression profile for CD11b, CD64, CD11c, and CX3CR1 on blood and lung CD1c+ DCs relative to their respective tissue CD141+ DCs and CD14+ monocyte or DCs. Both human lung CD1c+ DC and murine CD11b+ DCs are CD11b+, CD14−, CD11c+, BTLA+, and CX3CR1+ (Figures 7A and 7B; Figure S6A). We next tested whether human CD1c+ DCs expressed IRF4 protein, in line with our microarray data and the IRF4-dependent nature of murine tissue CD11b+ DCs. We assessed intracellular IRF4 expression on blood, lung, and SI DC subsets by flow cytometry and observed highest IRF4 expression on CD1c+ DCs from all three tissues (Figure 7C). Because human Th17 cell responses to pathogens including A. fumigatus have been documented (Chamilos et al., 2010; Sallusto et al., 2012), we assessed whether CD1c+ DCs were capable of inducing Th17 cell responses following A. fumigatus challenge. Unstimulated lung CD1c+ DCs expressed higher levels of the Th17 cell polarization cytokine IL-23p19 transcript compared to CD141+ DCs but at a comparable level to CD14+ DCs (Figure 7D). We also measured cytokine secretion by blood CD1c+ DCs and CD14+ monocytes in response to a range of stimuli (Figure S6B). Blood CD1c+ DCs consistently produced higher amounts of IL-23p19 upon stimulation with CD40L+LPS, the TLR3 agonist poly I:C and the TLR7-8 agonist CL075 in addition to A. fumigatus hyphae stimulation. IL-12p70 was detected only upon stimulation of CD1c+ DCs with CL075 but at a lower concentration to IL-23p19 by using the same stimulus.
Figure 7.
Human IRF4 Expressing CD1c+ DCs Induce IL-17 T Helper Response
Flow cytometry of peripheral blood and mechanically dispersed lung. Gating strategy used to identify three myeloid DC subsets within Lin−HLA-DR+ fraction (CD123+ pDCs were excluded from the CD14−CD16− fraction): (1) CD14+ monocytes (red gate), (2) CD14−CD1c+CD141− DCs (blue gate), (3) CD14−CD1c−CD141+ DCs (green gate).
(A) Representative from ten blood and three lung donors are shown. Relative expression of indicated markers by CD14+ monocytes/DCs (red), CD1c+ DCs (blue), and CD141+ DCs (green).
(B) Representative data from three blood and lung donors are shown.
(C) Same as (B) for IRF4 expression compared to isotype (black) from indicated tissues. Representative data from three blood, lung, and two SI donors are shown. mRNA expression of IL-23p19 by freshly sorted human DC subsets from lung (n = 3, mean ± SEM).
(D) Expression normalized to IL-23p19 mRNA expression in unstimulated CD141+ DCs. Intracellular IFN-γ and IL-17A expression by PMA/Ionomycin restimulated CD4+ T cells cultured with autologous lung indicated DC subsets pulsed with A. fumigatus hyphae.
(E) Representative dot plot with percentage of total IL-17+ cells and composite results shown in bar graph (n = 4, mean ± SEM fold increase [%+DC/%-DC]).
In order to assess Th17 cell induction by human DCs, we cultured A. fumigatus hyphae stimulated lung and blood DC subsets with autologous CD4+ T cells and assessed T cell IL-17A and IFN-γ production. Both lung and blood CD1c+ DCs were potent inducers of CD4+ T cell IL-17A production compared to CD141+ DCs and CD14+ DC and/or monocytes (Figure 7E; Figures S6C and S6D), demonstrating conserved interspecies IRF4-expressing DC function in relation to IL-17 immune responses following A. fumigatus challenge.
Discussion
This study describes the phenotype and function of human and murine IRF4+CD11b+ DCs found in mucosal tissues that control Th17 T cell differentiation through the secretion of IL-23 during steady state and inflammation. We have identified FLT3-dependent CD11b+CD24+CD64− bona fide DCs and CSF-1R-dependent CD11b+CD24−CD64+ MACs in steady state lung and SI. The separation of these two cell types clarifies previous suggestions of heterogeneity within tissue MHCII+CD11c+CD11b+ cells (Ginhoux et al., 2009). Supporting these data, the DC lineage-restricted TF Zbtb46 is highly expressed in the CD24+ fraction of lung MHCII+CD11c+CD11b+ and gut LP MHCII+CD11c+CD11b+CD103+ populations (Satpathy et al., 2012). Similarly, CD64, a recognized marker of human macrophages, was recently shown to identify murine macrophages (Gautier et al., 2012; Tamoutounour et al., 2012) and inflammatory lung (Plantinga et al., 2013) and muscle (Langlet et al., 2012) DCs. Gautier et al. also highlighted heterogeneity within lung CD11b+ DCs, describing CD24+CD64− cells potentially as DCs and CD24−CD64+ cells as macrophages (Gautier et al., 2012). We now propose a general gating strategy for analyzing the CD11b+ DCs in gut and lung, which will remove contaminating MHCII+ macrophages.
Investigating the parallel between NLT and LT DCs, we found that gut CD24+CD11b+CD103+ DCs and lung CD24+CD11b+ DCs expressed IRF4, similar to splenic CD4+CD11b+ DCs. However, unlike splenic CD4+CD11b+ DCs, both NLT DC populations were dependent on IRF4 as shown by the decreased contribution of the IRF4-deficient BM cells to these populations in mixed BM chimeras and their dramatic reduction in mice lacking IRF4 expression in the DC lineage. Importantly, DC specific IRF4 deficiency did not fully ablate lung or gut CD24+CD11b+ DCs, suggesting that IRF4 is not involved in their differentiation but rather in their survival or proliferation. In agreement with this hypothesis, we found no differences in the relative numbers of DC progenitors in Itgax-cre Irf4fl/fl versus WT mice suggesting that IRF4 involvement is not at the DC progenitor level. IRF4 appears to have a prosurvival effect on NLT CD11b+ DCs because the remaining lung CD11b+ DCs lacking IRF4 exhibited enhanced mitochondrial fragmentation compared to WT cells. This finding is in agreement with a study reporting that IRF4-inhibited apoptosis in myeloid cells (Shaffer et al., 2008). The exact molecular mechanisms by which IRF4 regulates mucosal CD11b+ DC survival remain to be investigated.
We could not identify similar IRF4-dependent CD11b+ DCs in the dermis, kidney, or liver. CD24 and CD64 did not separate the CD11b+ DC compartment and we did not detect any IRF4-dependent CD11b+ DCs in both IRF4 mixed BM chimeras and in mice lacking IRF4 in the DC lineage in these tissues. These data suggest that IRF4-dependent CD11b+ DCs are specific to the lung and the SI, suggesting the existence of a mucosal CD11b+ DC lineage. This implies that CD11b+ DCs may not be as homogeneous and conserved as the CD8α or CD103 lineage. It remains to be ascertained if such mucosal specificity is regulated at the tissue level by local conditioning of common circulating DC progenitors or through a distinct lineage of specific mucosal DC progenitors. Local microbiota present in mucosal tissues could control IRF4 expression that in turn promotes CD11b+ DC survival. However, we found no difference in the relative numbers of mucosal tissue CD11b+ DC composition, as well as IRF4 expression levels in germ-free mice compared to control SPF mice (data not shown). Evidence for a restricted progenitor potential was recently shown by BM CCR9− pDC progenitors specifically giving rise to CD11c+MCHII+CD11b+ cells in the lung upon adoptive transfer (Schlitzer et al., 2012). A gut-tropic migratory DC precursor preferentially giving rise to gut CD11b−CD103+ DCs was also recently described (Zeng et al., 2012).
Migratory CD11b+ DCs are reduced in skin draining LNs of the IRF4-deficient mice, whereas both CD11b+ and CD103+ DCs are increased in the dermis of these mice (Bajaña et al., 2012). We did not detect any major defect in the splenic or dermal DC network of IRF4 mixed BM chimeras or in mice lacking IRF4 in the DC lineage compared to the IRF4-deficient mice, suggesting that either the IRF4 molecular mechanism of action is different in these tissues or that an additional cell type is involved, which confers the observed phenotype in the dermis (Bajaña et al., 2012) and the spleen of the full IRF4-deficient mice (Suzuki et al., 2004; Tamura et al., 2005). However, we observed defective migration of CD11b+ DCs in the skin draining LN, MLN, and LLN. This observation suggests that IRF4, in addition to its specific role in the homeostasis of mucosal CD11b+ DCs, plays an additional role in the migration of peripheral tissue CD11b+ DCs.
IL-23 is a crucial cytokine for the induction of Th17 cells at mucosal sites such as the lung and the SI (Aggarwal et al., 2003; Ahern et al., 2010). IL-23 at mucosal sites can be induced by ligands derived from the microbiota (Ivanov et al., 2009). However, the cell type responding to such exogenous cues in mucosal tissues is ill understood. Previous data highlighted the specific expression of IL-23 by a subset of SI-LP TLR5+CD11b+ DCs promoting Th17 T helper cells (Uematsu et al., 2008), and by flagellin-stimulated CD11b+CD103+ DCs (Kinnebrew et al., 2012). Another mouse model devoid of SI-LP CD11b+CD103+ DCs exhibited a reduced fraction of Th17 cells (Lewis et al., 2011). This is also consistent with the recently reported capacity of CD11b+CD103+ DCs to induce Th17 cell differentiation in vitro (Denning et al., 2011). Moreover, clearance and protection of asthma induced by A. fumigatus infection is mediated by TLR6 expression on CD11b+ DCs, which ultimately leads to secretion of IL-23 and induction of protective Th17 immunity (Moreira et al., 2011). In line with these reports, our data show that CD11b+ DCs in the lung, as well as CD11b+CD103+ DCs in the SI, express IL-23 during steady state and that their selective loss leads to reduced IL-17A secreting CD4+ T cells and a concomitant increase in IFN-γ-producing T cells. Furthermore, upon A. fumigatus infection in mice specifically lacking lung CD11b+ DCs, Th17 T cells were reduced in numbers. However, Th17 polarization may not be exclusive to IRF4-dependent CD11b+ DCs, because we did not study the capacity of IRF4-independent CD11b+ DCs in other nonmucosal tissues and inflammatory DCs to promote Th17 immunity.
IRF4-dependent CD11b+ DCs may not only be specialized in Th17 polarization. In our hands, IRF4-dependent CD11b+ DCs did not contribute to Th1 polarization in a model of Influenza infection because we found no difference in the relative numbers of IFN-γ-secreting CD4+ T cells between Itgax-cre Irf4fl/fl and WT mice after Influenza challenge (data not shown). However, they can contribute to Th2 polarization when challenged with the adequate stimuli as shown recently in a model of low-dose house dust mite allergen (Plantinga et al., 2013).
The separation of murine CD11b+ DCs from CD11b+ MACs has now revealed the transcriptomic, phenotypic, and functional similarities between human CD1c+ DCs and the bona fide mouse CD11b+ DCs. Our previous transcriptomics approach to align human and mouse DCs employed gene signature generation for each DC subset followed by enrichment analysis to interrogate signature similarities and differences between subsets. By using the same approach, we found the lung CD24+CD11b+ DC gene signature to be related to the human CD1c lineage, but the association was not statistically significant. This was due to the modest signature list of 167 genes for the human CD1c+ DC lineage, and the lower power of the nonparametric Kolmogorov-Smirnov test used in the signature analysis (data not shown). However, by using the predictions of our previous analysis that CD11b+ MACs would be closer to CD14+ monocytes than CD1c+ DCs and the converse relationship for CD11b+ DCs with CD1c+ DCs rather than CD14+ monocytes, we were able to confirm the homology between CD11b+ DCs with CD1c+ DCs by analyzing shared DEG between the two mouse and human subsets. This strategy utilized a more powerful parametric (hypergeometric) analysis to enhance statistical sensitivity to test this hypothesis. We identified biologically significant genes shared by mouse CD11b+ DCs and human CD1c+ DCs compared to CD11b+ MACs and CD14+ monocytes respectively such as Irf4, Ccr7, and the cell-cycle genes Cdh1 and Cdca71. The homology identified between mouse CD11b+ DCs and human CD1c+ DCs through transcriptomics was further substantiated by phenotypic and functional similarities.
We found highest amount of IRF4 expression on human CD1c+ DCs. Lung CD1c+ DCs also expressed significantly higher levels of IL-23p19 transcript compared to CD141+ DCs, but at a comparable level to CD14+ DCs. IL-23 production has been described in monocyte and monocyte-related inflammatory DCs (Plantinga et al., 2013; Segura et al., 2013) and is in keeping with the close association of human CD14+ DCs to the monocyte and/or macrophage lineage (Haniffa et al., 2012). We also observed IL-23p19 and IL-12p70 coproduction by blood CD1c+ DCs upon CL075 stimulation. IL-12p70 secretion was not detected from human skin CD1c+ DCs (Haniffa et al., 2012) and LP mononuclear cells (Dillon et al., 2010).
Nevertheless, both human lung and blood CD1c+ DCs pulsed with A. fumigatus hyphae were superior at inducing IL-17A production by autologous CD4+ T cells. The majority of Th17 cells generated did not coproduce IFN-γ or IL-22 (data not shown), distinct to the IL-17 and IFN-γ-producing Th17 cells observed when naive CD4+ T cells were cultured with autologous monocytes pulsed with C. albicans (Zielinski et al., 2012). A variety of in vitro systems incorporating allogeneic and pathogen-induced models have been utilized to study human Th17 differentiation (Acosta-Rodriguez et al., 2007; Hansel et al., 2011). Blood CD14+ monocytes and Slan+ DCs have been previously shown to induce IL-17A-producing T helper cells in allogeneic models (Acosta-Rodriguez et al., 2007; Hansel et al., 2011). It is therefore possible that Th17 polarization is a specific function of, but not exclusive to human IRF4-expressing CD1c+ DCs. This is suggested by the reasonable capacity of CD14+ DCs to induce Th17 polarization ex vivo, although it remains to be proven whether this subset migrates to the draining LN and is capable of priming a naive response.
Finally, our observations advance the understanding of the parallel organization of the mononuclear phagocyte system in mouse and human mucosal tissues in steady state. Both murine and human tissues contain resident SSChi macrophages and two major DC subsets; mouse CD103+ DCs related to human CD141+ DCs with cross-presentation abilities and mouse CD11b+ DCs, which we now show to be related to human CD1c+ DCs with Th17 polarizing capabilities. In addition, human tissues also contain CD14+ DCs with monocyte and/or macrophage characteristics. The murine counterpart of human CD14+ DCs is currently unknown. It is tempting to speculate that the SSCloCD11b+CD64+CD14+ macrophage fraction we have characterized here (CD11b+ MACs) contains the functional equivalent of human CD14+ DCs, although this requires further validation. Functional alignment of mucosal mouse CD11b+ DCs and human CD1c+ DCs will facilitate the translation of mouse in vivo findings to human DC biology and aid the development of DC-based therapeutic strategies.
Experimental Procedures
Murine and Human Cell Suspension Preparation
Cell suspension from mouse organs and LN were prepared according to published methods (Ginhoux et al., 2009; Bogunovic et al., 2009). Human samples were obtained with informed consent in accordance with a favorable ethical opinion from Newcastle, Munich, and Singapore Singhealth and National Health Care Group Research Ethics Committees. PBMCs and lung DCs were isolated as described previously (Haniffa et al., 2012). Colonic mucosa was digested with Collagenase and DNase and passed through a 70 μm cell strainer. Autologous CD4+ T cells were isolated from fresh blood by using the RosetteSep human CD4+ T cell enrichment kit (StemCell Technologies). Autologous naive CD4+ T cells were isolated by using the EasySep human naive CD4+ T cell enrichment kit.
Infection Protocols
A. fumigatus (AF293, from ATCC) were cultivated for 5 days on Potato dextrose agar (Sigma Aldrich, USA) and washed with PBS 0.05% Tween 20 to harvest A. fumigatus conidia. For infection, mice were anesthetized prior to instillation of a suspension of 2 × 107 conidia/20 μl PBS intranasally (i.n.). For infection in langerin-DTR mice, DT was administered intraperitoneally (i.p.) at day −1 and day + 1 after live A. fumigatus conidia instillation and the proportion of lung and lung LN CD3+CD4+ T expressing IFN-γ or IL-17A was measured at day 7. Mice were also monitored for fungal growth (CFU/organ, mean ± SEM) by serially diluting homogenates and plating them in triplicate on Sabouraud agar plates.
Microarray and Analysis
Total cellular RNA was extracted by using the mirVana™ miRNA isolation kit (Ambion Inc, Austin, TX, USA) for mouse samples and Trizol (Invitrogen, Karlsbad, USA) for human samples and prepared for microarray (Illumina Mouse WG6, Illumina Inc, San Diego, CA, USA) according to manufacturer’s instructions. The human and mouse microarray expression results were quantile normalized and summarized from probe level to gene level by taking the mean expression across multiple probes mapping to the same gene. Signatures were obtained in the human data set by selecting genes that showed at least a 1.5-fold difference in mean expression between the blood CD1c+ DC and the blood CD14+ monocyte samples. Similarly, the mouse signatures were obtained by using the same methodology, but between lung CD11b+ DCs and lung CD11b+ MACs. Details of transcriptomic analysis are available in Supplemental Experimental Procedures.
T Cell Polarization Assay
FACS-sorted blood DCs were cultured at 5 × 104 cells/ml in 10% FCS IMDM medium in a 96 well round-bottom plate. DCs were treated with A. fumigatus hyphae (1 hyphae: 10 DCs) o/n. After o/n DC stimulation, 1 × 106/ml CD4+ T cells were added to DCs (1 DC: 20 T cells) or incubated alone ± stimuli. On day 6, fresh media containing 10 U/mL IL-2 was added and cells were transferred to a 96 well flat-bottom plate. On day 10, T cells were stimulated with phorbol myristate acetate (PMA; 10ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich) for 4.5 hr in the presence of 10 μg/ml Brefeldin A (10 μg/ml) after the first hour.
Statistical Analysis
Mann-Whitney, Kruskal Wallis, and unpaired t tests (with a 95% confidence) were performed by using Prism 6.0 (GraphPad Software, La Jolla, USA). All p values are two-tailed. *p < 0.05; **p < 0.01; and ***p < 0.001.
Acknowledgments
This work was supported by The Wellcome Trust, UK (WT088555MA; M.H. and N.M.); Singapore Immunology Network core grant (F.G., P.R.-C., L.G.N. and L.R.); Histiocytosis Association and Histiocytosis Research Trust (M.C.), by the National Institutes of Health grant (CA26504; E.R.S.), and by JGW Patterson Foundation (H.A.P. and C.M.U.H.). We thank R. Basu for technical assistance; L. Robinson for critical review and editing of the manuscript; M.L. Ng, S.H. Tan, and T.B. Lu from the Electron Microscopy Unit of the National University of Singapore; S. Pettersson of the Singapore General Hospital and I. Dimmick and D. McDonald at the Flow Cytometry Core Facility of the Faculty of Medicine Newcastle University; A. Larbi, I. Low, and N. Binte Shadan at the Flow Cytometry Core Service; J. Connolly and K. Ng at the Immunomonitoring platform, J. Lum and F. Zolezzi at the Functional Genomics Laboratory of SIgN; S. Stamenkovic and S. Barnard from the Freeman Hospital Cardothoracic Unit; J. Majo, N. Thampy, and F. Black from the Royal Victoria Infirmary Department of Cellular Pathology at Newcastle upon Tyne.
Published: May 23, 2013
Footnotes
Supplemental Information includes six figures, three tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2013.04.011.
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Accession Numbers
The microarray data have been deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession number GSE46680.
Supplemental Information
References
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- Aggarwal S., Ghilardi N., Xie M.H., de Sauvage F.J., Gurney A.L. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- Ahern P.P., Schiering C., Buonocore S., McGeachy M.J., Cua D.J., Maloy K.J., Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annacker O., Coombes J.L., Malmstrom V., Uhlig H.H., Bourne T., Johansson-Lindbom B., Agace W.W., Parker C.M., Powrie F. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J. Exp. Med. 2005;202:1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S., Roach K., Turner S., Paul J., Kovats S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 2012;189:3368–3377. doi: 10.4049/jimmunol.1102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., Briere F., Caux C., Davoust J., Lebecque S., Liu Y.J., Pulendran B., Palucka K. Immunobiology of dendritic cells. Annu. Rev. Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Bedoret D., Wallemacq H., Marichal T., Desmet C., Quesada Calvo F., Henry E., Closset R., Dewals B., Thielen C., Gustin P. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J. Clin. Invest. 2009;119:3723–3738. doi: 10.1172/JCI39717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V., Houston S.A., Scott C.L., Aumeunier A., Yrlid U., Mowat A.M., Milling S.W. Intestinal CD103(-) dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 2012;6:104–113. doi: 10.1038/mi.2012.53. [DOI] [PubMed] [Google Scholar]
- Chamilos G., Ganguly D., Lande R., Gregorio J., Meller S., Goldman W.E., Gilliet M., Kontoyiannis D.P. Generation of IL-23 producing dendritic cells (DCs) by airborne fungi regulates fungal pathogenicity via the induction of T(H)-17 responses. PLoS ONE. 2010;5:e12955. doi: 10.1371/journal.pone.0012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Contreras V., Feuillet V., Dutertre C.A., Ventre E., Vu Manh T.P., Baranek T., Storset A.K., Marvel J. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning T.L., Norris B.A., Medina-Contreras O., Manicassamy S., Geem D., Madan R., Karp C.L., Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S.M., Rogers L.M., Howe R., Hostetler L.A., Buhrman J., McCarter M.D., Wilson C.C. Human intestinal lamina propria CD1c+ dendritic cells display an activated phenotype at steady state and produce IL-23 in response to TLR7/8 stimulation. J. Immunol. 2010;184:6612–6621. doi: 10.4049/jimmunol.1000041. [DOI] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., Immunological Genome Consortium Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Willart M.A., van Rijt L.S., Muskens F., Kool M., Baas C., Thielemans K., Bennett C., Clausen B.E., Hoogsteden H.C. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J. Exp. Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.N., Malinarich F., Malleret B. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37:60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel A., Gunther C., Ingwersen J., Starke J., Schmitz M., Bachmann M., Meurer M., Rieber E.P., Schakel K. Human slan (6-sulfo LacNAc) dendritic cells are inflammatory dermal dendritic cells in psoriasis and drive strong TH17/TH1 T-cell responses. J Allergy Clin Immunol. 2011;127:787–794. doi: 10.1016/j.jaci.2010.12.009. e781–789. [DOI] [PubMed] [Google Scholar]
- Hashimoto D., Miller J., Merad M. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 2011;35:323–335. doi: 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl T.M., Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot. Cell. 2007;6:1953–1963. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew M.A., Buffie C.G., Diehl G.E., Zenewicz L.A., Leiner I., Hohl T.M., Flavell R.A., Littman D.R., Pamer E.G. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Langlet C., Tamoutounour S., Henri S., Luche H., Ardouin L., Grégoire C., Malissen B., Guilliams M. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J. Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou J.C., Youle R.J. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., Immunological Genome Consortium Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A.P., Cavassani K.A., Ismailoglu U.B., Hullinger R., Dunleavy M.P., Knight D.A., Kunkel S.L., Uematsu S., Akira S., Hogaboam C.M. The protective role of TLR6 in a mouse model of asthma is mediated by IL-23 and IL-17A. J. Clin. Invest. 2011;121:4420–4432. doi: 10.1172/JCI44999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka K., Banchereau J., Mellman I. Designing vaccines based on biology of human dendritic cell subsets. Immunity. 2010;33:464–478. doi: 10.1016/j.immuni.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga M., Guilliams M., Vanheerswynghels M., Deswarte K., Branco-Madeira F., Toussaint W., Vanhoutte L., Neyt K., Killeen N., Malissen B. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Poulin L.F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J.L., Keller A.M., Joffre O., Zelenay S., Nye E. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L. Immunity to fungal infections. Nat. Rev. Immunol. 2004;4:1–23. doi: 10.1038/nri1255. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Zielinski C.E., Lanzavecchia A. Human Th17 subsets. Eur. J. Immunol. 2012;42:2215–2220. doi: 10.1002/eji.201242741. [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., Murphy K.M. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Heiseke A.F., Einwächter H., Reindl W., Schiemann M., Manta C.P., See P., Niess J.H., Suter T., Ginhoux F., Krug A.B. Tissue-specific differentiation of a circulating CCR9- pDC-like common dendritic cell precursor. Blood. 2012;119:6063–6071. doi: 10.1182/blood-2012-03-418400. [DOI] [PubMed] [Google Scholar]
- Schulz O., Jaensson E., Persson E.K., Liu X., Worbs T., Agace W.W., Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Touzot M., Bohineust A., Cappuccio A., Chiocchia G., Hosmalin A., Dalod M., Soumelis V., Amigorena S. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013;38:336–348. doi: 10.1016/j.immuni.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Emre N.C., Lamy L., Ngo V.N., Wright G., Xiao W., Powell J., Dave S., Yu X., Zhao H. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortman K., Naik S.H. Steady-state and inflammatory dendritic-cell development. Nat. Rev. Immunol. 2007;7:19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- Steinman R.M., Hawiger D., Nussenzweig M.C. Tolerogenic dendritic cells. Annu. Rev. Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Honma K., Matsuyama T., Suzuki K., Toriyama K., Akitoyo I., Yamamoto K., Suematsu T., Nakamura M., Yui K., Kumatori A. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc. Natl. Acad. Sci. USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour S., Henri S., Lelouard H., de Bovis B., de Haar C., van der Woude C.J., Woltman A.M., Reyal Y., Bonnet D., Sichien D. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- Tamura T., Tailor P., Yamaoka K., Kong H.J., Tsujimura H., O’Shea J.J., Singh H., Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 2005;174:2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- Uematsu S., Fujimoto K., Jang M.H., Yang B.G., Jung Y.J., Nishiyama M., Sato S., Tsujimura T., Yamamoto M., Yokota Y. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- Zeng R., Oderup C., Yuan R., Lee M., Habtezion A., Hadeiba H., Butcher E.C. Retinoic acid regulates the development of a gut-homing precursor for intestinal dendritic cells. Mucosal Immunol. 2012 doi: 10.1038/mi.2012.123. Published online December 12, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski C.E., Mele F., Aschenbrenner D., Jarrossay D., Ronchi F., Gattorno M., Monticelli S., Lanzavecchia A., Sallusto F. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.