Abstract
The advent of massively parallel sequencing technologies has allowed the characterization of cancer genomes at an unprecedented resolution. Investigation of the mutational landscape of tumours is providing new insights into cancer genome evolution, laying bare the interplay of somatic mutation, adaptation of clones to their environment and natural selection. These studies have demonstrated the extent of the heterogeneity of cancer genomes, have allowed inferences to be made about the forces that act on nascent cancer clones as they evolve and have shown insight into the mutational processes that generate genetic variation. Here we review our emerging understanding of the dynamic evolution of the cancer genome and of the implications for basic cancer biology and the development of antitumour therapy.
Cancer is a disease of the genome. The classic model of carcinogenesis describes multiple, successive clonal expansions driven by the accumulation of genomic changes or ‘mutations’ that are preferentially selected by the tumour environment1,2,3. This traditional picture of linear cancer genome evolution has become more nuanced over the past decade as the research scalpel allows ever-sharper prosection of the underlying biology (FIG. 1; BOX 1).
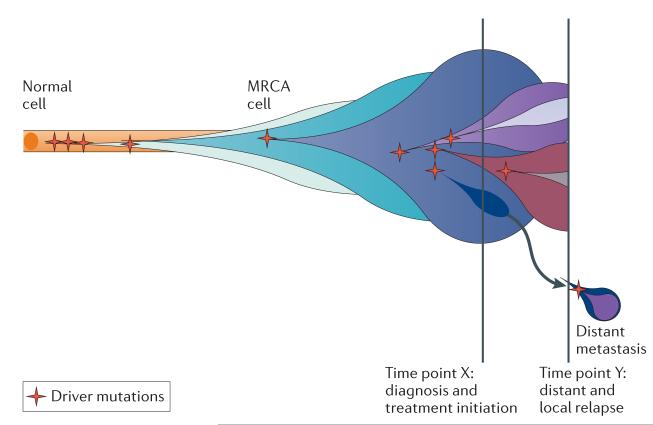
Figure 1. The evolution of clonal populations.
Cancers are genomically diverse and dynamic entities. Unique clones (represented by different coloured bubbles) emerge as a consequence of accumulating driver mutations in the progeny of a single most recent common ancestor (MRCA) cell. Ongoing linear and branching evolution results in multiple simultaneous subclones that may individually be capable of giving rise to episodes of disease relapse and metastasis. The dynamic clonal architecture is shaped by mutation and competition between subclones in light of environmental selection pressures, including those that are exerted by cancer treatments.
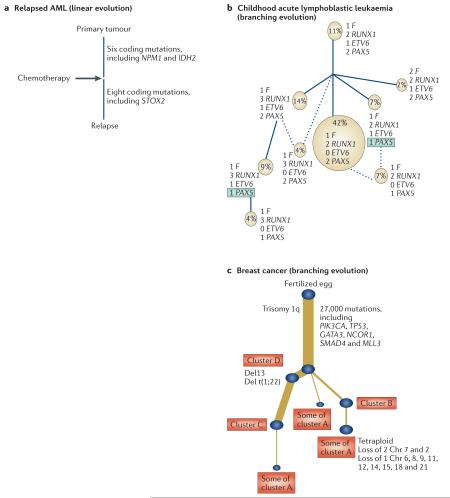
Box 1. Phylogenetic cancer trees.
A phylogenetic tree is a pictorial representation of how a tumour is inferred to have evolved. As discussed in the text, these inferences can be based on a wide range of molecular biology and sampling techniques coupled with existing and new bioinformatics algorithms for reconstructing the tree. Several key properties of the evolution of a tumour are coded in the tree and provide important biological information about the genetic diversity of a cancer and clonal mix.
All trees have a shared ‘trunk’, which represents the complement of mutations shared by all malignant cells within the cancer. Because these mutations are fully clonal, there must have been a single ancestral cell that carried all of these mutations and through which all extant tumour cells can trace their lineage; we denote this cell the ‘most recent common ancestor’, borrowing the term from population genetics. Emergence of this cell initiated the final complete selective sweep within the cancer: all clonal expansions thereafter are, by definition, incomplete. All mutations that occur after the most recent appearance of a common ancestor are subclonal.
The length of individual branches (and the trunk) denotes the number of mutations that occurs in that lineage: a so-called ‘molecular clock’. If mutation rates per unit time were constant, then this would correlate with chronological time. However, for many cancers, this assumption is probably invalid (as discussed in the text), and molecular time is likely to be a poor proxy for chronological time.
The branching structure of the tree captures the number of subclonal populations within the cancer samples and their genetic relationships. For example, both linear and branching patterns of evolution have been described in a range of cancers. Linear evolution (panel a of the figure) was described in acute myeloid leukaemia (AML) and identifies the post-treatment relapse clone as a direct descendant of the major clone. The tree in panel b demonstrates branching evolution and specifically convergent evolution, in which the same genetic consequence independently emerges in separate clades of the phylogenetic tree highlighted by green boxes containing recurrently mutated genes. Brown circles represent cytogenetically distinct populations, and the numbers represent the number of copies of each adjacent gene. Solid lines represent the most likely ancestral origin of subclones, whereas dashed lines suggest alternative origins.
As sequencing goes genome-wide, phylogenies have been constructed for single-tumour samples that are composed of multiple constituent cellular subclones. The identification of tens of thousands of mutations genome-wide permits the delineation of distinct clusters of mutations — these clusters consist of groups of mutations that share similar mutant allele frequencies (corrected for local copy number). In the tree in panel c, we present a phylogenetic tree in which the variable thicknesses of the branches reflect the numbers of mutations within each distinct mutation ‘cluster’. This gives an indication of the patterns of subclonal importance and dominance within the cancer population. Chr, chromosome; ETV6, ETS variant 6; F, ETV6–RUNX1 fusion gene; GATA3, GATA-binding protein 3; IDH2, isocitrate dehydrogenase 2; PAX5, paired box 5; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha; NCOR1, nuclear receptor co-repressor 1; MLL3, myeloid/lymphoid or mixed-lineage leukaemia 3; NPM1, nucleophosmin (nucleolar phosphoprotein B23, numatrin); RUNX1, runt-related transcription factor 1; SMAD4, SMAD family member 4; STOX2, storkhead box 2. Panel a is adapted, with permission, from REF. 21 © (2012) Macmillan Publishers Ltd. Panel b is adapted, with permission, from REF. 15 © (2011) Macmillan Publishers Ltd. All rights reserved. Panel c is adapted, with permission, from REF. 24 © (2012) Cell Press.
Recent advances in sequencing technologies have delivered, for the first time, the opportunity to scrutinize all expressed genes (‘transcriptomes’), all exons (‘exomes’) and whole cancer genomes at base-pair resolution4. A number of different sequencing platforms now exist, including pico-titre plate pyrosequencing and ligation-based sequencing. From the viewpoint of understanding cancer genome evolution, the key aspect of this generation of sequencing technologies is that billions of independent sequencing reads are generated in parallel, with each read deriving from a single molecule of DNA. Thus, albeit with some sampling biases, the data represent a random sample of DNA molecules (and hence the genomes of individual cells) contained in the tumour sample. By contrast, the data derived from conventional genomic approaches, such as capillary sequencing or copy number arrays, are aggregate signals from many thousands of DNA molecules (BOX 2). Harnessing the attractive statistical properties of massively parallel data thus enables us to draw robust inferences about the mutational mix of a tumour sample, generating unprecedented insights into the fundamental genomic events that underlie the development of cancers and the order, rate and mechanisms by which they occur5–7.
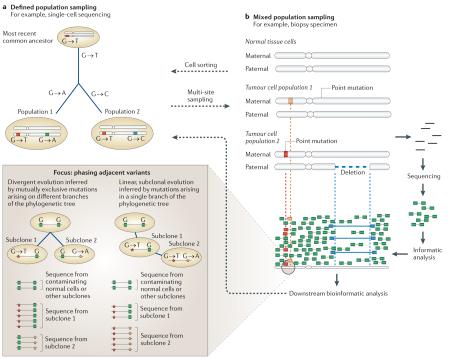
Box 2. Methodologies for understanding cancer genome evolution.
The existing methods used to hypothesize the order of evolutionary events, or phylogeny, derive from molecular genetics. These methods use multiple sampling techniques and assume that individuals can be isolated in a population. The evolutionary connections between organisms are calculated using a range of mathematical models, including parsimony, maximum likelihood, Markov chain Monte Carlo and Bayesian inference. Cancer genomics has adopted these tools to reconstruct the relationships between mixed populations of cells in individual cancers10,13–15. This approach is suitable when individual subclonal populations in a cancer can be reliably isolated, such as through single-cell sequencing or cytogenetics. A simplified example of this is represented in panel a of the figure, in which two different mutations result in evolutionary divergence from a presumed most recent common ancestor.
However, as illustrated in panel b of the figure, most cancer samples consist of mixed populations of normal cells and tumour cells, and next-generation sequencing data therefore provide a composite view of a random sample of DNA molecules from these different populations. Mutations in the data follow defined probability distributions that are dictated by coverage and the underlying allele frequency. There remains a paucity of statistical algorithms for analysing these data, but some useful techniques have recently been developed, such as mutational clustering, using kernel density analysis21 and Bayesian Dirichlet process modelling24,107, digital karyotyping26 and phasing adjacent somatic mutation pairs or adjacent somatic mutations and germline variants24. The phasing technique is summarized in the focus box (panel b).
The above methodologies may be amalgamated to handle the data from multi-site sampling studies that include defined populations that are nonetheless genetically heterogeneous7,9,12.
These approaches have been used to generate comprehensive catalogues of somatic mutations by comparing the genomic sequence of DNA taken from a patient’s cancer cells to the sequence of their germline DNA7,8. In particular, these studies have given an indication of the heterogeneity in cancer genome evolution, across tumour types, across individuals within a given tumour type and even within a single individual’s tumour9,10. In this Review, we provide an initial overview of recent strategic and methodological developments in cancer genomics. Heterogeneity is central to cancer genome evolution, and we describe this at the level of cancer genes and within individual patients. We consider the evidence for the role of different processes, gradual and abrupt, by which heterogeneity may arise. Finally, we present the evidence for an elevated mutation rate in shaping cancer evolution.
Recent strategic and methodological advances
Tumour multi-sampling strategies
With the objective of understanding how the cancer genome varies over space and time, various groups have carried out studies of tumours and their respective non-malignant tissues obtained from an individual patient. These approaches may be broadly divided into ‘geographical’ and ‘longitudinal’ sampling strategies. ‘Geographical sampling’ encompasses those studies that compare multiple samples from an individual cancer that have been obtained at a single point in time. These samples may be derived from geographically distinct areas within a single large tumour mass and/or may include metastatic deposits in lymph nodes or distant organs11,12. ‘Longitudinal sampling’, by contrast, compares samples obtained at different time points in the life history of a cancer: for example, at diagnosis, relapse and metastasis7. A limited number of published studies have included samples that are separated by both space and time9. The biological question posed and the clinical feasibility largely determine the sampling strategy.
Single-cell sequencing
Single-cell sequencing is a potentially useful approach towards the study of cancer evolution and is the ultimate resolution of the multi-sampling approach. In proof-of-principle studies, this approach has been successfully applied to generate catalogues of point mutations in protein-coding regions and copy number changes10,13,14. These approaches have a requirement for whole-genome amplification of the genome of each cell, and this introduces several biases, with the potential for both false-positive and false-negative mutation calls. For haematological malignancies, in situ hybridization techniques allow single cells to be studied for cytogenetic abnormalities15, and it is feasible that in the future, microfluidic techniques will allow cells to be isolated and analysed in one step for solid tumour samples as well16,17. The ability to make inferences about phylogenetic structure using single-cell sequencing will, however, still be fundamentally limited by how representative the biopsy sample is of the whole-tumour bulk and by how many cells are individually analysed.
Mathematical algorithms
Mathematical models have been widely applied in an attempt to unpick the complex and multifactorial influences on cancer progression18–20. Massively parallel sequencing data are particularly amenable to mathematical analysis because they represent a random sample of DNA molecules, and hence of individual cancer cell genomes, within a tumour specimen (BOX 2). Statistical algorithms for exploiting these properties have been developed, providing important insights into the clonal mix of the sample sequenced. For example, using the fraction of reads reporting a point mutation, the copy number at that locus and the level of normal cell contamination, we can work out whether the mutation is likely to be clonal or subclonal and whether the mutation has been duplicated by a subsequent copy number change7,21,22–24. Within a given copy number segment, this mandates a clear temporal precedence. The earliest mutations are those that are subsequently duplicated, followed by those that are clonal but that are present on a single copy of the locus and then by those that are subclonal. This allows inferences about the relative timing of the copy number gain and about the changing mutational signatures that are operative in the different epochs22,25.
With the exception of more complex processes such as chromothripsis (discussed below), genomic rearrangements generally represent simple events (such as deletions or inversions), occurring over the evolutionary time course of a cancer. Mathematically, these rearrangements can be considered as sequential selections from a known library of genomic transformations — remarkably, the constraints imposed by the simplicity of the repertoire of possible rearrangement types, the genome-wide, allelespecific copy number data and the observed breakpoints mean that even deeply complex clusters of rearrangements can be disentangled to yield both the final genomic configuration of segments and the temporal order in which the rearrangements occurred26.
Mutations occur in a given genomic context, and this can also be exploited to understand cancer evolution. In particular, mutations can be ‘phased’ with nearby heterozygous germline SNPs, allowing haplotypespecific analysis of clonal and subclonal mutations24. Furthermore, pairs of mutations can be phased relative to one another, allowing patterns of branching and subclonal evolution to be delineated5,24 (BOX 2). Although such approaches are currently limited to samples with hypermutable regions or with a high mutation burden, the increasing read lengths coming in future generations of single-molecule sequencers will vastly expand the power of this approach.
The heterogeneous cancer genome
The cancer genome is characterized by heterogeneity that is seen across tumour types, among cases of a particular tumour type and even within an individual cancer. This heterogeneity reflects the action of the twin evolutionary forces of variation generation and selection. The extent of genomic variability is testament to the diverse and dynamic nature of these forces.
The heterogeneity of cancer genes
Massively parallel sequencing has enabled us to construct nearly comprehensive catalogues of every mutation within an individual cancer genome at a single point in time6. To date, using conventional and newer technologies, almost 500 cancer genes have been identified27. In a handful of cancer types, specific underlying cancer genes are consistently mutated, such as the oncogenic fusion protein BCR–ABL in chronic myeloid leukaemia (CML) or inactivating mutations in the tumour suppressor gene retinoblastoma 1 (RB1) in retinoblastomas28. Specific cancer genes have also been implicated in the development of the same rare cancer type in different tissues. The oncogenic fusion gene MYB–NFIB, for example, drives the development of adenoid cystic carcinomas that arise in both breast and salivary tissues29.
These examples, however, remain the exception rather than the rule. Most common cancers are associated with many diverse cancer genes that are mutated at a low frequency. One of the most striking observations from large cancer databases is the genetic heterogeneity between cancers and even within individual cancer types. The Cancer Genome Atlas Project analysed 489 high-grade serous ovarian cancers, and among the thousands of somatic mutations identified, only 10 of these were recurrently mutated cancer genes, and all but TP53 were present in less than 10% of cases. The recent genomic analysis of 77 oestrogen-receptor-positive breast cancers also identified that most recurrent mutations occur infrequently, but they do cluster within a limited number of cellular pathways that are central to tumour cell biology30.
Long-standing evidence indicates that breast cancer exhibits heterogeneity in terms of clinical behaviour and response to therapy. More recently, the genomic diversity underlying this heterogeneity has been documented25,30–32. For example, identification of a TP53 mutation in breast cancer correlated with a higher proliferation index before therapy and less dramatic suppression of proliferation during therapy with an aromatase inhibitor30. The conventional subclasses of breast cancers are based on histopathological type and grade, immunohistochemical analysis of hormone receptors and overexpression of human epidermal growth factor receptor 2 (HER2; also known as ERBB2). However, in the past few years, these categories have been extended by molecular profiling studies that use expression analysis to reclassify breast cancers with unique biological and prognostic features33. These categories, which can be identified on gene expression profiles, reflect to some extent the underlying genomic profiles of the tumours31, and it will be interesting to see how integrative transcriptional and genomic studies define this further in the whole-genome sequencing era.
The heterogeneity of the mutational landscape
In addition to the heterogeneity of cancer genes, there is considerable diversity in the nature, number and distribution of mutations within and across different cancer histologies25. Recent studies have revealed, for example, that the childhood cancers retinoblastoma and medulloblastoma contain substantially fewer somatic substitutions than do common adult-onset solid tumours and haematological malignancies, such as breast cancer or acute myeloid leukaemia (AML)7,34–37. This extends even to specific subtypes of tumours — for example, the number of mutations among individual HER2-positive breast cancers differed by a factor of six in a recent study25.
Patterns of structural variants differ across tumour types: breast and ovarian cancers show many more tandem duplications than other tumour types do38,39; pancreatic cancer is characterized by frequent breakage–fusion–bridge cycles of chromosomal rearrangement12; prostate cancer shows balanced chains of rearrangements40,41; and various cancers, especially sarcomas and neuroblastomas, demonstrate chromothripsis (discussed below)37,42,43. Similarly, patterns of base substitutions differ extensively across tumours, depending on DNA repair defects and carcinogenic exposures8,44,45. Many of the pathways underlying the acquisition of somatic mutations in these cancers are poorly understood. For example, at least six or seven distinct point mutational signatures can be identified in breast cancers, of which only one or two can currently be attributed to known biological processes25.
It is, in many ways, remarkable that this degree of heterogeneity in the routes to cancer can lead to such convergent phenotypes. Although detailed genotype–phenotype studies in the massively parallel sequencing era are lacking, it is nonetheless the case that, for example, a histologically typical ER-positive breast cancer can result from a wide array of different cancer genes that have been mutated through many different processes. It is also conceivable that diverse sets of genes will also give rise to cancers with similar behaviours and sensitivities to certain treatments. Optimizing the clinical benefit of cancer genomics for the future therefore demands the systematic integration of genomic data with meaningful clinical information in large databases.
Heterogeneity within an individual cancer
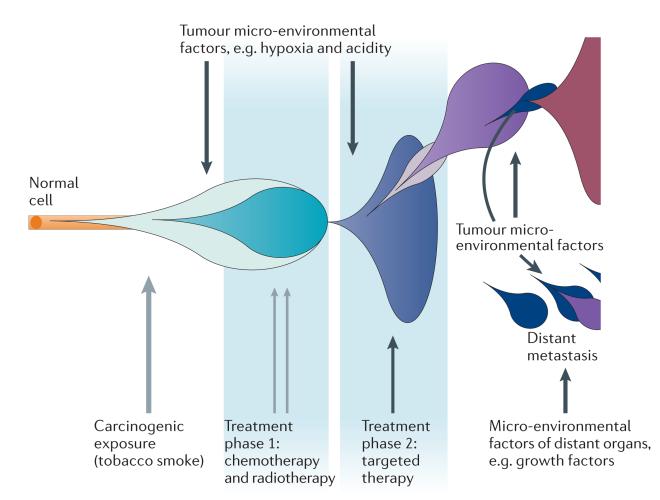
A number of external forces can act on the cancer genome to generate heterogeneity and to influence the subclonal structure (FIG. 2). The tumour microenvironment has an important role in selecting the cells that are best adapted to the (often hostile) environments in which they exist46. The identification of organ-specific branches within phylogenetic trees in metastatic studies is also indicative of environmental factors that select and drive specific genomic changes11,12 (BOX 1). Carcinogenic exposures, such as tobacco smoke, ultraviolet light and even some cancer treatments, may also have an important direct role in driving cancer heterogeneity6,45,47–49. The selective ‘environment’ also includes anticancer treatments. For example, verumafenib — an inhibitor of the serine/threonine protein kinase BRAF — has revolutionized the treatment of metastatic melanoma by providing a targeted therapy for patients with the V600E BRAF point mutation. However, patients usually relapse within a few months as a result of emerging resistance. It is postulated that resistant clones are selected on the basis of either pre-existing or de novo abnormalities arising in alternative pathways50,51. This implies that genomic heterogeneity supports cancer survival in response to a changing environment.
Figure 2. The role of the environment in evolutionary adaptation.
A multitude of environmental factors may shape the evolutionary processes within a single cancer. Blue and purple bubbles represent successive cancer clones, the expansion of which is altered by directly mutagenic factors (grey arrows) and non-mutagenic factors (black arrows).
Cellular ground state and cancer evolution
The observation that specific genes are associated with certain types of cancer in some tissues but not others indicates that the cell of origin may be an important factor in dictating the evolutionary trajectory. Every cell in the body is clonal, having arisen from a single zygote. There is some ‘physiological’ genomic change within organ systems, such as rearrangement and mutation of immunoglobulins in lymphocytes and somatic retrotransposition of long interspersed elements (LINEs) in the brain52, but it is largely the case that the huge phenotypic variability among cells in a human is dictated by the epigenome, transcriptome and proteome of those cells. It follows that this ‘ground state’ of a cell in which a somatic mutation arises will strongly influence how that mutation plays out, as the early life of a somatic mutation is fraught with the threat of extinction through random genetic drift. This concept is exemplified by the BCR–ABL fusion gene, which is frequently associated with a range of haematological malignancies. Studies have identified that the activation of ABL kinases in breast cancer cell lines promotes invasiveness; however, BCR–ABL has not been implicated in the pathogenesis of solid tumours53. Reasons for this specificity may include the low transcriptional activity of the BCR promoter in non-haematological cells, a lack of interacting partners needed for full oncogenic effects of the fusion protein or failure to induce a sustaining population of cancer cells.
The importance of the ‘ground state’ is exemplified by the specific ‘oncogenicity’ of KIT mutations in gastrointestinal stromal tumours (GISTs). KIT is a receptor tyrosine kinase that is activated by stem cell factor binding (also known as mast cell growth factor binding), resulting in a signalling cascade that promotes cell survival, differentiation and proliferation, and germline KIT-activating mutations are associated with hyperplasia of interstitial cells of Cajal (ICCs) and GISTs54. In mouse models, it has recently been demonstrated that GISTs exclusively arise in a subset of ICCs that expresses high levels of endogenous ETS variant 1 (ETV1)55,56. ETV1 is a member of the ETS family of transcription factors, which are involved in various key cellular processes, including cell cycling, proliferation and differentiation. In ICCs, ETV1 acts as both a survival factor and as a master regulator of a specific transcription programme that is co-opted by and required for transformation by activated KIT56. The implication is that in the absence of high levels of endogenous ETV1 expression, KIT mutations fail to drive the emergence of GIST cancers.
However, in most situations, the link between the cell of origin and the cancer phenotype appears to be less clear-cut. As an increasing number of genomic studies report broad catalogues of cancer genes, it is becoming apparent that many of the same genes are implicated across a broad range of tissue types — albeit at different frequencies. For example, two independent studies identified that cancer genes that are historically associated with haematological malignancy, such as runt-related transcription factor 1 (RUNX1) and core-binding factor, beta subunit (CBFB), are also recurrently mutated in breast cancer30,57.
The role of epistasis in cancer genome evolution
A new mutation in a cancer gene does not occur in isolation but rather enters into an established genomic landscape. This existing gene network may have a profound effect on the fate of the cell, determining whether there is a cell death or clonal expansion. The ground state of a cell can be considered to represent interactions with cellular identity, whereas epistasis, by contrast, represents interactions among oncogenic mutations.
Three major lines of evidence drawn from recent studies have demonstrated the probable importance of epistatic factors in cancer genome evolution. First, the large cancer gene databases have shown that, despite extensive heterogeneity in common cancers, particular combinations of somatic mutations may co-occur more than expected by chance, such as TP53 and breast cancer 1, early onset (BRCA1) and breast cancer 2, late onset (BRCA2) mutations in breast cancer58 or the oncogenic KRAS and serine/threonine kinase 11 (STK11; also known as LKB1) mutations in lung cancer59,60. Second, activation of many oncogenes, including KRAS, can lead to a state of ‘oncogene-induced senescence’61,62. This is an acute and telomere-independent form of senescence that can occur in response to the expression of oncogenes and is protective against cancer. It is widely believed that second hits, such as cyclin-dependent kinase inhibitor 2A (CDKN2A) inactivation, are required to ameliorate these effects63,64. Third, convergent evolution among subclones within the malignant tumour (or tumours) of a particular patient also implies cooperativity among somatic mutations12,15,65, and this is exemplified by patients with renal cancer9 (BOX 1). A recent multi-region sampling study identified that after ubiquitous von Hippel–Lindau tumour suppressor, E3 ubiquitin protein ligase (VHL) loss, driver mutations inactivating histone modifiers can independently arise in different branches of the phylogenetic tree. Even more strikingly, independent phosphatase and tensin (PTEN) mutations occurred twice in one patient in different subclones, despite PTEN mutations being found in only 1% of renal cancers overall9. This implies that some specific feature of the genomic landscape of this patient’s cancer was particularly dependent on the inactivation of PTEN — an event that is not required for most renal cancers.
Little is known about whether the order of mutation acquisition is important. The renal cancer studies described above do suggest a pre-requisite for early VHL loss in renal cancer, but the extent to which this is a general rule is unclear. Ancestral gene reconstruction and protein-engineering studies demonstrate that epistatic interactions can limit the potential mutational trajectories that are available and can also enforce ratchet-like constraints by inhibiting the reversibility of the evolutionary process66. Certain oncogenic mutations may mandate that specific cellular pathways be targeted by subsequent mutations. Such an effect would restrict the set of potential driver mutations that could occur after the initial event67.
The strands of data discussed above imply cooperativity among cancer-causing mutations, and this cooperativity can include mutations that ameliorate negative effects of other variants (synthetic viability) or mutations that, when combined, result in synergistic effects (greater than the sum of their individual effects). This is exemplified by the interaction between the proto-oncogene MYC (also known as c-MYC) and B cell CLL/lymphoma 2 (BCL2) in cell lines. The overexpression of MYC induces apoptosis, but the co-expression of BCL2 overrides this effect and permits MYC to drive the cell into cycling68,69.
From the clinical standpoint, discovering and understanding epistatic interactions such as synthetic lethality is proving useful in the design of targeted therapies70. The sensitivity of BRCA1−/− cells to poly(ADP-ribose) polymerase (PARP) inhibitors is already a widely cited ‘synthetic lethality’ interaction in the clinic71. BRCA1 is essential for homologous recombination repair of dsDNA breaks. BRCA1−/− cells are able to survive despite this defect, but it comes at the cost of critical dependence on alternative repair pathways involving PARP function. Therefore, in BRCA1−/− cells that have been treated with PARP inhibitors, DNA breaks that arise from collapsed replication forks cannot be repaired, resulting in cell arrest and death. However, in the face of PARP inhibitor therapy, ‘reversion’ mutations in BRCA1 and BRCA2 can result in the restoration of a partially functional protein homologous recombination72,73, leading to the escape of the clone from the detrimental effects of the treatment. This example shows the clinical potential to exploit epistatic interactions but also the complexity of these networks and the problems posed by the dynamic and rapidly evolving cancer genome.
The role of genomic crises in tumorigenesis
Recent lines of evidence derived from directly studying cancer genomes indicate that, in some cases, a huge number of mutations can occur in a timescale that is considerably shorter than that on which clonal selection operates (FIG. 3). These mutational processes can take several forms.
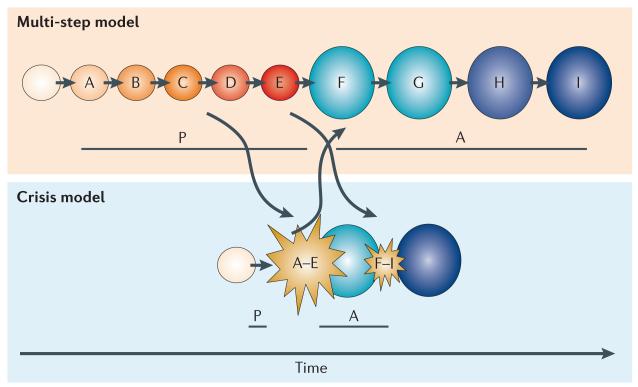
Figure 3. Stepwise versus crisis-driven mutation accumulation.
Multi-step and crisis event models of carcinogenesis are represented. It is thought that these pathways are not necessarily mutually exclusive but that they may coincide and overlap. In this example, mutations A–E (orange to red circles) are those that are required to initiate clonal expansion and malignant transformation, whereas mutations F–H (blue circles) drive ongoing evolution and the acquisition of aggressive clinical characteristics. The pre-malignant phase (P) and the time from malignancy onset to acquisition of an aggressive phenotype (A) are reduced in the crisis event model compared to the multi-step model. This indicates that standard screening techniques that aim to detect pre-invasive and early malignancies may be inadequate in cancers that develop through crisis events.
Telomere attrition is associated with end-to-end chromosome fusions, and this can drive massive genomic disruption through repeated breakage–fusion–bridge cycles74. An end-to-end chromosome fusion generates a dicentric chromosome (that is, a chromosome with two centromeres), and the two centromeres are pulled to opposite daughter cells during mitosis, generating further DNA breaks. This process can be repeated with every cell cycle until a telomere is restored to the naked DNA ends. Within a few cell cycles, and certainly on a much faster timescale than natural selection can operate, widespread chromosomal deletions and exponential genomic amplification can develop75,76.
Balanced chains of somatically acquired genomic rearrangements have been observed in prostate cancer40 and some haematological malignancies. These chains can show up to ten genomic regions involved in a mutual exchange of DNA segments without copy number loss. In some cases, these generate oncogenic fusion genes (for example, in the TMPRSS2–ERG loci) or gene disruptions. Curiously, regions that are involved in these chains show a propensity to involve highly transcribed genes. In one example, breakpoints were in close proximity to four potential cancer genes: TANK-binding kinase 1 (TBK1), TP53, mitogen-activated protein kinase kinase 4 (MAP2K4) and ABL1.
Approximately 2–3% of cancers show evidence for a catastrophic mutational process that has been coined chromothripsis41. A process of genome shattering and reassembly occurs in a one-off crisis, resulting in a characteristic pattern of oscillating DNA copy number and up to several hundred genomic rearrangements localized to one or a few chromosomes. This localization may be the result of physically isolating the damaged chromosomes in micronuclei created during anaphase77. Chromothripsis has been observed at a low frequency in a diverse range of cancers, including chronic lymphocytic leukaemia (CLL), neuroblastomas, myelomas, breast cancer, small and non-small-cell lung cancers and renal, thyroid and gastrointestinal malignancies8,41,43. It is notable that a recent study identified high rates of chromothripsis in medulloblastomas and AML in the presence of mutant TP53 (100% and 47% of cancers, respectively) but not wild-type TP53 (0% and 1%, respectively)42. Bone sarcomas also seem to have a particularly high rate of chromothripsis41.
In addition to clusters of structural variants, multiple point mutations can also be acquired in one-off bursts. In a sizable proportion of breast cancers, we have observed clusters of cytosine mutations near sites of genomic rearrangement: a process that we termed kataegis24. These clusters can represent up to 10–20 base substitutions in one or two kilobases, all occurring at cytosines in a TpC context, all collinear (that is, in linkage) and all occurring on either the forward or reverse strand of DNA. The mechanism underlying such events remains mysterious, although the mRNA-editing APOBEC proteins may be involved25. This phenomenon has not yet been reported in other types of cancer.
All of these catastrophic mutational processes imply that cancer genome evolution may not always be a gradual stepwise progression (FIG. 3). In one of the patients with chromothripsis, the crisis simultaneously disrupted three tumour-suppressor genes — the E3 ubiquitin protein ligase FBXW7, CDKN2A and the RecQ helicase WRN41. This suggests that the clone would have taken a substantial leap along the path to malignancy after the catastrophic event. In multiple myelomas, samples with evidence for chromothripsis were associated with reduced survival, indicating that the large-scale genomic disruption may have rendered the myeloma cells more malignant, with similar data emerging for neuroblastoma78,43. These mechanisms of mutation accumulation are not mutually exclusive. Gradual mutation accumulation occurs to some degree in all cancers, representing perpetual adaptation to the tumour environment but may be punctuated by highly disruptive episodes.
The role of mutation rate in cancer evolution
The number of driver mutations required for a cancer to become fully malignant is debated, but it is generally considered to be between 2 and 20 in most types of common solid malignancy79,80. Some cancers, such as certain subtypes of AML, accumulate a sufficient complement of mutations to transform to a malignant phenotype in the presence of an apparently normal mutation rate81. There are several lines of evidence, however, indicating that many cancers achieve the required complement of driver mutations by means of an elevated mutation rate.
‘Mutator mutations’ in carcinogensis
‘Mutator mutations’ are mutations within cancer genes that increase the mutation rate across the cancer genome. The effects of such mutations can be broadly categorized as: reduced ability to detect and/or repair DNA damage; failure of genomic surveillance mechanisms; and increased susceptibility to DNA damage by exogenous and endogenous carcinogens (FIG. 4). It has long been recognized that inherited cancer syndromes, including ataxia telangiectasia, xeroderma pigmentosum, Bloom’s syndrome and hereditary non-polyposis colorectal cancer, are caused by germline defects in specific DNA repair genes82. They are associated with an elevated mutation rate and are characterised by early onset cancers.
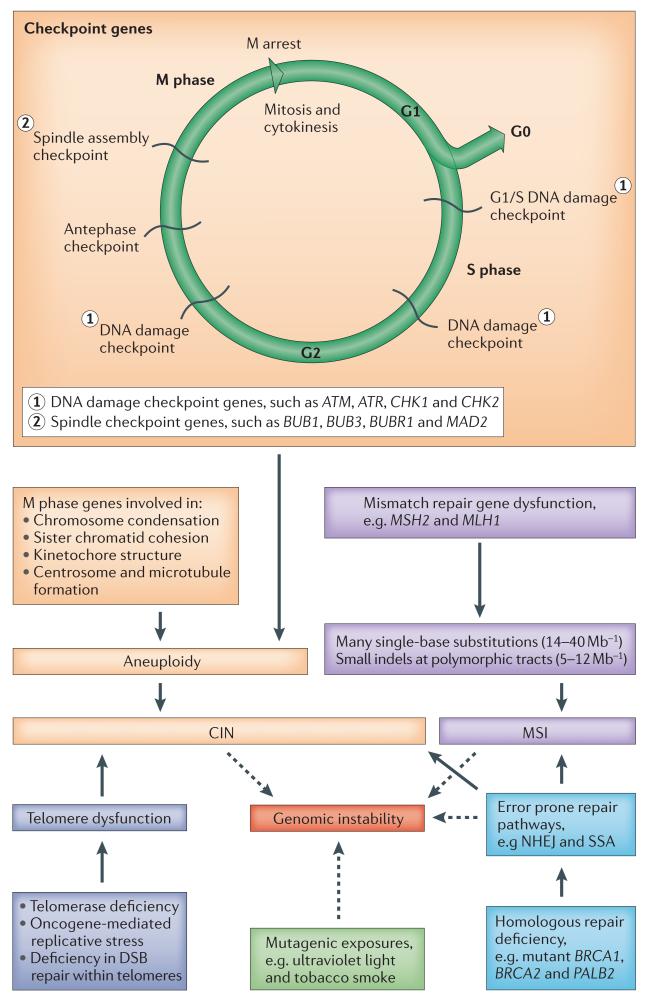
Figure 4. ‘Mutator mutations’ drive genomic instability in cancers.
There are two major recognized routes by which genomic instability may arise. Chromosomal instability (CIN) is common across all types of cancer and may be numeric (aneuploidy) or structural. CIN may arise through mutations in a wide range of genes involved in cell cycling and division (orange boxes) or through other diverse mechanisms, such as telomeric dysfunction or as a consequence of failure in homologous repair. Microsatellite instability (MSI) is less common and occurs as a result of mutations in the mismatch repair genes (purple boxes). Instability may also directly arise as a consequence of defects in homologous repair, necessitating the use of alternative error prone pathways, such as non-homologous end joining (NHEJ) and single-strand annealing (SSA). Error-prone pathways may result in both chromosomal instability and genomic instability through frequent small deletions or substitutions. Mutagenic exposures may also contribute to genomic instability. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related; BUB1, budding uninhibited by benzimidazoles 1; BUBR1, budding uninhibited by benzimidazoles 1 beta; BRCA1, breast cancer 1, early onset; BRCA2, breast cancer 2, early onset; DSB, double-strand break; indel, insertion or deletion mutation; MAD2, MAD2 mitotic arrest deficient-like 1; MSH2, mutS homologue 2, colon cancer, nonpolyposis type 1; MLH1, mutL homologue 1, colon cancer nonpolyposis type 2; PALB2, partner and localizer of BRCA2.
Microsatellite instability (MSI) is characterized by a high rate of substitutions and small insertions and deletions, and it arises from mutations in mismatch repair genes, including MSH2 and MCH1 (REF. 44). MSI occurs in less than 20% of colorectal cancers and has also been reported at low frequency in a diverse range of other tumour types including gastric, endometrial and sebaceous cancers and lymphomas83. A much more frequent pattern of genomic instability that is seen in nearly all types of common solid malignancy, including breast and colorectal cancer, is chromosomal instability (CIN), which is a process whereby whole-chromosome segregation abnormalities during mitosis result in aneuploidy84–86.
Epigenetic instability is also common in a wide range of cancers (BOX 3). Aberrant methylation of CpG islands in promoter regions is correlated with silencing of multiple tumour suppressor genes, resulting in the CpG island methylator phenotype (CIMP). This has been observed in many cancer types and is associated with aetiologically and clinically distinct types of colorectal cancer87 and glioma88.
Box 3. The interplay of the epigenome and genome in cancer evolution by natural selection.
For epigenetic factors to be important in cancer evolution, three criteria must be met: stochastic variation must exist among competing clones in a given epigenetic locus; this variation must be heritable; and there should be phenotypic consequences of epigenetic variation for natural selection to act on.
Within cancers, individual tumour suppressor genes may be epigenetically silenced through promoter CpG island hypermethylation108. However, in contrast to mutation, little is known about the stochastic acquisition and loss of epigenetic changes. Nonetheless, bisulphite sequencing of individual haplotypes of CpGs has shown that some regions do show variability across different cells in a sample109,110. There is a robust machinery of cytosine methylases that faithfully copy methylation at CpG dinucleotides from the template strand to the newly synthesized DNA strand during DNA replication, indicating that these changes are heritable. It is less clear how histone marks are transmitted to the daughter cells, although such pathways are presumed to exist.
An emerging theme of recent genomic discoveries in cancer has been the frequent mutation of genes that are involved in epigenetic regulation, further highlighting the importance of interactions between genetic and epigenetic changes. This is exemplified by mutations in AT-rich interactive domain 1A (ARID1A) in ovarian cancer111,112, inactivation of polybromo 1 (PBRM1), lysine-specific demethylase 5C (KDM5C), KDM6A (also known as UTX) and SET domain containing 2 (SETD2) in renal cancer113 and the remarkable observation of activating mutations of the Polycomb group gene EZH2 in follicular lymphoma54 but inactivating mutations of the same gene in chronic myeloid malignancies114,115. Chromatin studies have indeed shown epigenetic consequences of these mutations, but we lack a detailed understanding of the particular target genes involved or the Darwinian evolution of the epigenetic landscape after these genomic aberrations appear. Nonetheless, a recent genome-wide methylation profiling study in acute myeloid leukaemia identified that genetically distinct subtypes of disease carried characteristic epigenetic profiles116. This implies that a particular driver gene may promote the evolution of and may cooperate with an epigenetic landscape that is ‘optimal’ for that genomic change.
Elevated mutation rate
Many of the aggressive clinical characteristics of cancers, such as the abilities to resist treatment, to relapse and to metastasize, depend on the continued generation of genetic variation that permits adaptation7,12. However, whether all cancers have elevated mutation rates compared to normal cells has been controversial81,89,90. In the era of whole-cancergenome sequencing, however, it is becoming clear that the overwhelming majority of tumours carry hundreds to hundreds of thousands of somatic mutations, which is suggestive of an elevated mutation rate.
A preponderance of a specific type of base substitution in a given context, such as C-to-T mutations in a CpG context, can be viewed as a ‘mutational signature’ that reflects an underlying mutational process6,12,45. Analysing these mutational signatures in 21 breast cancer genomes identified several major processes involving base substitutions, small insertions and deletions and genomic rearrangements25. Mutations acquired early in the development of the cancer were dominated by C-to-T transitions, especially in a CpG context, that are likely to represent spontaneous deamination of methylated cytosines. This is a rather generic mutational process, is similar to that seen in the germ line and reflects that seen by exome sequencing of normal haematopoietic stem and/or progenitor cells in healthy people81. However, nearly all tumours demonstrated a substantial shift in the contribution from individual processes over time with several novel mutational processes reported generally emerging late in the development of the cancer24. Taken together, this information implies that the vast majority of breast cancers have an elevated, cancer-specific increase in mutation rate. Environmental exposures, including the traditional cancer treatments (namely, chemotherapy and radiotherapy), also influence mutation rate and spectrum. Chemotherapy, for example, is associated with an increase in transversions21. The functional effect of endogenous and external factors that increase mutation rate on cancer progression remains to be elucidated.
There may be examples of tumours in which mutation rate per cell division is not increased. AML, for example, does not have an excessive mutation burden, and when compared with age-matched normal haematopoietic cells, mutation numbers are broadly similar81. In a similar fashion, there is a correlation between age and mutation burden in the childhood tumour, medulloblastoma34,91, suggesting that mutation accumulation in this disease is more a function of time than it is an acquisition of specific mutational processes.
Mutation rate distribution across the cancer genome
Mutation is generally modelled as a random process, but there is increasing evidence that the distribution of somatic mutations shows variegation across the genome in both the rate and the type of variation. The most extreme example of this is somatic hypermutation in lymphoid malignancies. In normal B lymphocyte ontogeny, the immunoglobulin gene is subjected to targeted mutation to increase antibody diversity in response to infection. Sometimes, however, the tightly controlled genomic localization of the hypermutation machinery can be loosened, and other genes that are highly expressed during lymphoid differentiation may be subjected to this process. This aberrant somatic hypermutation has only been described in association with a handful of genes. It preferentially targets the 5′ untranslated region and the first coding exon of the gene and can repeatedly occur during lymphoma development, driving much subclonal diversity just at these specific loci92. In particular, the oncogene BCL6 is commonly mutated in this way in diffuse large B cell lymphoma93,94.
Less extreme examples of variable mutation rates across the genome abound. Chromosomal fragile sites have been documented in cytogenetic studies for some years, and cancers show increased rates of heterozygous and homozygous deletion at these sites compared to other regions of the genome95,96. This increased rate of genomic rearrangement may in part result from these regions having fairly sparse origins of replication and being late replicating during the cell cycle97,98. In many cases, these deletions are of no biological consequence to the cell, but there is some evidence that cancer genes may reside in these loci. For example, Parkinson protein 2, E3 ubiquitin protein ligase (PARK2) can be recurrently deleted and mutated in gastrointestinal tumours99, and knockout of the gene in mice increases the rate of APC-induced colorectal tumours100.
The underlying chromatin state may also contribute to genomic rearrangement. In lymphomas, genes that are frequently fused with the immunoglobulin locus are often geographically proximate during interphase101, and in prostate cancer androgen can induce intra- and interchromosomal proximity between the ETS fusion gene partners102,103. In breast cancer, 50% of somatically acquired genomic rearrangements involve a gene footprint compared with 40% expected by chance104.
With regard to point mutations, mismatch repair deficiency causes a specific distribution and signature of mutations across the genome. For example, in microsatellite-unstable colorectal cancer, genes such as the type 2 TGFβ receptor gene are particularly prone to mutation owing to their specific nucleotide mix, whereas this gene is almost never mutated in microsatellite-stable colorectal cancer105. Similarly, the distribution of oncogenic point mutations in TP53 and KRAS in tobacco-induced lung cancer differs from lung cancers that develop in people who have never smoked45,49. The efficacy of DNA repair processes also leads to genomic variation in mutation rate: in carcinogen-induced tumours especially, lower rates of mutation are seen in highly expressed genes compared with non-expressed genes106.
How does this variegation in mutation rate across the genome impact on our understanding of cancer evolution? Clearly, cancer can arise from a vast array of different possible driver mutations. These data indicate that the observed distribution of driver mutations seen in a given tumour type depends not only on the oncogenicity of the given genes in that cellular context (namely, the selective advantage associated with the mutation) but also on the probability with which a given change can arise in the population of competing clones.
Conclusions
In the not too distant future, genomic features of every patient’s cancer type will be characterized at the point of diagnosis. A list of implicated cancer genes and mutational processes will be generated, and a personalized therapeutic regimen will be chosen. One of the major challenges to this vision is how to sample the cancers to attain an accurate view of the underlying complexity and to address the fact that cancers are highly dynamic evolutionary processes9. A single sample is a ‘snapshot’ in space and time. Multi-region sampling and sampling of distinct metastatic sites will help to reduce the problem posed by geographical heterogeneity but will have to be balanced with clinical risk and patient choice. It is necessary to acknowledge that even with the most sensitive and accurate of genomic technologies, clinically important mutations that are confined to subclones may be missed on account of inadequate sampling. The clinical approach towards sampling will therefore be guided by multi-sampling studies within all cancer types, and in particular important insights may be gained from studies that use sequential time-ordered sampling of cancers with well-defined precursor lesions, such as cervical intra-epithelial neoplasia in cervical cancer and Barrett’s oesophagus in oesophageal cancer.
Understanding how the cancer genome responds to treatment and promotes metastasis presents a further challenge, requiring longitudinal sampling strategies incorporated into long-term clinical trials. Furthermore, the optimal targeted therapeutic approaches to cancers with branching evolutionary architectures remains unclear. The observation that any individual cancer may contain both clonal driver mutations (that is, mutations that occur within the phylogenetic tree trunk) and subclonal driver mutations, which are linked through epistatic interactions, indicates that cancer eradication may well demand complex combinations of drugs.
Finally, the heterogeneity of cancer genes and cancer pathways mutated across human malignancy mandates the development of large-scale, publicly available databases with carefully annotated clinical outcome data linked to detailed genomic analyses. After sample sizes in these databases have reached numbers in the tens of thousands, we will have the raw material with which to build algorithms for personalized decision support for oncologists.
Acknowledgements
We would like to thank the Wellcome Trust for funding.
Glossary
- Mutational signatures
Patterns of mutations that are characteristic of a type of cancer or that are indicative of a specific process.
- Chromothripsis
A single event that causes genome shattering and reassembly, resulting in a characteristic pattern of oscillating copy number and up to several hundred genomic rearrangements localized to one or a few chromosomes
- Driver mutations
Somatic mutations within cancer genes that confer a clonal advantage, that are causally implicated in oncogenesis and that are positively selected for during cancer evolution.
- Synthetic lethality
Two genes are synthetically lethal if mutation of either in isolation is compatible with viability, but mutation of both to cell death.
- Kataegis
A localized hypermutation that often colocalizes with somatic rearrangements.
- Microsatellite instability
(MSI). Microsatellites are repeating sequences within DNA of 2–6 base pairs in length; defects in mismatch repair can give rise to genomic instability within these regions.
- Chromosomal instability
(CIN). A form of genomic instability that is common in cancers and is characterized by large chromosomal losses by as of yet undefined mechanisms.
Footnotes
Competing interests statement The authors declare no competing financial interests.
FURTHER INFORMATION Peter J. Campbell’s homepage: http://www.sanger.ac.uk/research/projects/cancergenome
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 4.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl Acad. Sci. USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turajlic S, et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012;22:196–207. doi: 10.1101/gr.125591.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 13 and 14 provide proof of principle that next-generation sequencing technologies can be combined with single-cell approaches can be used to investigate intra-tumoural heterogeneity.
- 15.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 16.Baker M. Digital PCR hits its stride. Nature Methods. 2012;9:541–544. [Google Scholar]
- 17.Wang J, et al. Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin. Chem. 2010;56:623–632. doi: 10.1373/clinchem.2009.134973. [DOI] [PubMed] [Google Scholar]
- 18.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]; This is a comprehensive review of the literature from the field of mathematical modelling in cancer evolution.
- 19.Michor F, Iwasa Y, Nowak MA. Dynamics of cancer progression. Nature Rev. Cancer. 2004;4:197–205. doi: 10.1038/nrc1295. [DOI] [PubMed] [Google Scholar]
- 20.Attolini CS, Michor F. Evolutionary theory of cancer. Ann. NY Acad. Sci. 2009;1168:23–51. doi: 10.1111/j.1749-6632.2009.04880.x. [DOI] [PubMed] [Google Scholar]
- 21.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotech. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durinck S, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenman CD, et al. Estimation of rearrangement phylogeny for cancer genomes. Genome Res. 2012;22:346–361. doi: 10.1101/gr.118414.110. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this article, a mathematical framework is presented for reconstructing temporal sequences of rearrangements and hence evolutionary selection.
- 27.Futreal PA, et al. A census of human cancer genes. Nature Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanquet V, et al. Spectrum of germline mutations in the RB1 gene: a study of 232 patients with hereditary and non hereditary retinoblastoma. Hum. Mol. Genet. 1995;4:383–388. doi: 10.1093/hmg/4.3.383. [DOI] [PubMed] [Google Scholar]
- 29.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl Acad. Sci. USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]; This was one of the first studies to correlate somatic mutation changes identified through next-generation sequencing, with treatment responses. Somatic mutations are also mapped to distinct pathways of relevance to tumour cell biology.
- 31.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is an integrated analysis of copy number and gene expression data with long-term clinical follow-up providing a novel molecular stratification of the breast cancer population.
- 32.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perreard L, et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng CK, et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J. Pathol. 2012;226:703–712. doi: 10.1002/path.3980. [DOI] [PubMed] [Google Scholar]
- 39.McBride DJ, et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J. Pathol. 2012;227:446–455. doi: 10.1002/path.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 44.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 45.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gatenby RA, et al. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br. J. Cancer. 2007;97:646–653. doi: 10.1038/sj.bjc.6603922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz JL, Jordan R, Sun J, Ma H, Hsieb AW. Dose-dependent changes in the spectrum of mutations induced by ionizing radiation. Radiat. Res. 2000;153:312–317. doi: 10.1667/0033-7587(2000)153[0312:ddcits]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 48.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat. Res. 2004;567:447–474. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Le Calvez F, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076–5083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
- 50.Arkenau HT, Kefford R, Long GV. Targeting BRAF for patients with melanoma. Br. J. Cancer. 2011;104:392–398. doi: 10.1038/sj.bjc.6606030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su X, et al. Cascading adoptive cell therapy for metastatic melanoma. Cancer Biother. Radiopharm. 2011;26:401–406. doi: 10.1089/cbr.2010.0947. [DOI] [PubMed] [Google Scholar]
- 52.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasan D, Plattner R. Activation of ABL tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 2006;66:5648–5655. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- 54.Antonescu CR. Gastrointestinal stromal tumor (GIST) pathogenesis, familial GIST, and animal models. Semin. Diagn. Pathol. 2006;23:63–69. doi: 10.1053/j.semdp.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 55.Kwon JG, et al. Changes in the structure and function of ICC networks in ICC hyperplasia and gastrointestinal stromal tumors. Gastroenterology. 2009;136:630–639. doi: 10.1053/j.gastro.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chi P, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–853. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 2001;61:4092–4097. [PubMed] [Google Scholar]
- 59.Matsumoto S, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahoney CL, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br. J. Cancer. 2009;100:370–375. doi: 10.1038/sj.bjc.6604886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones CJ, et al. Evidence for a telomere-independent “clock” limiting RAS oncogene-driven proliferation of human thyroid epithelial cells. Mol. Cell. Biol. 2000;20:5690–5699. doi: 10.1128/mcb.20.15.5690-5699.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 63.Jacobs JJ, et al. Senescence bypass screen identifies TBX2, which represses CDKN2A (p19(ARF)) and is amplified in a subset of human breast cancers. Nature Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 64.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005;65:2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 65.Notta F, et al. Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 66.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moore JH. A global view of epistasis. Nature Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- 68.Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Apoptotic cell death induced by c-MYC is inhibited by BCL-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 69.Fanidi A, Harrington EA, Evan GI. Cooperative interaction between c-MYC and BCL-2 proto-oncogenes. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- 70.Rehman FL, Lord CJ, Ashworth A. Synthetic lethal approaches to breast cancer therapy. Nature Rev. Clin. Oncol. 2010;7:718–724. doi: 10.1038/nrclinonc.2010.172. [DOI] [PubMed] [Google Scholar]
- 71.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 72.Swisher EM, et al. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Artandi SE, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 75.Bignell GR, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nature Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 77.Crasta K, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118:675–678. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beerenwinkel N, et al. Genetic progression and the waiting time to cancer. PLoS Comput. Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 81.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study presents mutational data from normal haematopoietic stem cells, which were found to show similar mutational burden and signatures to those seen in acute leukaemias.
- 82.Martin SA, Hewish M, Lord CJ, Ashworth A. Genomic instability and the selection of treatments for cancer. J. Pathol. 2010;220:281–289. doi: 10.1002/path.2631. [DOI] [PubMed] [Google Scholar]
- 83.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer—the stable evidence. Nature Rev. Clin. Oncol. 2010;7:153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solomon DA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nature Rev. Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 87.Cheng YW, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin. Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- 90.Bodmer W, Bielas JH, Beckman RA. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68:3558–3560. doi: 10.1158/0008-5472.CAN-07-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 93.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Migliazza A, et al. Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc. Natl Acad. Sci. USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gandhi M, Dillon LW, Pramanik S, Nikiforov YE, Wang YH. DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene. 2010;29:2272–2280. doi: 10.1038/onc.2009.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 98.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol. Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Veeriah S, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nature Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Poulogiannis G, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl Acad. Sci. USA. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARα genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- 102.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stephens PJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Markowitz S, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 106.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dunson DB. Nonparametric Bayes Applications to Biostatistics. Cambridge Univ. Press; 2010. [Google Scholar]
- 108.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 109.Flanagan JM, et al. Intra- and interindividual epigenetic variation in human germ cells. Am. J. Hum. Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 116.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]