Abstract
The correct outgrowth of axons is essential for the development and regeneration of nervous systems. Axon growth is primarily driven by microtubules. Key regulators of microtubules in this context are the spectraplakins, a family of evolutionarily conserved actin-microtubule linkers. Loss of function of the mouse spectraplakin ACF7 or of its close Drosophila homolog Short stop/Shot similarly cause severe axon shortening and microtubule disorganization. How spectraplakins perform these functions is not known. Here we show that axonal growth-promoting roles of Shot require interaction with EB1 (End binding protein) at polymerizing plus ends of microtubules. We show that binding of Shot to EB1 requires SxIP motifs in Shot's C-terminal tail (Ctail), mutations of these motifs abolish Shot functions in axonal growth, loss of EB1 function phenocopies Shot loss, and genetic interaction studies reveal strong functional links between Shot and EB1 in axonal growth and microtubule organization. In addition, we report that Shot localizes along microtubule shafts and stabilizes them against pharmacologically induced depolymerization. This function is EB1-independent but requires net positive charges within Ctail which essentially contribute to the microtubule shaft association of Shot. Therefore, spectraplakins are true members of two important classes of neuronal microtubule regulating proteins: +TIPs (tip interacting proteins; plus end regulators) and structural MAPs (microtubule-associated proteins). From our data we deduce a model that relates the different features of the spectraplakin C terminus to the two functions of Shot during axonal growth.
Introduction
The correct outgrowth of axonal projections is essential for the development and regeneration of nervous systems. Axonal extension is primarily executed by microtubules (MTs). MT dynamics are regulated through the processes of MT stabilization, MT polymerization, MT-based transport, and the linkage of MTs to F-actin networks (Conde and Cáceres, 2009; Dent et al., 2011). However, we still have little understanding of how such processes contribute to axon extension.
Spectraplakins, are a family of large actin-MT linker molecules that are key regulators of axonal growth (as well as many other clinically relevant processes; Sonnenberg and Liem, 2007). In the absence of spectraplakins, axons are short and MTs lose their orderly bundled appearance. These phenotypes are found in mouse neurons deficient for the spectraplakin ACF7 as well as in Drosophila neurons lacking the close ACF7 homolog Short stop/Shot (Sánchez-Soriano et al., 2009), consistent with the general assumption that spectraplakins are functionally conserved (Röper et al., 2002; Sonnenberg and Liem, 2007). However, the molecular mechanisms through which spectraplakins perform these roles are poorly understood.
Studies in non-neuronal cells have suggested that spectraplakins interact with MTs using two conserved C-terminal domains, the Gas2 (growth arrest specific 2)-related domain (GRD) and the adjacent C-terminal tail (Ctail). GRDs in fibroblasts associate along MT shafts and protect them against the MT-destabilizing drug nocodazole (Sun et al., 2001; Lee and Kolodziej, 2002). Ctails in non-neuronal cells have been reported to associate with either MT shafts (Sun et al., 2001) or EB1 (end binding protein 1) at polymerizing MT plus ends (Honnappa et al., 2009; Applewhite et al., 2010). These data suggest that spectraplakins may work as MT-stabilizing factors similar to classical MAPs (microtubule-associated proteins; Chilton and Gordon-Weeks, 2007), or they might have roles as regulators of MT plus ends similar to +TIPs (tip interacting proteins; Gouveia and Akhmanova, 2010). However, so far we lack proof for the relevance of any of these mechanisms for reported biological functions of spectraplakins, especially in neurons where MT networks are very differently organized compared with non-neuronal cells (Conde and Cáceres, 2009).
Here we focused on the genetically amenable Drosophila Shot to investigate the molecular mechanisms through which spectraplakins regulate neuronal MTs during axon extension. We found that both GRD and Ctail are essential for two parallel roles of Shot, one in MT stabilization and one in guiding polymerizing MT plus ends in the direction of axonal growth. Therefore, spectraplakins act as both MAPs and +TIPs in developing neurons. The +TIP function requires interaction with EB1 and establishes the Shot-EB1 complex as an important determinant of axonal growth.
Materials and Methods
Fly strains
Specimens used for these studies were primary neurons or embryos of Drosophila that were of either sex. For loss-of-function analyses we used the strongest available alleles of short stop (shot3, shotsf20), the chromosomal deficiency Df(2R)MK1 uncovering the shot locus (Strumpf and Volk, 1998; Sánchez-Soriano et al., 2009) and eb1 (eb104524; Bloomington Drosophila Stock Center, #11379; Elliott et al., 2005). Driver lines for targeted gene expression: eve-Gal4RN2D+O (motor neurons in vivo; Sánchez-Soriano and Prokop, 2005), elav-Gal4 (eb1iRNA expression in primary neurons cultured for 6 d; Luo et al., 1994), scabrous-Gal4 (sca-Gal4; targeted gene expression in primary neurons cultured for 6 h; courtesy of J. Urban; P-element insertion into the sca locus), and stripe-Gal4 (tendon cells in vivo; courtesy of T. Volk, Weizmann Institute of Science, Rehovot, Israel; Subramanian et al., 2003). Transgenic fly lines for targeted gene expression: UAS-eb1-GFP (courtesy of P. Kolodziej, Vanderbilt University, Nashville, TN; Sánchez-Soriano et al., 2010), UAS-eb1iRNA (Vienna Drosophila iRNA Center, #24451), UAS-EGC (EF-GRD-Ctail; courtesy of T. Volk; synonymous to UAS-EGG described by Subramanian et al., 2003), UAS-shot-RE-GFP (Shot-FL) and UAS-shot-RE-ΔGRD-GFP (Shot-ΔGRD, synonymous to Shot-ΔGas2; both courtesy of P. Kolodziej; Lee and Kolodziej, 2002). The newly generated constructs UAS-Ctail, UAS-Ctail-3MtLS*, UAS-shot-RE-ΔCtail-GFP,UAS-shot-RE-3MtLS*-GFP and UAS-eb1-mCherry were used for the establishment of transgenic fly lines (outsourced to BestGene Inc.) via PhiC31-mediated site-specific insertion of M-6-attB-UAS-1-3-4-born constructs onto the third chromosome (PBac{y+-attP-3B}CG13800VK00031; Bloomington line #9748). See Figure 1R for details of all UAS-shot constructs.
Figure 1.
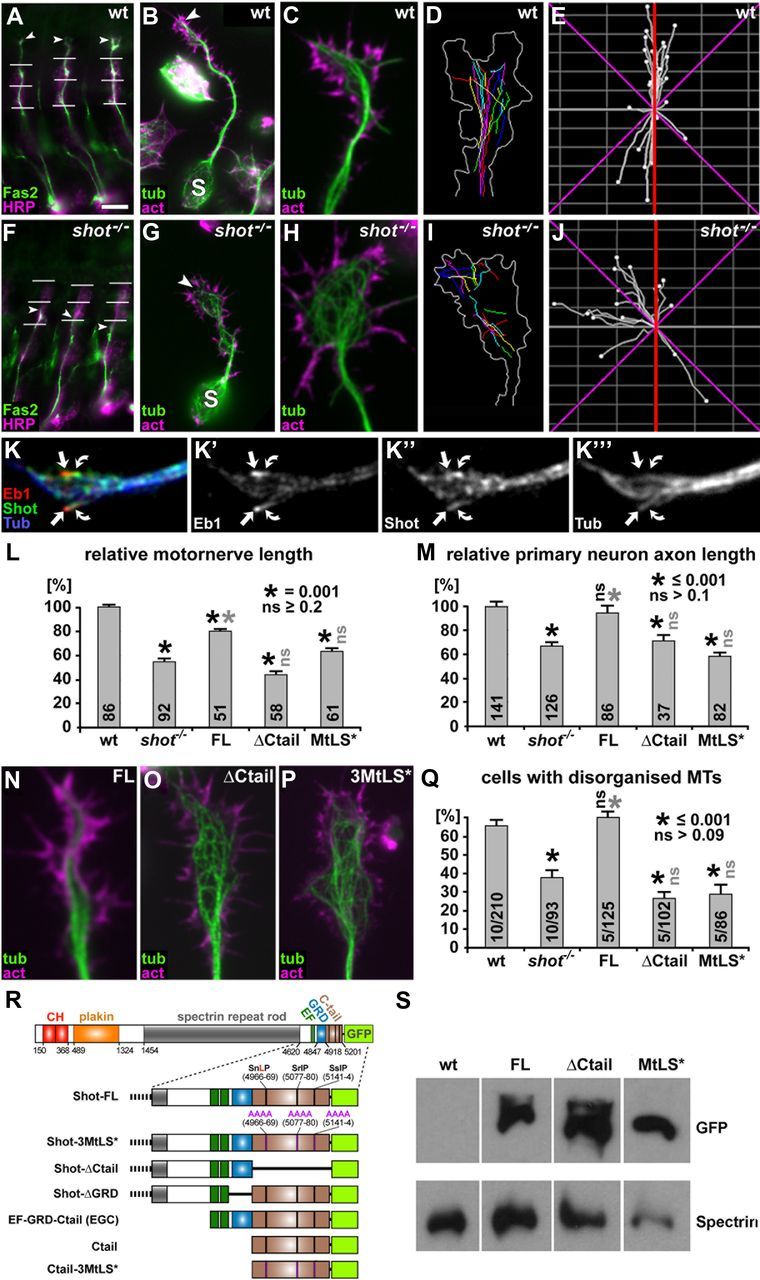
Ctail and MtLS motifs are required for axon growth and axonal MT organization in embryonic motor neurons in vivo and in primary embryonic neurons in culture. A–J, Illustrations of axonal phenotypes of wild-type (top) and shot−/− mutant neurons (bottom); stainings in A–C and F–H as indicated: act, phalloidin-labeled filamentous actin; Fas2, motor axonal marker Fasciclin 2; HRP, neuronal marker horseradish peroxidase; tub, tubulin. A and F show intersegmental motor nerves in three consecutive segments of the embryo which are shorter in the mutant (arrowheads, nerve tips; horizontal lines, indicators of ventrodorsal position as described in Materials and Methods); B and G show primary neurons after 6 h in culture with shorter axons in the mutant (S, somata; arrowheads, axon tips); C and H show close-ups of axonal growth cones of wt and shot−/− mutant primary neurons with disorganized MTs; D and I show traces of trajectories of polymerizing EB1-labeled MT plus ends (growth cone outline in white); E and J show directionality plots of the same growth cones with a higher degree of abaxial projections in the mutant (red lines indicate the axon axis, magenta lines represent 45° from the axon axis). K–K‴, Growth cones fixed with a specific protocol for MT plus end-associated proteins (Rogers et al., 2002) showing Shot at MT plus ends (green, curved arrows) trailing slightly behind EB1 (red, arrows) at the plus ends of MTs (blue). L, M, Quantifications of motor nerve lengths in embryos (see A, F) and axon lengths in primary neurons (see B, G) normalized to wild-type (wt); rescue experiments in shot−/− mutant embryos or neurons were performed with Shot-FL, Shot-ΔCtail and Shot-3MtLS*, as indicated; numbers in columns indicate the pooled numbers of assessed nerves or neurons; quantifications were statistically assessed by Kruskal–Wallis one-way ANOVA on Ranks (H = 181.593, 4 degrees of freedom, p ≤ 0.001 in L; H = 83.791, 4 degrees of freedom, p ≤ 0.001 in M) and Mann–Whitney rank sum test (black asterisks, significant when compared with wt; gray asterisks, significant when compared with shot−/−; black ns, not significant when compared with wt; gray ns, not significant when compared with shot−/−; values as indicated in insets). Note that rescues mediated by Shot-ΔCtail and Shot-3MtLS* show different trends in embryos and primary neurons, likely caused by distinct properties of these constructs in MT stabilization; Figure 2). N–P, Growth cones of 6 h shot−/− mutant primary neurons (compare H) expressing Shot-FL, Shot-ΔCtail or Shot-3MtLS*, as indicated. Q, Quantification of MT disorganization in 6 h primary neurons; numbers in columns indicate numbers of independent experiments (before slash) as well as overall numbers of assessed neurons (after slash); statistics as in L and M (Variance: H = 26.671, 4 degrees of freedom, p ≤ 0.001 in Q). R, Transgenic constructs of Shot-FL and its derivatives used for rescue experiments and localization studies (Figs. 1–4, 7; numbers refer to Ensembl ID FBpp0086744). S, Western blot analysis of embryos, wild-type or with sca-Gal4-mediated expression of Shot-FL, Shot-ΔCtail or Shot-3MtLS*, probed with anti-GFP and with anti-α-Spectrin as a loading control. Scale bars: (in A) A and F, 20 μm; (in B) B and G, 5 μm; C, D, H, I, N–P, 1 μm.
For analyses of MT polymerization events in shot−/− mutant background, primary neurons were generated from double-recombinant embryos (Df(2R)MK1, sca-Gal4/shotsf20, UAS-eb1-GFP) where only homozygously mutant specimens display fluorescence (sca-Gal4/UAS-eb1-GFP as controls). For rescue experiments, cultures were produced from embryos carrying the following combinations: UAS-shot-FL-GFP/+; Df(2R)MK1, sca-Gal4/shotsf20; UAS-eb1-mCherry/+ or Df(2R)MK1, sca-Gal4/shotsf20; UAS-eb1-mCherry/UAS-shot-RE-3MtLS*-GFP (scabrous-Gal4/+; UAS-eb1-mCherry/+ as controls), using CyO, twi::GFP-balancers to select for shot−/− mutant embryos.
DNA constructs
Recombineering was used to generate UAS-shot-RE-ΔCtail-GFP and UAS-shot-RE-3MtLS*-GFP. First, using recombination, the sequence of full-length Shot::GFP from p{UAST}-shot-RE-GFP (Lee and Kolodziej, 2002) was inserted into M-6-attB-UAS-1-3-4 (see below). Then the GalK positive/negative selection strategy (Warming et al., 2005) was used to delete the Ctail or replace it with an MtLS* mutated version. We used pcDNA3.1 vector (Invitrogen) to generate all Shot constructs transfected into fibroblasts and the primers used to amplify each construct/insert are listed in Table 1. We also modified the multiple cloning site of pcDNA3.1 introducing AscI and PacI restriction sites (5′ and 3′, respectively) to allow simple digestion/ligation transfers of the large inserts from p[acman] constructs.
Table 1.
List of primers used in this work
| New name/Position in shot CDS | Nucleotide sequence* (5′_3′) |
|---|---|
| shot_14491–14508.fw | GGCGGAAAGCTTGGCATGGCCCTTCGTCCCGATTGG |
| shot_14747–14769.bw | CATACCACTACCACTACCCTCTATGTTAGTGCGTCCTTTGG |
| shot_15031–15047.fw | GAGGGTAGTGGTAGTGGT ATGGTGAGCAAGGGCGAGGAG |
| shot_14756–14774.fw | GGCGGAAAGCTTGGCATGGGCACTAACATAGAGCTACG |
| GFP.bw | GCCGCCCTCGAGTCACAAAGATCCTCTAGGCG |
| shot_14284–14304.fw | GGCGGAAAGCTTGGCATGCTCAAGTACATGAACCACAAG |
| shot_14881–14919_Ala.fw | CACAATGGCGGCAGCGCCGCCGCGGCCCCATATATGAGT |
| shot_15214–15254_Ala.fw | GACGATCACACGGCCGCAGCCGCTGCGCAACGCAAGCCTTC |
| shot_15409–15444_Ala.fw | ATGAGTAGATCAGCCGCTGCAGCAGCACTAACAGGC |
| MCS_AscI + PacI_mod.fw | AATTAAGCTTGGCGCGCCGGT ACCGAGCTCGGATCC |
| MCS_AscI + PacI_mod.bw | TTAATCTAGATGCTTAATTAAATGCTCGAGCGGCCGC |
| eb1-mCherry-fw1 | AATTGGCGCGCCATGGCTGTAAACGTCTACTCC |
| eb1-mCherry-bw1 | CTCGCCCTTGCTCACCATGTTAATTAAATACTCCTCG |
| eb1-mCherry-fw2 | CGAGGAGTATTTAATTAACATGGTGAGCAAGGGCGAG |
| eb1-mCherry-bw2 | TTAACTTAATTAAATTACTTGTACAGCTCGTCCATG |
The name of each primer refers to its position in the shot-RE coding sequence (Ensembl ID: FBtr0087618). UAS-shot-RE-GFP was used as template for the PCRs. The primer GFP.bw was used in combination with shot_14756–14774.fw, and shot_14491–14508.fw to amplify Ctail-GFP and GRD-Ctail-GFP, respectively. The alanine substitution of MtLS sites was generated with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) using the primers shot_14881–14919_Ala, shot_15214–15254_Ala, and shot_15409–15444_Ala. Two PCRs were used to selectively amplify the GRD (primers: shot_14491–14508.fw with shot_14747–14769.bw) and the GFP tag (primers: shot_15031–15047.fw with GFP.bw). A third PCR was performed using the primers shot_14491–14508.fw and GFP.bw to link the two initial products into a GRD-GFP insert. The primers MCS_AscI + PacI_mod.fw and MCS_AscI + PacI_mod.bw were used to insert the AscI and PacI restriction sites into the MCS of pcDNA3.1. Links of 5 (GSGSG) or 10 (GSGPGSGPG) amino acids were used to separate Shot domains and GFP-tags. Restriction enzyme recognition sites are highlighted in bold and Shot-specific sequences are underlined. A Ctail variant in which all arginine residues were substituted by asparagines was synthesized in vitro by GenScript. This cDNA was used as template to generate Ctail-R* and GRD-Ctail-R* using the primer pair combinations described above for Ctail-GFP and GRD-Ctail-GFP, respectively. The UAS-eb1-mCherry construct was built using the primers eb1-mCherry-fw1, eb1-mCherry-bw1, eb1-mCherry-fw2, eb1-mCherry-bw2 to amplify the coding sequence of Drosophila EB1 linked to an mCherry fluorescent tag. The final PCR product was cloned into the M-6-attB-UAS-1-3-4 vector.
Engineering of M-6-attB-UAS-1-3-4 (P[acman]-1-3-4- chloramphenicol)
The new vector, M-6-attB-UAS-1-3-4 (or P[acman]-1-3-4-chloramphenicol) for the generation of site-specific transgenic flies, was developed in 3 steps.
Step 1: construction of P[acman] M-6.
The multiple cloning site was released from MCS-2 (Venken et al., 2006) as a BamHI fragment, gel purified, blunt-ended with Klenow polymerase, and subcloned into a Klenow-blunt-ended and CIAP dephosphorylated 6.4 kb SalI fragment from the vector pJW360 (a gift from Waclaw Szybalski, University of Wisconsin, Madison, WI) (Wild et al., 2002). The ligation product was electroporated into the EPI300 strain, and selected on LB with chloramphenicol (12.5 μg/ml, Cl12.5), resulting in P[acman] M-6.
Step 2: construction of P[acman] M-6-attB.
The ΦC31 attB site was amplified from pTA-attB (Groth et al., 2000) with primers attB1-NheI-F (CCTAGCTAGCCTCGACGATGTAGGTCACGGTC) and attB1-SalI-NsiI-R (CCAATGCATGTCGACCTCGACATGCCCGCCGTGAC), gel purified and subcloned as a NheI/NsiI fragment into a NheI/NsiI cut P[acman] M-6. The ligation product was electroporated into the EPI300 strain, and selected on LB (Cl12.5), resulting into P[acman] M-6-attB.
Step 3: construction of M-6-attB-UAS-1-3-4 (P[acman]-1-3-4-chloramphenicol).
The existing NotI site in MCS-P5-P3-w+-7 (Venken et al., 2006) was replaced by cutting out the existing polylinker with AscI and AsiSI and moving in a novel multiple cloning site encoded by two annealed oligos: MCS-NEW-F (CGCGCCGGCCTTAATGGCCTTAATTAACGAT) and MCS-NEW-R (CGTTAATTAAGGCCATTAAGGCCGG). The ligation product was transformed into the PIR1 strain, and selected on LB (K30), resulting into plasmid MCS-P5-P3-w+-7-NEW.
A triple SV40 polyA signal was amplified from pBS-3pA (a gift from Chuan-Wei Jang and Richard Behringer, The University of Texas MD Anderson Cancer Center, Houston, TX) with primers 3pA-Late-F (GGGCTAGACTAGCTAGAACTAGTGATC) and 3pA-Late-R (CCCCCTCAGTCCTCACAGTCTGTTC) phosphorylated with T4 Polynucleotide Kinase, gel purified and blunt subcloned into the EcoRI and Klenow-blunt-ended and CIAP dephosphorylated MCS-P5-P3-w+-7-NEW. The ligation product was transformed into the PIR1 strain, and selected on LB with kanamycin (30 μg/ml, K30), resulting into plasmid MCS-P5-P3-w+-7-NEW-3pA-Late.
A 5XUAS fragment was obtained from pUAST (Drosophila Genomics Resource Center) (Brand and Perrimon, 1993) with primers 5xUAS-AgeI-F (CCGACCGGTAAGCTTGCATGCCTGCAGGTC) and 5xUAS-XhoI-R (CCGCTCGAGTCGCTAGAGTCTCCGCTC), and PCR purified. A 5xUAS/Hsp70-TATA fragment was obtained from pUAST with primers 5xUAS-Hsp70-TATA-XhoI-F (CCGCTCGAGAAGCTTGCATGCCTGCAGGTC) and 5xUAS-Hsp70-TATA-SpeI-AgeI-R (GGACTAGTACCGGTCTATTCAGAGTTCTCTTCTTG), and PCR purified. The AgeI/XhoI cut 5XUAS fragment and the XhoI/SpeI cut 5xUAS/Hsp70-TATA fragment were triple ligated into the AgeI/SpeI cut MCS-2 plasmid (Venken et al., 2006). The ligation product was transformed into the PIR1 strain, and selected on LB (K30), resulting into plasmid 10xUAS.
The 10xUAS fragment was cut out of plasmid 10xUAS with AgeI, gel purified and cloned into AgeI- and SAP-treated MCS-P5-P3-w+-7-NEW-3pA-Late. The ligation product was transformed into the PIR1 strain, and selected on LB (K30), resulting into plasmid 3pA-Late-UAS.
The entire P element, containing P3-10xUAS-3pA-white+-P5, was released from 3pA-Late-UAS as a BamHI fragment, gel purified, blunt-ended with Klenow polymerase, and subcloned into the Klenow-blunt-ended and SAP dephosphorylated SalI cut plasmid P[acman] M-6-attB. The ligation product was transformed into the EPI300 strain, and selected on LB (Cl12.5), resulting into plasmid M-6-attB-UAS-1-3-4 or P[acman]-1-3-4-chloramphenicol.
Drosophila primary neuron cultures
The generation of primary cell cultures was performed as described previously (Sánchez-Soriano et al., 2010). In brief, cells were collected with micromanipulator-attached needles from stage 11 wild-type or mutant embryos (6–7 h after egg lay at 25°C; Campos-Ortega and Hartenstein, 1997), treated for 5 min at 37°C with dispersion medium, washed and eventually resuspended in the final volume of Schneider's medium (Schneider, 1964; Invitrogen; 5–6 μl/donor embryo), plated to coverslips, kept as hanging drop cultures in air-tight special culture chambers (Küppers-Munther et al., 2004) usually for 6 h at 26°C. Cells were grown directly on glass, or on coverslips coated with 0.5 mg/ml Concanavalin A (Sigma) which is favorable for the analysis of MT disorganization phenotypes (Sánchez-Soriano et al., 2009). Dilutions of the MT-destabilizing drug nocodazole (20 μm; Sigma) and of the MT-stabilizing drug taxol (1 nm; Sigma) in Schneider's medium were prepared from stock solutions in DMSO. For controls, equivalent concentrations of DMSO were diluted in Schneider's medium. To deplete maternal levels of EB1 in eb1−/− mutant or eb1iRNA-expressing neurons, they were kept for several days in culture medium in centrifuge tubes before they were suspended and plated (Sánchez-Soriano et al., 2010).
Fibroblast cell culture
NIH3T3 mouse fibroblasts were cultured in DMEM (Sigma-Aldrich), supplemented with 1% l-glutamine (Invitrogen), 1% penicillin/streptomycin (Invitrogen), and 10% FCS in a humidified incubator at 5% CO2 and passaged in a 1:10 dilution every 3 d. Lipofectamine and Plus reagent (Invitrogen) were used for transient DNA transfections according to the manufacturer's instructions. Cells were replated 5 h after transfection at ∼40% confluence in glass-bottom dishes (MatTek Corporation) coated with 10 μg/ml bovine plasma fibronectin (Sigma-Aldrich). For live imaging (24 or 48 h post-transfection), cells were maintained in Ham's F-10 medium (Sigma-Aldrich) supplemented with 4% FCS. Nocodazole was applied at 10 μm in Ham's F-10 medium for 3 h at 37°C, followed by immediate fixation.
Coimmunoprecipitation and Western blot analyses
Cos-7 cells (African Green Monkey Kidney Fibroblast Cells) were transfected with Ctail::GFP or GRD-Ctail::GFP. Cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS and 1% glutamine in a humidified incubator at 5% CO2 and passaged in a 1:10 dilution every 3 d. Lipofectamine and Plus reagent (Invitrogen) were used for transient DNA transfections of 70% confluent cultures in 10 mm culture dishes, according to the manufacturer's instructions. Forty-eight hours after transfection, cells were lysed on the dish with 2 ml of lysis buffer (50 mm Tris pH7.4; 120 mm NaCl; 2.5 mm EGTA; 10 mm MgCl; 1% NP40) supplemented with 1 mm PMSF (Sigma) and complete mini EDTA-free Protease Inhibitor Cocktail (Roche). Cell extracts were subjected to Immunoprecipitation (IP) using GFP-Trap A kit (ChromoTek) following the manufacturer's instructions. Cell extracts and IP samples were resolved in 10% Bis-Tris NuPage gels (Invitrogen) and then transferred to Hybond-P PVDF membranes (GE Healthcare). Membranes were stained with anti-human EB1 (Santa Cruz Biotechnology; diluted 1:1000), anti-GFP (Invitrogen; diluted 1:2000) and anti-Vinculin (Sigma; diluted 1:2000) antibodies, followed by HRP-conjugated secondary antibodies, and developed using Pierce ECL Plus Western Blotting substrate kit).
For Western blot analysis of Shot-FL, Shot-ΔCtail and Shot-3MtLS* expression levels stage 16 wild-type embryos and embryos expressing the constructs under the control of scabrous-Gal4 were collected in 2× sample Laemmli buffer (Sigma), at a concentration of 1 embryo/μl. The samples were homogenized with a pestle and heated for 5 min at 96°C before electrophoresis. Twenty microliters of each embryonic extract were resolved in 3–8% Tris-acetate gradient NuPage gels (Invitrogen) and Western blot was performed as described above using anti-GFP (Invitrogen; diluted 1:2000) and anti-α-spectrin [Developmental Studies Hybridoma Bank (DSHB); diluted 1:2000].
Fixation, staining, microscopy, and documentation
Procedures for the dissection, immunohistochemistry and motor axon length measurements of embryos, as well as standard staining procedures for primary neurons were described previously (Bottenberg et al., 2009; Sánchez-Soriano et al., 2010). For anti-EB1 stainings, cells were treated for 10 min with −80°C +TIP fixative (90% methanol, 3% paraformaldehyde, 5 mm sodium carbonate, pH 9; Rogers et al., 2002). Fibroblasts were fixed either with −20°C methanol for 5 min (anti-tubulin staining) or with 4% paraformaldehyde in 0.05 m phosphate buffer for 30–60 min (phalloidin stainings). Before adding antibodies, cells were washed 3 times (10 min each) with PBT (PBS, 0.3% Triton-X). Staining reagents: anti-tubulin (clone DM1A, mouse, 1:1000, Sigma; alternatively, clone YL1/2, rat, 1:500, Millipore Bioscience Research Reagents); anti-DmEB1 (rabbit, 1:2000; Elliott et al., 2005); anti-FasII (clone ID4, mouse, 1:20, DSHB); anti-GFP (goat, 1:500, Abcam); anti-Shot (1:200, guinea pig; Strumpf and Volk, 1998); Cy3-conjugated anti-HRP (goat, 1:100, Jackson ImmunoResearch); FITC-, Cy3- or Cy5-conjugated secondary antibodies (donkey, purified, 1:100–200; Jackson ImmunoResearch); TRITC/Alexa647-coupled Phalloidin (1:100 or 1:500; Invitrogen). Standard documentation was performed with AxioCam monochrome digital cameras (Carl Zeiss Ltd.) mounted on BX50WI or BX51 Olympus compound fluorescent microscopes.
Live imaging was performed at 26°C on a Delta Vision RT (Applied Precision) restoration microscope using a 100×/1.3 Ph3 Uplan Fl objective and the Sedat filter set (Chroma 89000). The images were collected using a Coolsnap HQ (Photometrics) camera. For time lapse recording, images were taken every 4 s for 2 min with an exposure time of 0.8–1 s. To generate velocity and lifetime measurements of MTs, comets of fluorescently tagged EB1 were tracked manually using the manual tracking plugin for ImageJ. Anterograde comet paths that did not become merged with other comets were used for analysis. To determine the directionality of MT growth, MT plus ends labeled with EB1-GFP were manually traced using the manual tracking plugin for ImageJ and the traces were analyzed using the ImageJ Chemotaxis plugin.
Axon length and staining intensity measurements were performed using ImageJ software (ImageJ 1.42, http://rsb.info.nih.gov/ij). To quantify MT disorganization, relative numbers of primary neurons were counted that showed areas of curled non-coalescent microtubules in either the axon or growth cone. To quantify MT stability upon nocodazole-treatment, relative numbers of primary neurons were counted in which MTs (stained with anti-tubulin) were absent in parts of the axons. To measure MT association, fibroblasts were transfected with same amounts of cDNA encoding GFP-tagged proteins. Based on the rationale that stronger MT affinity of a protein leads to a higher density of molecules associated with MTs (reflected in the intensity of staining), ImageJ was used to draw lines across cells, crossing the shaft or plus ends of MTs, and maximal gray values were determined (two regions for each cell). Measurements were performed on live images taken in the green channel with identical exposure times of cells expressing similar overall levels of the GFP-fusion proteins. The cytoplasmic fluorescence intensity was subtracted by bandpass 5:1 filtering, resulting in images of GFP-fusion protein associated with MTs. All data are shown as mean ± SEM. Statistical analyses were performed in Sigma Stat 3.0 using χ2 tests or Kruskal–Wallis one-way ANOVA on Ranks followed by Mann–Whitney Rank Sum Tests.
Results
Shot regulates the direction of MT polymerization
Previous work had shown that neurons lacking Shot (i.e., carrying two loss-of-function mutant alleles of shot, referred to as shot−/−) extend much shorter axons (illustrated in Fig. 1A,B vs F,G,L,M; Lee and Luo, 1999; Lee and Kolodziej, 2002; Bottenberg et al., 2009; Sánchez-Soriano et al., 2009). In these experiments, axon shortening correlated with a marked increase in curled non-coalescent microtubules which frequently cross one another (from here on this is referred to as MT disorganization; illustrated in Fig. 1H,Q; Sánchez-Soriano et al., 2009). However, the molecular mechanisms of Shot function that explain these axon growth and MT disorganization phenotypes are unknown.
Two possible explanations have previously been suggested to account for the appearance of looped MTs in growth cones (Purro et al., 2008). Curled MTs might arise from continued MT polymerization at the axonal or growth cone membrane, forcing MTs to bend backward or buckle, as similarly described for primary mouse endoderm cells lacking ACF7 (Kodama et al., 2003). Alternatively, curled organization would be expected if MTs lose their tendency to perform guided extension in the direction of axon growth. In the latter scenario they would polymerize along random trajectories with reduced probability to reach the leading edge. To distinguish between these two possibilities, we assessed the directionality of MT extension in growth cones of wild-type and shot−/− mutant neurons. To this end, we labeled polymerizing MT plus ends with EB1::GFP and followed their growth using time lapse. We found that, in control neurons, 93.3% of microtubules polymerized in the direction of axonal growth, and only 6.7% (n = 206 microtubules from 11 distinct cells) showed transverse, abaxial polymerization (i.e., deviated >45° from the axon axis; Fig. 1D,E). In contrast, the fraction of abaxial MT trajectories was increased threefold to 19.1% in shot−/− mutant growth cones (n = 235 microtubules from 11 distinct cells; Fig. 1I,J; χ2 = 11.503 with 1 degrees of freedom; p ≤ 0.001). This increase in transverse MT trajectories suggested MT guidance to be a likely mechanism through which Shot contributes to the organization of axonal MTs and thus promotes axon extension.
Ctail and its MtLS motifs are essential for axon growth and MT organization
If MT guidance is an important aspect of Shot function during MT organization and axon growth, its function should require interaction with MT plus ends, which are the sites of MT polymerization. We found Shot localization to be in agreement with this hypothesis. When using specific fixation protocols for MT plus end-associated proteins (Rogers et al., 2002), we found Shot at MT plus ends trailing slightly behind EB1 (Fig. 1K–K‴). This localization was in agreement with findings in non-neuronal Drosophila S2 cells (Slep et al., 2005).
Next we assessed whether plus end localization of Shot is functionally relevant. To this end, we tested the requirement of three putative MtLS (MT tip localization sequence) motifs located in the Ctail of Shot (Fig. 1R), based on the rationale that this type of motif is required for MT plus end localization in a number of proteins (Honnappa et al., 2009). We generated two derivatives of the Shot full-length construct which either lacked Ctail completely (Shot-ΔCtail; Fig. 1R) or carried targeted mutations replacing each of the three MtLS motifs by four alanines (Shot-3MtLS*; Fig. 1R). We used the Gal4/UAS system to target expression of these constructs to shot−/− mutant Drosophila neurons and tested their functional ability to rescue the shot−/− mutant axonal phenotypes. In control experiments, normal full-length Shot (Shot-FL) achieved significant rescue of motor nerve stall in shot−/− mutant embryos in vivo, and a complete rescue of axon extension and MT organization phenotypes in shot−/− mutant primary neurons in culture (Fig. 1L–N,Q; see also Bottenberg et al., 2009; Sánchez-Soriano et al., 2009). In contrast, we found that both Shot-ΔCtail and Shot-3MtLS* failed to rescue either of these defects both in vivo and culture (Fig. 1L,M,O–Q). Western blot analyses and localization studies in primary neurons revealed that all constructs were expressed at comparable levels in neurons and localized to growing axons (Figs. 1S, 2A′,B′,C′).
Figure 2.
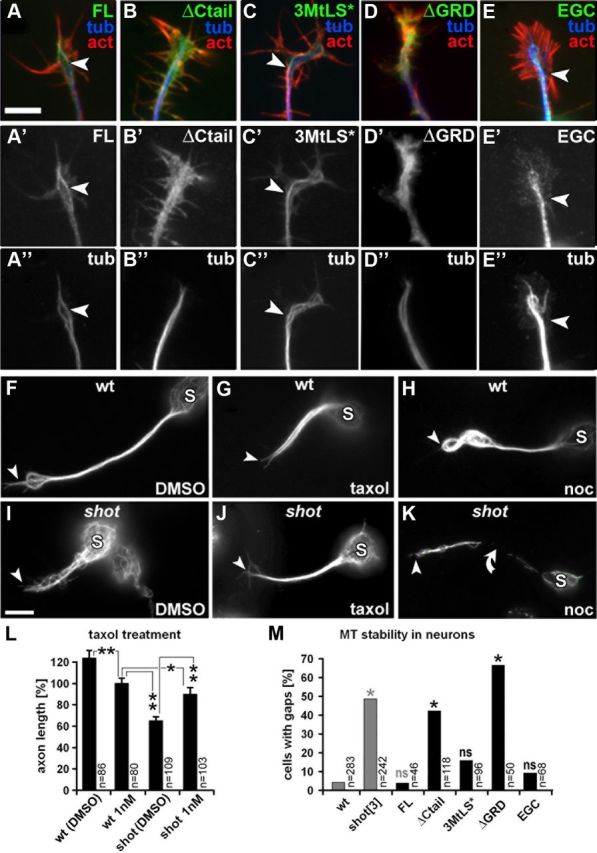
Shot confers stability to MTs through its C terminus. A–E″, Primary embryonic Drosophila neurons with targeted expression of transgenic Shot constructs (illustrated in Fig. 1R), stained for F-actin (red), tubulin (blue) and GFP (green); arrowheads point at obvious MT-association of the GFP-tagged Shot-FL and Shot-3MtLS* constructs. Note, that A′–C′ were taken with identical camera settings from parallel cell cultures, indicating that all constructs are equally well expressed. F–K, Anti-tubulin-stained primary neurons treated with taxol or nocodazole (noc), or control-treated with the vehicle DMSO. Somata are indicated by S, the tips of the axons by arrowheads, and the curved arrow points to a region of the axon devoid of microtubules. L, Quantification of axon lengths upon taxol treatment (**p ≤ 0.001, *p = 0.03, as determined by Mann–Whitney rank sum test). M, Quantification of destabilizing effects of nocodazole on axonal MTs of wt and shot−/− mutant primary neurons without targeted expression (gray) or of shot−/− mutant neurons expressing Shot constructs (black; constructs illustrated in Fig. 1R); χ2 tests (*p ≤ 0.004; ns, not significant p ≥ 0.01) were performed relative to wt (gray asterisks) or relative to FL (black asterisks). Scale bar (in A) A–E″, 2 μm; F–K, 5 μm.
We conclude that the Ctail and its MtLS motifs are required for MT guidance and the growth-promoting function of Shot, suggesting that Shot acts as a +TIP in this context.
Shot stabilizes axonal MTs in growing neurons
Apart from localizing at MT plus ends, endogenous Shot and Shot-FL were described to localize along the shafts of MTs in neurons (Sánchez-Soriano et al., 2009; compare Fig. 2A′). We tested whether also this other fraction of Shot at MT shafts is functionally required. Localization along axonal MT shafts is a typical feature of structural MAPs (such as Tau, MAP2 or MAP1b) which act to stabilize MTs against depolymerization, thus positively impacting on axon growth (Chilton and Gordon-Weeks, 2007; Riederer, 2007; Morris et al., 2011). We tested therefore whether Shot likewise stabilizes axonal MTs and whether this function could contribute to axon growth regulation.
For this, we treated shot−/− mutant neurons with low doses of the MT-stabilizing drug taxol for 4 h. We found that treatment with taxol, but not vehicle, significantly rescued the short axon phenotype of shot−/− mutant neurons (Fig. 2I,J,L). In contrast, the same taxol treatment of wild-type neurons modestly inhibited axon growth (Fig. 2F,G,L), which is in agreement with previous reports for both Drosophila and vertebrate neurons (Letourneau et al., 1987; Sánchez-Soriano et al., 2010; Hur et al., 2011) and may be due to the suppression of MT dynamics through overstabilization. Therefore, the right balance of MT stabilization seems essential for axon growth, and taxol might help to bring the low MT stability levels in shot−/− mutant neurons back to levels which better support axon growth.
To directly assess potential MT-stabilizing roles of Shot, we treated primary neurons for 2.5 h with the MT destabilizing drug nocodazole. We found that ∼50% of nocodazole-treated shot−/− mutant neurons displayed areas in their axons which were devoid of MTs (Fig. 2K,M), whereas such effects were hardly ever seen in wild-type neurons (Fig. 2H,M).
Together, these results clearly demonstrate that Shot stabilizes axonal MTs and suggest it is functionally related to structural MAPs.
The C terminus of Shot mediates MT stabilization in an MtLS-independent manner
We next asked which domains of Shot are required for MT stabilization. For this, we expressed different Shot constructs in primary neurons and assessed their localization as well as their ability to reinstate MT resistance against nocodazole in the absence of endogenous Shot. We found that Shot-FL prominently localizes along MTs and fully reestablishes MT stability to shot−/− mutant neurons, confirming that MT stabilization is a true function of Shot (Fig. 2A′,M). We next tested the C-terminal GRD, which is essential for Shot function in axon growth and has been suggested to stabilize MTs in mouse fibroblasts (Lee and Kolodziej, 2002; Bottenberg et al., 2009; Sánchez-Soriano et al., 2009). Accordingly, a Shot-FL derivative lacking GRD (Shot-ΔGRD; Fig. 1R) failed to protect axonal MTs of shot−/− mutant primary neurons against nocodazole treatment (Fig. 2M). Notably, Shot-ΔGRD also failed to display obvious MT association, but was diffusely distributed throughout neurons instead (Fig. 2D′). To our surprise we found that Shot-ΔCtail similarly showed a diffuse localization pattern and failed to protect axonal MTs (Fig. 2B′,M). In contrast, Shot-3MtLS* displayed normal MT association and fully rescued MT resistance to nocodazole (Fig. 2C′,M). Therefore, Ctail is important for MT-stabilizing functions of Shot, but its MtLS motifs are dispensable for this. Finally, we tested the EGC construct which consists of only the Shot C terminus including GRD and Ctail (Fig. 1R). EGC displayed homogeneous localization along MTs and completely restored nocodazole resistance to shot−/− mutant neurons (Fig. 2E′,M), indicating that only the C terminus of Shot is sufficient for this function.
Together, both the GRD and Ctail are essential for MT shaft localization and stabilization, but the MtLS motifs are not required for this function.
Tendon cells confirm context-specific requirements of MtLS motifs for Shot function
We next tested whether our findings of MTLS-dependent and independent functions of Shot in neurons might be of relevance also in other cellular context. To this end, we investigated tendon cells as a further example of cell types that require Shot function (Alves-Silva et al., 2008). Tendon cells are specialized cells of the epidermis to which muscles attach, and which display prominent apicobasal MT arrays that are highly resistant to nocodazole (Fig. 3C). In shot−/− mutant embryos, tendon cells are disrupted, and this phenotype can be rescued through targeted expression of Shot-FL (Fig. 3B,E; Prokop et al., 1998; Alves-Silva et al., 2008; Bottenberg et al., 2009). We have shown previously that GRD is absolutely required for Shot function in tendon cells (Bottenberg et al., 2009). Here we assessed the role of Ctail and MtLS motifs. We found that Shot-ΔCtail and Shot-3MtLS* both localized normally in tendon cells when assessed in wild-type larvae (Fig. 3I,L). However, when expressed in shot−/− mutant embryos, only Shot-3MtLS* fully restored tendon cell integrity, whereas Shot-ΔCtail completely failed to rescue the shot−/− mutant tendon cell phenotype (Fig. 3H,K). Therefore, Shot function in tendon cells absolutely requires Ctail but not its MtLS motifs.
Figure 3.
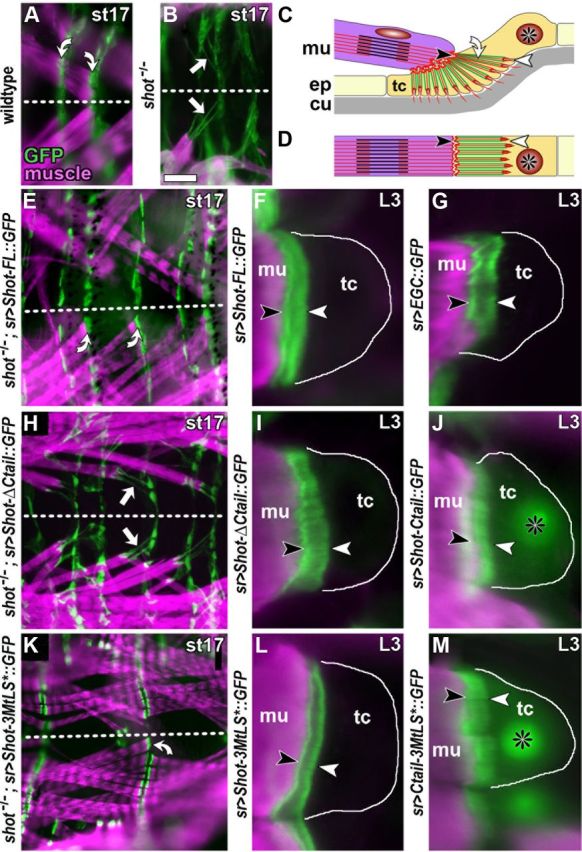
Localization and rescue capability of mutant Shot constructs in tendon cells. A, A late stage 17 (st17) wild-type embryo in plain view (anterior left; dashed line indicates midline) displaying muscles (stained with phalloidin, magenta) which attach with their tips to tendon cells with prominent cytoskeletal arrays (curved arrows; stained with actin::GFP, green). B, In shot−/− mutant embryos, tendon cell integrity is affected as reflected by abnormal elongation of actin::GFP-labeled cytoskeletal arrays (white arrows). C, D, Diagrams illustrating tendon cell morphology in lateral (C) and plain (D) view; muscles (mu, magenta) attach to basal surfaces (black arrowheads) of tendon cells (tc; asterisks indicate nuclei), which are specialized cells of the epidermis (ep); apical tendon cell surfaces (white arrowhead) link to the exoskeleton called cuticle (cu, gray); apical and basal tendon cell surfaces are connected through cytoskeletal arrays (curved arrow) which are composed of parallel actin fibers (red) and MTs (green) and appear as a continuous band in horizontal view (D). E, H, K, Plain views of shot−/− mutant embryos with targeted expression of Shot-FL, Shot-ΔCtail or Shot-3MtLS (as indicated on the left); successful rescue of tendon cell integrity by Shot-FL and Shot-3MtLS is indicated by curved arrows in E and K, respectively; arrows point at stretched cytoskeletal arrays reflecting failed rescue through Shot-ΔCtail; a similar lack of rescue was observed for Shot-ΔGRD (Bottenberg et al., 2009). F, G, I, J, L, M, Images from late L3 larvae show muscle tips (magenta) attached to tendon cells (outlined with white line) which express different GFP-tagged constructs (green; as indicated on the left of each panel; all symbols and abbreviations as in C); Shot-FL, Shot-ΔCtail, Shot-3MtLS* and EGC all show strong association with cytoskeletal arrays with a slightly higher concentration at apical and basal ends. Strong MT association is surprising especially for Shot-ΔCtail, but similar observations were made for Shot-ΔGRD (Bottenberg et al., 2009); they might be explained through dimerization of these deletion constructs with endogenous Shot or interactions of N-terminal or central domains with other constituents proteins of cytoskeletal arrays. In contrast, Ctail and Ctail-3MtLS* show weaker and homogeneous localization at cytoskeletal arrays, and higher cytoplasmic and nuclear levels. Scale bar (in A) A, B, E, H, K, 40 μm; F, G, I, J, L, M, 7 μm.
Ctail supports GRD-mediated MT stabilization by enhancing MT association of Shot
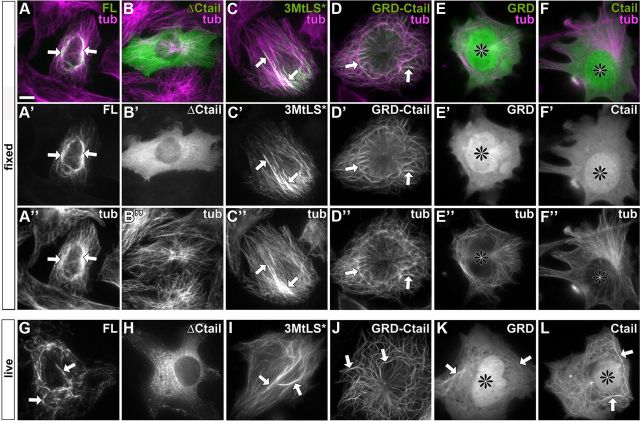
To uncover the functional contributions of GRD and Ctail to MT-stabilizing functions of Shot, we used mouse NIH3T3 fibroblasts which are ideally suited for the imaging of cytoskeletal networks and have been used successfully for the study of the C termini of mammalian spectraplakins (Sun et al., 2001). We first expressed equivalent Shot variants in fibroblasts and found the same correlation as in neurons. Shot-FL, Shot-3MtLS* and a C-terminal GRD-Ctail construct (see Fig. 6K) all strongly associated with MTs (Fig. 4A′,C′,D′,G,I,J) and conferred stability of MTs against nocodazole treatment in all transfected cells (see Notes). In contrast, Shot-ΔCtail failed to associate with MTs and stabilize them in ∼70% of transfected fibroblasts (Fig. 4B′,H; see Notes). These results clearly confirmed our findings in neurons and suggested that fibroblasts represent a suitable cellular model for our studies.
Figure 6.
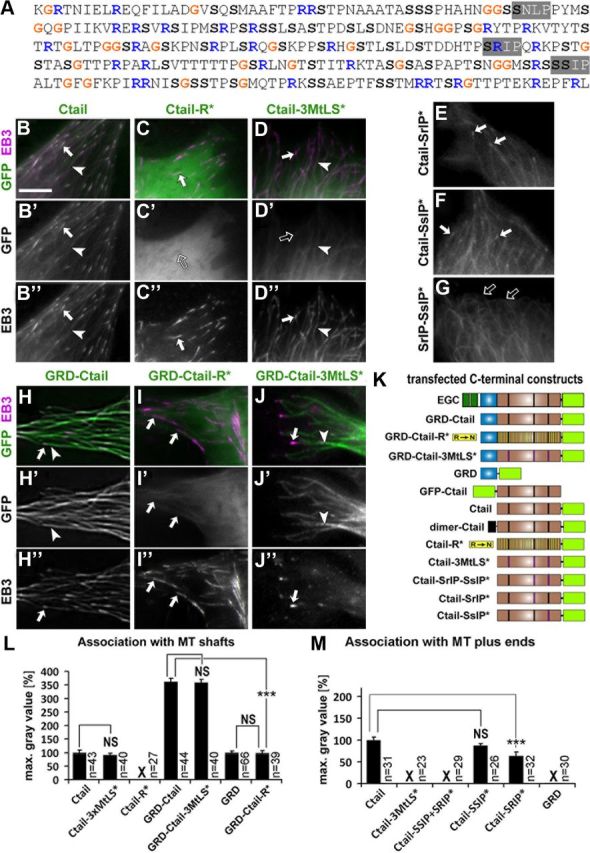
Association to MT shaft or plus end is determined by positive charge and MtLS motifs in the Ctail. A, Sequence of Ctail (Ensembl ID: FBpp0086744) illustrating the distribution of glycines (G), arginines (R) and serines (S), and the location of the three putative MtLS motifs (highlighted in gray). B–J″, Stills taken from live movies of fibroblasts expressing Shot domain constructs (as indicated); arrows point at MT plus end localization of GFP-constructs and/or EB3::RFP, open arrows indicate lack of MT plus end localization, and arrowheads show association along MT shafts; E–G, Ctail constructs with mutations of only subsets of their MtLS motifs imaged in the absence of additional markers showing that EB3 coexpression had no obvious impact on the localization of GFP-tagged constructs. K, C-terminal constructs of Shot that were analyzed in fibroblasts. L, M, Quantification of construct associations with MT shafts or MT plus ends; X indicates absence of any detectable association; Statistics were performed using Kruskal–Wallis one-way ANOVA on Ranks (H = 320.309 with 5 degrees of freedom, p ≤ 0.001 in H; H = 19.104 with 2 degrees of freedom, p ≤ 0.001 in I) and Mann–Whitney rank sum test (***p ≤ 0.001; NS, not significant; p > 0.05). Scale bar in B, 4 μm in all images.
Figure 4.
Localization of GFP-tagged Shot constructs in fibroblasts. A–F″, Fixed fibroblasts transfected with GFP-tagged Shot constructs (green in A–F and gray in A′–F′) and stained for Tubulin (magenta in A–F and gray in A″–F″). G–L, Still images taken from live movies of cells expressing the same constructs as shown above: Shot-FL (G) and Shot-3MtLS* (I) usually associate with MTs in a discontinuous way; GRD-Ctail (J) prominently decorates MTs; GRD and Ctail (K, L) mildly associate with MTs although relatively high levels of proteins seem to be available in the cytoplasm (and tend to enrich also in nuclei; asterisks); Shot-ΔCtail shows no obvious MT association (H). Note that MT localization of GRD and Ctail is lost after fixation (E′, F′), further indicating their weak tendency to associate with MTs. Arrows indicate MT association throughout. Scale bar in A, 10 μm in all images.
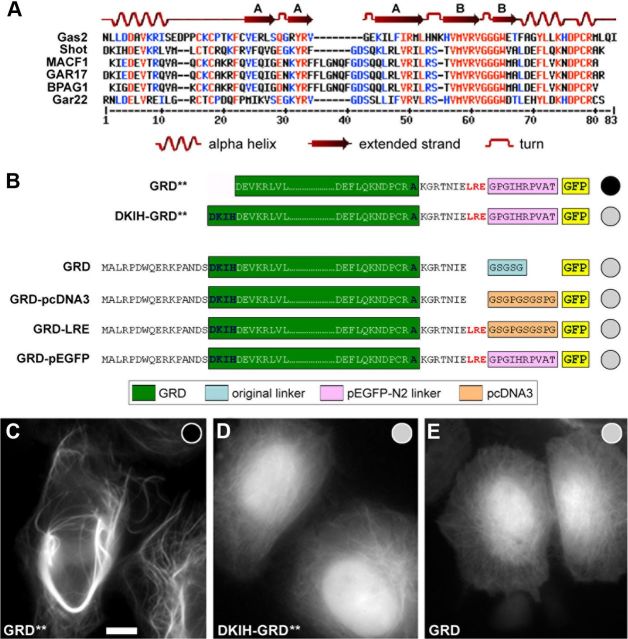
To understand how GRD and Ctail mediate MT association and stabilization, we investigated their properties in isolation. We found that each domain alone only weakly associated with MTs, and this is in contrast with the very strong localization of the GRD-Ctail tandem construct (Fig. 4D′–F′, J–L). Furthermore, GRD and Ctail displayed high cytoplasmic levels and prominent nuclear localization (Fig. 4, black asterisks), suggesting that MT association was too weak to efficiently sequester the available protein. As a third indicator for their weak MT association, the localization to MTs observed by live imaging was frequently lost during immunohistochemical fixation and staining procedures (Fig. 4E′,F′ vs K,L). Finally, the weak association is in agreement with reports for GRDs and Ctails of mammalian spectraplakins or Gas2-like proteins (Sun et al., 2001; Goriounov et al., 2003). Notably, the weak MT association of our GRD construct contradicted previous reports which demonstrated that the GRD of Shot strongly localizes to MTs (Lee and Kolodziej, 2002; Applewhite et al., 2010). However, we were able to verify our findings by showing that previously used GRD constructs were incomplete and lacked four N-terminal amino acids within the first α helix. This deletion caused the enhanced MT localization in former studies (see explanations in the legend of Fig. 5).
Figure 5.
N-terminal residues of the GRD significantly influence its MT association. A, Alignment of GRDs from various spectraplakins and Gas2-like molecules (taken from Ensembl) with the resolved structure of the GRD from mouse Gas2 (growth arrest specific 2; http://www.rcsb.org/pdb/explore/explore.do?structureId=1V5R). B, A different GRD construct (GRD**) used in previous publications (Lee and Kolodziej, 2002; Applewhite et al., 2010; construct kindly provided by S. Lee, Seoul National University, Seoul, Republic of Korea) shows much stronger MT association (indicated by black circle) than our GRD construct (gray circle). The GRD** construct lacks the 4 amino acids D-K-I-H of the first α helix, and adding these residues back (DKIH-GRD**) is sufficient to reduce the strength of MT association (gray circle). In contrast, the following modifications of the GRD construct did not change MT association: extending the linker region (GRD-pcDNA3), adding LRE from the Shot protein sequence (GRD-LRE), adding LRE and using the linker present in GRD** (Gas2-pEGFP). C–E, Images of fibroblasts expressing some of the above mentioned constructs. Scale bar in C, 10 μm in all images.
We next tested GRD and Ctail for their ability to protect against nocodazole. We found that GRD displayed high stabilizing properties (see Notes), whereas Ctail completely failed to protect MTs (see Notes). We wondered whether Ctail's inability to protect MTs was due to its weak MT association. However, when triggering strong MT association of Ctail (through fusing a dimerization domain of the yeast transcriptional activator GCN4 to its N terminus; Fig. 6K; Viñals et al., 2002) this was still not sufficient to protect MTs against nocodazole treatment (data not shown).
We conclude that Ctail is itself not a MT-stabilizing element of the Shot C terminus. However, it enhances MT association and thus helps to maintain full-length Shot on MTs as prerequisite for its GRD-mediated role in MT stabilization.
MT association of Ctail requires its net positive charge
We next asked what properties of Ctail are required to associate with MT shafts. Ctails of all spectraplakins and Gas2-like proteins are poorly conserved at the sequence level. Instead, a characteristic commonality is their high content in glycines, serines, and arginines (Sun et al., 2001; Goriounov et al., 2003; Stroud et al., 2011). Shot Ctail contains 10% glycines, 19% serines and 11% arginines, which are distributed throughout the sequence (Fig. 6A). In particular the positive charge conferred by the abundant arginine residues might be expected to contribute to MT association (Wolff, 1998; Wu et al., 2011). To test this possibility, we generated a Ctail derivative in which all arginines were substituted by uncharged polar asparagine residues (Fig. 6K, Ctail-R*). When expressed in fibroblasts, Ctail-R* failed to display any obvious MT association (Fig. 6C′,L). Furthermore, when Ctail-R* was fused to GRD (GRD-Ctail-R*) it displayed weak MT association similar to GRD alone (Fig. 5E vs 6I′), indicating that Ctail-R* fails to enhance MT association of GRD. Therefore, positively charged arginines contribute to Shot association with MTs.
MtLS motifs mediate binding to EB1 and recruit the Shot C terminus to MT plus ends
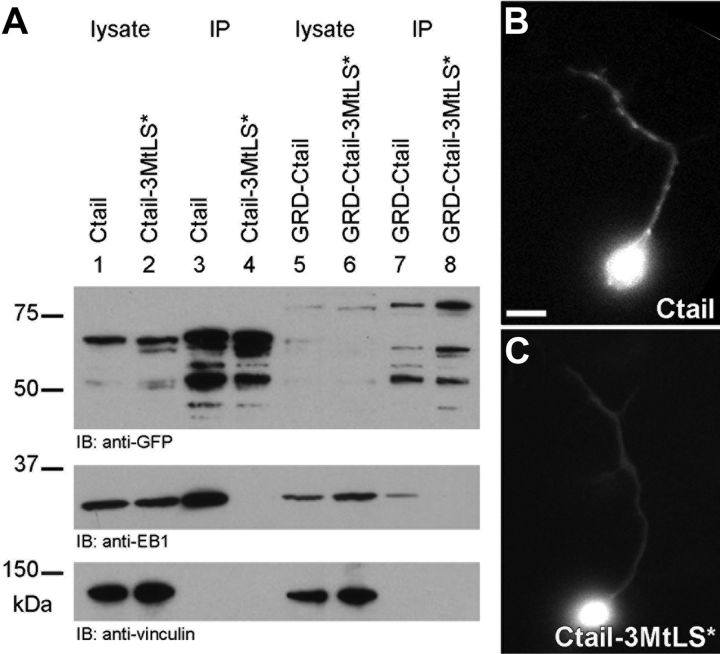
Having understood how Ctail contributes to MT-stabilizing roles of Shot, we next addressed how its MtLS motifs contribute to functions in axonal extension and MT organization. We observed that Ctail, in addition to localizing along MT shafts in fibroblasts, strongly accumulates at their polymerizing plus ends (Fig. 6B′, arrows). Such localization was abolished when the three MtLS motifs were mutated (Ctail-3MtLS*; Fig. 6D′,M). In support of this finding, endogenous EB1 was coimmunoprecipitated with Ctail in extracts of Cos-7 cells, but not with Ctail-3MtLS* (Fig. 7A). Importantly, in Drosophila primary neurons, Ctail also tracked polymerizing MT plus ends, whereas Ctail-3MtLS* failed to do so (Fig. 7B,C), suggesting that MtLS motifs might also mediate MT plus end localization of Shot in neurons.
Figure 7.
Validating the requirement of MtLS motifs for EB1-mediated MT plus end localization. A, Coimmunoprecipitation analyses in Cos-7 cells. Top, Extracts of Cos-7 cells expressing GFP-tagged Ctail, Ctail-3MtLS*, GRD-Ctail or GRD-Ctail-3MtLS*, before (lysate) or after (IP) enrichment through immunoprecipitation with GFP-Trap as indicated. Ctail-GFP and GRD-Ctail-GFP are expected to be 68 and 79 kDa in size, respectively, and a tendency of the protein to degrade could be suppressed to tolerable levels; middle, coimmunoprecipitation of endogenous EB1 is observed only in IP samples of Ctail (lane 3) and GRD-Ctail (lane 7), but not in the corresponding MtLS-mutant versions (lanes 4 and 8, respectively); bottom, anti-α-Vinculin reveals comparable amounts of loaded cell lysate extracts and also serves as an unspecific control for the coimmunoprecipitation. B, C, In primary Drosophila neurons, Ctail but not Ctail-3MtLS* tracks MT plus ends. Scale bar (in B) B, C, 5 μm.
There are three putative MtLS motifs in Ctail, two of which match the SxIP consensus sequence, whereas the third one is a related SnLP motif (Figs. 1R, 6A). To assess which of these motifs are required for MT plus end localization, we first generated constructs in which only the two SxIP motifs were substituted by alanine residues (Ctail-SrIP-SsIP*; Fig. 6K). This construct completely failed to accumulate at MT plus ends (Fig. 6G,M), suggesting that the SnLP motif is dispensable for MT plus end localization of Ctail. If SrIP or SsIP were mutated singly (Ctail-SrIP* and Ctail-SsIP*; Fig. 6K), only minor reductions in MT plus end association were observed (Fig. 6E,F,M). Therefore, each of the two SxIP motifs alone is sufficient to mediate plus end localization, and these two motifs do not display strong cooperative effects (as was previously described for the two MtLS motifs of CLASP; Honnappa et al., 2009).
However, our data also suggest that MtLS motifs are not sufficient for MT plus end localization. For example, MT plus end localization was lost by the Ctail-R* construct (which contains an intact SsIP motif; Fig. 6C′, open arrow) as well as by a second construct in which all arginines were substituted except in islands of 14 residues centered around each of the three MtLS motifs (to preserve their functional integrity; Honnappa et al., 2009; data not shown). These data suggest that the overall positive charge of Ctail is important for both MT shaft and plus end localization of Ctail.
Finally, we assessed whether MtLS motifs also had an influence on the localization of GRD-Ctail constructs. Like Shot-FL in neurons, GRD-Ctail in fibroblasts displays strong association along MT shafts, providing an opportunity to study how MT shaft and plus end localization relate to each other. For this, fibroblasts were transfected with GRD-Ctail or GRD-Ctail-3MtLS* together with the EB1 homolog EB3::RFP (as a live tracker of MT plus ends). We found that MT association of GRD-Ctail along MTs reached to their polymerizing plus ends, overlapping with the tails of EB3 comets (Fig. 6H–H″). In contrast, GRD-Ctail-3MtLS* did not reach as far to the MT plus ends and failed to overlap with EB3 comets (Fig. 6J–J″). Accordingly, GRD-Ctail but not GRD-Ctail-3MtLS* was able to coimmunoprecipitate EB1 in extracts of Cos-7 cells (Fig. 7A), although in lower quantities than Ctail alone (potentially because of the fact that Ctail fully overlaps with EB1 comets, whereas GRD-Ctail only overlaps with the comet tail).
We conclude that EB1 interaction via the two MtLS motifs mediates MT plus end localization, even under conditions where Shot is strongly associated along MT shafts.
Shot regulates MT polymerization dynamics in Drosophila neurons
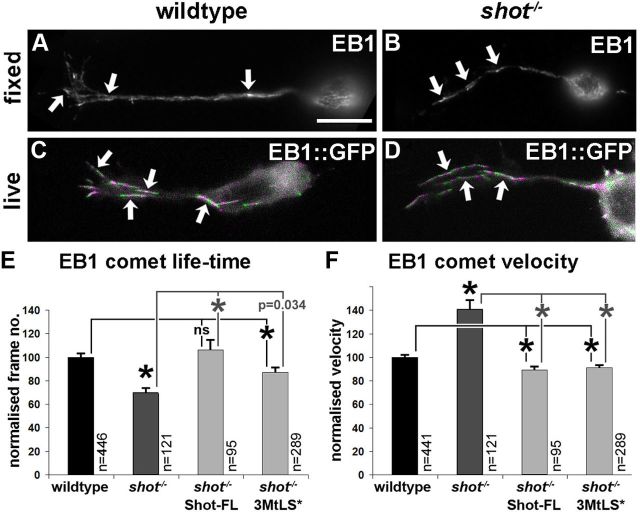
We next tested whether Shot at MT plus ends influences the dynamics of MT polymerization in neurons. To this end, we labeled the plus ends of polymerizing MTs with EB1::GFP (Fig. 8C,D) and measured their dynamics in wild-type and shot−/− mutant primary neurons (speed and life-time of EB1 comets). We found that EB1::GFP comets traveled 40% faster in shot−/− than in wild-type neurons, whereas the life-time of comets was reduced by 25.5% (Fig. 8E,F).
Figure 8.
Shot regulates MT polymerization in primary neurons. A–D, Endogenous EB1 (A, B) and transgenic EB1::GFP (C, D) can be seen as comets (arrows) in wild-type (left) and shot−/− mutant neurons (right) alike; to generate the images C and D which illustrate examples of growing MTs, frames from time laps of EB1::GFP-expressing neurons were color-coded alternating in green and magenta and then merged together. E, F, Measurements of the life-time and velocity of EB1::GFP comets in wild-type (black) and shot−/− mutant neurons (dark gray), and in mutant neurons with targeted expression of Shot constructs (light gray), all normalized to wild-type. The 100% life-time equates to 4.31 frames ± 0.13 SEM (= 17.3 ± 0.52 s) and 100% velocity to 0.156 μm/s ± 0.003 SEM. Statistics were performed using Kruskal–Wallis one-way ANOVA on Ranks (H = 31.987, 3 degrees of freedom, p < 0.001 in F; H = 70.828, 3 degrees of freedom, p < 0.001 in G) and Mann–Whitney rank sum test (*indicates p ≤ 0.004; ns, not significant p > 0.05). Scale bar (in A) A, B, 8 μm; C, D, 4 μm.
To validate these findings we performed rescue experiments. Notably, both Shot-FL and Shot-3MtLS* rescued the shot−/− mutant velocity phenotype from 141% (relative to wild-type) to ∼90% (Fig. 8F). These rescues confirmed that Shot inhibits the movement of EB1::GFP comets, and they showed that Shot does not require interaction with EB1 to this end. Therefore, Shot in its function as a MAP seems to negatively impact on the speed of MT polymerization, as is in agreement with reports for other classical MAPs which influence MT polymerization through mechanisms not yet understood (Tymanskyj et al., 2012). We cannot rule out that changes in MT sliding or translocation might contribute to this phenotype, but it seems unlikely that such sliding would primarily occur in the direction of MT polymerization (as would be required to enhance net speed of comets).
In contrast, only Shot-FL rescued the life-time phenotype, whereas Shot-3MtLS* achieved only a partial rescue (Fig. 8E). Therefore, Shot regulates the life-time of MT polymerization in an MtLS-dependent manner. A potential explanation could be that MtLS-dependent functions of Shot in guiding MT plus ends also protect them from collapse-inducing factors at the neuronal cortex (Letourneau, 2009).
EB1 is essential for axonal MT organization and axonal growth and interacts with Shot
The requirement of MtLS motifs for axonal functions of Shot and for its physical interaction with EB1 led us to predict that EB1 is also essentially required for axonal growth and MT organization. Previous work had established that Shot localization to MT plus ends depends on EB1 (Slep et al., 2005). As expected, our analyses with fluorescently tagged EB1 and endogenous EB1 in shot−/− mutant neurons (Fig. 8B,D) demonstrated that MT plus end localization of EB1 does not depend on Shot. Therefore, Shot requires EB1 to localize at MT plus ends but not vice versa.
Next, we tested potential axonal phenotypes caused by loss of EB1 function. To this end, we used fly strains carrying the strongest reported loss-of-function mutant allele eb104524, which had been shown to cause depletion of EB1 at the larval stage (Elliott et al., 2005). Accordingly, we found that primary neurons extracted from eb1−/− mutant embryos still showed high levels of EB1 staining after 6 h in culture (data not shown), but these levels were severely reduced when cells were precultured for several days to deplete maternally contributed proteins (Fig. 9B; Materials and Methods). Using our established readouts, we assessed the phenotypes of eb1−/− mutant primary neurons. Compared with parallel wild-type controls, we found that eb1−/− mutant neurons displayed a significant shortening of axons to 62% and frequent patches of non-coalescent, criss-crossed MTs. These EB1-deficient phenotypes were reminiscent of the shot−/− mutant axonal defects (Fig. 9B,E,I,J). EB1 depletion, MT disorganization and reduced axon length were also observed on knocking down EB1 through targeted expression of an eb1iRNA construct in primary neurons during a 6 d preculture period (Fig. 9C,F,I,J). Furthermore, rescue of mutant phenotypes was achieved with targeted expression of EB1::GFP in eb1−/− mutant neurons (Fig. 9I,J). Therefore, our results clearly demonstrated that, like Shot, EB1 promotes axonal growth and is required for the organization of neuronal MTs.
Figure 9.
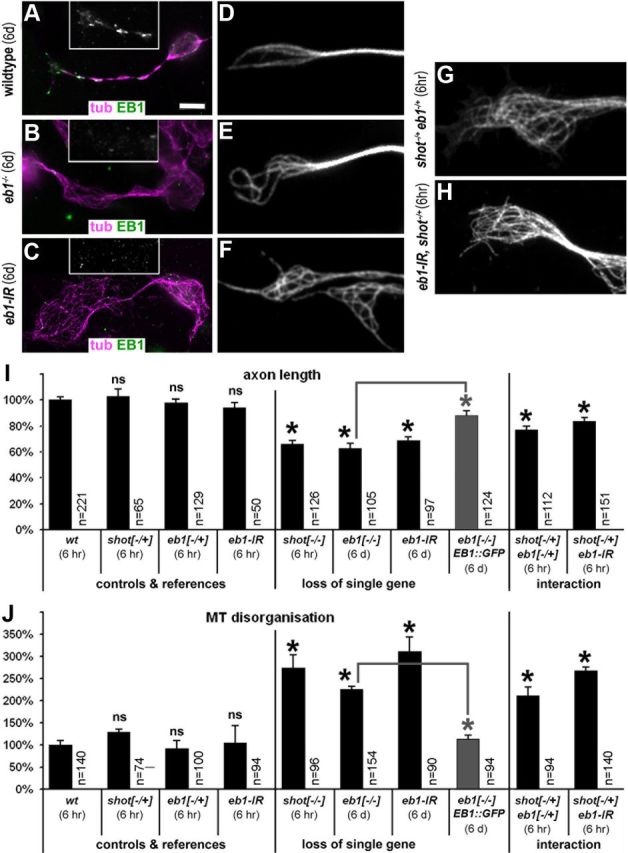
EB1 is required for axonal growth and functionally interacts with Shot. A–C, EB1 protein levels in neurons after 6 d in culture are still high in wild-type neurons (A), whereas they are strongly reduced in eb104524 mutant (B) or eb1iRNA-expressing primary neurons (C). D–F, A high degree of curling, criss-crossed MTs is seen only in EB1-depleted neurons after 6 d in culture (compare E, F to D). G, H, However, if eb104524/04524 mutant or eb1iRNA-expressing neurons in addition carried one mutant copy of shot (shot−/+), axon shortening and MT disorganization were already identified after 6 h in culture. I, Quantification of axonal length. J, Quantification of neurons with disorganized microtubules. Statistics were assessed by Kruskal–Wallis one-way ANOVA on Ranks (H = 194.602, 9 degrees of freedom, p ≤ 0.001 in I; H = 56.061, 9 degrees of freedom, p ≤ 0.001 in H) and Mann–Whitney rank sum test (highly significant *p ≤ 0.001; ns, not significant; p > 0.3). Gray bars represent rescues of eb104524 mutant phenotypes with EB1::GFP. Scale bar (in A) A–C, 5 μm; D–H, 2.5 μm.
The eb1 mutant phenotypes provided us with means to assess potential functional links between Shot and EB1 through genetic interaction studies. Neurons cultured from embryos carrying one mutant and one wild-type copy of the two genes (shot−/+ eb1−/+), displayed disorganized MT networks and shorter axons compared with shot−/+ or eb1−/+ heterozygously mutant neurons (Fig. 9G,I,J). In support of these findings, genetic interaction was also observed when shot−/+ was combined with eb1iRNA. Thus, eb1iRNA-expressing neurons still contained relatively high levels of protein after 6 h in culture (data not shown), and failed to display obvious phenotypes (Fig. 9I,J). However, when eb1iRNA expression was combined with one mutant copy of shot (eb1iRNA shot−/+), neurons already displayed strong MT disorganization and axon length phenotypes after 6 h in culture (Fig. 9H–J). Hence, modestly reducing the amount of either EB1 or Shot is without effect, but modestly reducing levels of both proteins together in the same cells becomes limiting for their function, indicating a functional link between them. In addition, we found that axonal phenotypes in shot−/− eb1−/− double mutant neurons were not enhanced over phenotypes observed in neurons mutant for shot−/− alone, consistent with a view that EB1 and Shot work in the same pathway (data not shown).
Together, the requirement of EB1 for axon growth and MT organization and the genetic interaction between shot and eb1 in this context support a model in which EB1 recruits Shot to MT plus ends as a prerequisite for its role in MT organization and axon growth promotion.
Discussion
Spectraplakins are key regulators of neuronal MTs
The regulation of MT networks is essential for many neuronal functions and processes, ranging from axonal growth to neurodegeneration. However, understanding how MTs are regulated in neurons remains a major challenge. Many MT-binding proteins have been identified and their potential roles in regulating the key processes of MT stabilization, MT polymerization, MT-based transport and MT-actin cross talk have been highlighted (Chilton and Gordon-Weeks, 2007; Conde and Cáceres, 2009). But how these different functions merge into coordinated MT network regulation is poorly understood.
Our work indicates spectraplakins as key integrators of different MT regulatory processes (Fig. 10A). First, Shot stabilizes MTs, a role generally assigned to structural MAPs, such as Tau, MAP2 or MAP1b (Fig. 10C, MAP function; Chilton and Gordon-Weeks, 2007; Riederer, 2007; Morris et al., 2011). Second, Shot regulates MT polymerization dynamics and guides them in the direction of axonal growth. In this function, Shot interacts with EB1, firmly establishing Shot-EB1 interaction as an important determinant of microtubule guidance as a crucial mechanism underpinning axon growth (Fig. 10C, +TIP function). In addition, we previously demonstrated that Shot acts as an actin-MT linker during axonal growth (Lee and Kolodziej, 2002; Sánchez-Soriano et al., 2009, 2010), adding a third crucial MT regulatory role to the list. Therefore, spectraplakins work at the cross-roads of structural MAPs, +TIPs and actin-binding proteins, strongly suggesting that work on spectraplakins will provide exciting new opportunities to unravel the complexity of MT regulation in neurons.
Figure 10.
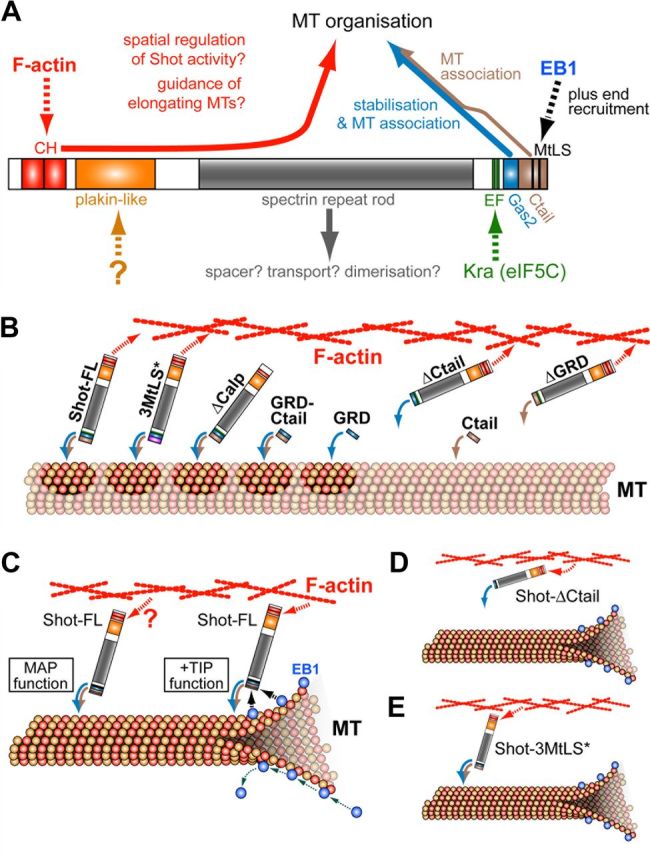
Model of Shot function in neuronal MT network organization. A, Molecular interactions (dashed arrows) and functions (solid arrows) of different Shot domains, as described here and previously (color code of arrows consistently used throughout the figure); apart from the C terminus, especially the calponin homology domains (CH) are crucial for Shot function in axonal growth and MT organization, and the EF hand domains are required for F-actin regulation in the context of pathfinding (Lee and Kolodziej, 2002; Bottenberg et al., 2009; Sánchez-Soriano et al., 2009, 2010). B, Although GRD (blue arrow) and Ctail (beige arrow) display MT association as isolated domains, their combined presence is required to maintain full-length Shot on MTs, since interactions of other domains might recruit Shot away (e.g., F-actin affinity; red arrow). GRD stabilizes MTs (darker patches), but can do so only when Shot is associated with MTs. C, Shot executes two functions in neurons. First, Shot stabilizes MTs via its C terminus (MAP function) requiring strong MT association (blue-brown curved arrows) but no obvious dependence on actin linkage (red arrow). Second, EB1 predominantly tracks polymerizing MT plus ends (green dotted arrows) and recruits some Shot activity to this location (black dashed arrows); this +TIP function of Shot requires F-actin linkage and is likely to guide MT polymerization events along F-actin structures, for example along the axonal cortex. D, The loss of MT association in Shot-ΔCtail, disturbs both MAP and +TIP functions and no rescue of any neuronal shot−/− mutant phenotype can be achieved (shown in Figures 1, L, M, O, Q, and 2M). E, Shot-3MtLS* can no longer interact with EB1, but can still associate with MTs; therefore, only +TIP function is abolished (leading to disorganized MTs and axonal growth defects shown in Fig. 1L, M, P, Q), whereas MAP functions are maintained reflected in the ability to stabilize MTs (Fig. 2M).
At the molecular level, our work revealed the importance of the Ctail for Spectraplakin function. While GRD had previously been established as the domain of Shot which has the widest functional requirements (Bottenberg et al., 2009), our current work suggested Ctail to be similarly important, as deduced from its crucial roles both in MT stabilization and guidance and its requirements in both neurons and tendon cells.
Shot executes MAP-like functions in growing neurons
We show that Shot localization along the shafts of axonal MTs is important for MT-stabilization, suggesting that spectraplakins might functionally overlap with structural MAPs (Riederer, 2007; Morris et al., 2011). This finding might have important implications. For example, loss of structural MAP functions has relatively mild phenotypes even in double knock-out mice (Takei et al., 2000; Teng et al., 2001), and this could be caused by functional compensation through spectraplakins. It remains to be seen whether such potential functional overlap is general or context-specific. For example, different MAPs and spectraplakins might display individual traits, such as dependence on different MT modifications (Janke and Kneussel, 2010) or selective antagonism to different destabilizing factors (Qiang et al., 2006).
In support of our findings with Shot, MT-stabilizing roles of spectraplakins appear conserved in mammals. Thus, MT-stabilizing roles in neurons have similarly been reported for the Shot homolog BPAG1 (Yang et al., 1999). Furthermore, axon shortening caused by knock-down of ACF7 in primary cortical neurons or N2A cells (Sánchez-Soriano et al., 2009) was rescued by taxol application (our unpublished data). In agreement with these findings, also the domain requirements underlying MT stabilization are conserved (summarized in Fig. 10B). Thus, studies in fibroblasts have shown that related C termini of the mammalian spectraplakin ACF7/MACF1 and of human Gas2-like1/hGAR22 and Gas2-like2/hGAR17 all display MT-stabilizing properties that are dependent on their GRDs, and in each case the Ctails enhance their MT association (Sun et al., 2001; Goriounov et al., 2003). Therefore, this mechanistic principle appears conserved across a range of homologous proteins, and our work has provided first experimental proof that it is functionally relevant in vivo, such as in growing axons and tendon cells.
Despite their obvious functional conservation, Ctails are not conserved at the protein sequence level but display other commonalities instead. They are all unlikely to form an ordered secondary or tertiary structure, they all display a high content of arginines, serines and glycines, and most contain MtLS motifs (Sun et al., 2001; Stroud et al., 2011). Here we identified a role for the arginines of Ctail and propose that this positive net charge attracts Ctail to negatively charged MTs. Such a mechanisms is consistent with other models for MT association (Wolff, 1998) and can explain why Ctails are not conserved at the amino acid sequence level. In support of this model, the C terminus of mouse ACF7 has recently been demonstrated to detach from MTs, when negative surface charges of MTs were enzymatically removed (Wu et al., 2011).
Shot acts as a +TIP during axonal growth
The second subcellular role of Shot in MT guidance establishes EB1 and the EB1-Shot complex as important determinants of axonal growth. In the absence of Shot-EB1 complex function, MTs are disorganized and axons extend shorter. Similar phenotypes of curled, criss-crossed MTs correlating with axon shortening have been described in mammalian neurons lacking ACF7 function (Sánchez-Soriano et al., 2009) but also defective for Dynein/Lis1 (Ahmad et al., 2006; Grabham et al., 2007), GSK3 (as a regulator of APC downstream of Wnt3a/Dvl1 and of CLASP; Purro et al., 2008; Hur et al., 2011), or Spinophilin/Doublecortin (Bielas et al., 2007). Our favored explanation for why MT disorganization attenuates axon growth is that MTs are less efficient in pushing along the axonal axis in the direction of axon growth. Notably, we have shown previously that roles of Shot in MT guidance and axon growth also essentially require its linkage to actin (Sánchez-Soriano et al., 2009, 2010). We therefore propose that Shot performs its MT guiding roles by linking MT plus ends to actin structures, such as the actin cortex in the axons or actin networks or bundles in growth cones. Such a function of spectraplakins is likely to coexist with parallel mechanisms of actin-MT linkage. For example, the F-actin-binding factor drebrin was shown to interact with EB3, and this interaction is likewise believed to be required to steer MT polymerization events in elongating axons (Geraldo et al., 2008).
Current models propose that +TIPs compete among each other for interaction with EB1 at MT plus ends (Akhmanova and Steinmetz, 2008; Gouveia and Akhmanova, 2010). Other +TIPs, such as CLASP, APC, Dynein/Lis and CLIPs, are present in neurons and contribute to axonal growth regulation (Chilton and Gordon-Weeks, 2007). Therefore, the +TIP functions of spectraplakins identified here can now be analyzed in the context of other +TIPs, providing new opportunities to gain an understanding of regulatory +TIP networks in the context of axonal growth.
Applying models of Shot function in axonal growth to other cellular contexts
We have shown that Shot works at the cross-roads of different mechanisms of MT regulation, and we were able to unravel the underpinning mechanisms using a genetically and experimentally amenable and functionally well conserved Drosophila model of axon growth (Sánchez-Soriano et al., 2010; Gonçalves-Pimentel et al., 2011). These findings do not only advance our principal understanding of cytoskeletal regulation during axonal growth, but can be extrapolated to other functions of spectraplakins. Thus, spectraplakins play roles in clinically relevant cellular processes including neurodegeneration, skin blistering and cell migration in wound healing and brain development (Sonnenberg and Liem, 2007; Goryunov et al., 2010). The two modes of Shot function we have proposed here may very well be applicable. For example, our model for MT guidance displays interesting commonalities with models for ACF7 function in migrating keratinocytes during wound healing, where ACF7 is suggested to guide MTs along actin stress fibers to focal adhesions (Wu et al., 2008). Furthermore, MT-stabilizing roles are likely to explain Shot function in Drosophila tendon cells. Just like MT-stabilizing roles of Shot in axons and fibroblasts, GRD and Ctail are essential in tendon cells, where they enhance each others localization (Fig. 3G vs J), whereas MtLS motifs (Fig. 3K,M) and actin-binding calponin-homology domains of Shot (Bottenberg et al., 2009) are dispensable. Notably, tendon cells have been proposed as a cellular model for support cells of the vertebrate inner ear that are known to express the Shot homolog BPAG1 (Alves-Silva et al., 2008). Therefore, the subcellular mechanisms of spectraplakins described here do not only have implications for the understanding of axonal growth but also for their other functions in neurons and non-neuronal cells.
Notes
Supplemental material for this article is available at https://www.escholar.manchester.ac.uk/uk-ac-man-scw:156942. The supplemental material shows images of nocodazole-treated fibroblasts transfected with GFP-tagged C-terminal constructs of Shot to assess the ability of these constructs to protect MTs against depolymerization. A graph is shown plotting quantification and statistical significance of nocodazole resistance conveyed by these Shot constructs. This material has not been peer reviewed.
Footnotes
This work was funded through grants by the Wellcome Trust to A.P., C.B., and N.S.-S. (077748/Z/05/Z) and to A.P. (078593/Z/05/Z, 087820/Z/08/Z, 092403/Z/10/Z), a grant of the Biotechnology and Biological Sciences Research Council (BBSRC) to A.P. (BB/C515998/1, BB/I002448/1), a BBSRC studentship to R.B. (BB/D526561/1), and a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science to R.A.K. (074343/Z/04/B). The Bioimaging Facility microscopes used in this study were purchased with grants from BBSRC, The Wellcome Trust, and the University of Manchester Strategic Fund, and the Fly Facility is supported by funds from The University of Manchester and the Wellcome Trust (087742/Z/08/Z). We are grateful to colleagues and stock centers for providing fly stocks and antibodies, as detailed in Materials and Methods. We thank the Drosophila Genomics Resource Center, Richard Behringer, Michelle Calos, Chuan-Wei Jang, and Waclaw Szybalski for plasmids, and Seungbok Lee for the GRD** construct, and John Humphries, Alex Carisey, Simon Woodcock, Chris Thompson, and Douda Bensasson for help and advice.
References
- Ahmad FJ, He Y, Myers KA, Hasaka TP, Francis F, Black MM, Baas PW. Effects of dynactin disruption and dynein depletion on axonal microtubules. Traffic. 2006;7:524–537. doi: 10.1111/j.1600-0854.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Alves-Silva J, Hahn I, Huber O, Mende M, Reissaus A, Prokop A. Prominent actin fibre arrays in Drosophila tendon cells represent architectural elements different from stress fibres. Mol Biol Cell. 2008;19:4287–4297. doi: 10.1091/mbc.E08-02-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Grode KD, Keller D, Zadeh AD, Slep KC, Rogers SL. The Spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol Biol Cell. 2010;21:1714–1724. doi: 10.1091/mbc.E10-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, Ellisman MH, Gleeson JG. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell. 2007;129:579–591. doi: 10.1016/j.cell.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottenberg W, Sánchez-Soriano N, Alves-Silva J, Hahn I, Mende M, Prokop A. Context-specific requirements of functional domains of the Spectraplakin Short stop in vivo. Mech Dev. 2009;126:489–502. doi: 10.1016/j.mod.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. Berlin: Springer; 1997. The embryonic development of Drosophila melanogaster. [Google Scholar]
- Chilton J, Gordon-Weeks P. Role of microtubules and MAPs during neuritogenesis. In: de Curtis I, editor. Intracellular mechanisms for neuritogenesis. New York: Springer; 2007. pp. 57–88. [Google Scholar]
- Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol. 2011;3:a001800. doi: 10.1101/cshperspect.a001800. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott SL, Cullen CF, Wrobel N, Kernan MJ, Ohkura H. EB1 is essential during Drosophila development and plays a crucial role in the integrity of chordotonal mechanosensory organs. Mol Biol Cell. 2005;16:891–901. doi: 10.1091/mbc.E04-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol. 2008;10:1181–1189. doi: 10.1038/ncb1778. [DOI] [PubMed] [Google Scholar]
- Gonçalves-Pimentel C, Gombos R, Mihály J, Sánchez-Soriano N, Prokop A. Dissecting regulatory networks of filopodia formation in a Drosophila growth cone model. PLoS One. 2011;6:e18340. doi: 10.1371/journal.pone.0018340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriounov D, Leung CL, Liem RK. Protein products of human Gas2-related genes on chromosomes 17 and 22 (hGAR17 and hGAR22) associate with both microfilaments and microtubules. J Cell Sci. 2003;116:1045–1058. doi: 10.1242/jcs.00272. [DOI] [PubMed] [Google Scholar]
- Goryunov D, He CZ, Lin CS, Leung CL, Liem RK. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol Cell Neurosci. 2010;44:1–14. doi: 10.1016/j.mcn.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia SM, Akhmanova A. Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int Rev Cell Mol Biol. 2010;285:1–74. doi: 10.1016/B978-0-12-381047-2.00001-3. [DOI] [PubMed] [Google Scholar]
- Grabham PW, Seale GE, Bennecib M, Goldberg DJ, Vallee RB. Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J Neurosci. 2007;27:5823–5834. doi: 10.1523/JNEUROSCI.1135-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, Jelesarov I, Winkler FK, Wüthrich K, Akhmanova A, Steinmetz MO. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Hur EM, Saijilafu, Lee BD, Kim SJ, Xu WL, Zhou FQ. GSK3 controls axon growth via CLASP-mediated regulation of growth cone microtubules. Genes Dev. 2011;25:1968–1981. doi: 10.1101/gad.17015911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Kneussel M. Tubulin post-translational modifications: encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 2010;33:362–372. doi: 10.1016/j.tins.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7. An essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Küppers-Munther B, Letzkus JJ, Lüer K, Technau G, Schmidt H, Prokop A. A new culturing strategy optimises Drosophila primary cell cultures for structural and functional analyses. Dev Biol. 2004;269:459–478. doi: 10.1016/j.ydbio.2004.01.038. [DOI] [PubMed] [Google Scholar]
- Lee S, Kolodziej PA. Short stop provides an essential link between F-actin and microtubules during axon extension. Development. 2002;129:1195–1204. doi: 10.1242/dev.129.5.1195. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible neurotechnique cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Letourneau PC. Actin in axons: stable scaffolds and dynamic filaments. Results Probl Cell Differ. 2009;48:65–90. doi: 10.1007/400_2009_15. [DOI] [PubMed] [Google Scholar]
- Letourneau PC, Shattuck TA, Ressler AH. “Pull” and “push” in neurite elongation: observations on the effects of different concentrations of cytochalasin B and taxol. Cell Motil Cytoskeleton. 1987;8:193–209. doi: 10.1002/cm.970080302. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, Uhler J, Roote J, Bate M. The kakapo mutation affects terminal arborisation and central dendritic sprouting of Drosophila motorneurons. J Cell Biol. 1998;143:1283–1294. doi: 10.1083/jcb.143.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purro SA, Ciani L, Hoyos-Flight M, Stamatakou E, Siomou E, Salinas PC. Wnt regulates axon behavior through changes in microtubule growth directionality: a new role for adenomatous polyposis coli. J Neurosci. 2008;28:8644–8654. doi: 10.1523/JNEUROSCI.2320-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Yu W, Andreadis A, Luo M, Baas PW. Tau protects microtubules in the axon from severing by katanin. J Neurosci. 2006;26:3120–3129. doi: 10.1523/JNEUROSCI.5392-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer BM. Microtubule-associated protein 1B, a growth-associated and phosphorylated scaffold protein. Brain Res Bull. 2007;71:541–558. doi: 10.1016/j.brainresbull.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K, Gregory SL, Brown NH. The ‘Spectraplakins’: cytoskeletal giants with characteristics of both spectrin and plakin families. J Cell Sci. 2002;115:4215–4225. doi: 10.1242/jcs.00157. [DOI] [PubMed] [Google Scholar]
- Sánchez-Soriano N, Prokop A. The influence of pioneer neurons on a growing motor nerve in Drosophila requires the neural cell adhesion molecule homolog FasciclinII. J Neurosci. 2005;25:78–87. doi: 10.1523/JNEUROSCI.2377-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Soriano N, Travis M, Dajas-Bailador F, Gonçalves-Pimentel C, Whitmarsh AJ, Prokop A. Mouse ACF7 and Drosophila Short stop modulate filopodia formation and microtubule organisation during neuronal growth. J Cell Sci. 2009;122:2534–2542. doi: 10.1242/jcs.046268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Soriano N, Gonçalves-Pimentel C, Beaven R, Haessler U, Ofner-Ziegenfuss L, Ballestrem C, Prokop A. Drosophila growth cones: a genetically tractable platform for the analysis of axonal growth dynamics. Dev Neurobiol. 2010;70:58–71. doi: 10.1002/dneu.20762. [DOI] [PubMed] [Google Scholar]
- Schneider I. Differentiation of larval Drosophila eye-antennal discs in vitro. J Exp Zool. 1964;156:91–103. doi: 10.1002/jez.1401560107. [DOI] [PubMed] [Google Scholar]
- Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Stroud MJ, Kammerer RA, Ballestrem C. Characterization of G2L3 (GAS2-like 3), a new microtubule- and actin-binding protein related to spectraplakins. J Biol Chem. 2011;286:24987–24995. doi: 10.1074/jbc.M111.242263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Volk T. Kakapo, a novel Drosophila protein, is essential for the restricted localization of the neuregulin-like factor, Vein, at the muscle-tendon junctional site. J Cell Biol. 1998;143:1259–1270. doi: 10.1083/jcb.143.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Prokop A, Yamamoto M, Sugimura K, Uemura T, Betschinger J, Knoblich JA, Volk T. Short stop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr Biol. 2003;13:1086–1095. doi: 10.1016/s0960-9822(03)00416-0. [DOI] [PubMed] [Google Scholar]
- Sun D, Leung CL, Liem RK. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): identification of a novel group of microtubule associated proteins. J Cell Sci. 2001;114:161–172. doi: 10.1242/jcs.114.1.161. [DOI] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng J, Takei Y, Harada A, Nakata T, Chen J, Hirokawa N. Synergistic effects of MAP2 and MAP1B knockout in neuronal migration, dendritic outgrowth, and microtubule organization. J Cell Biol. 2001;155:65–76. doi: 10.1083/jcb.200106025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymanskyj SR, Scales TM, Gordon-Weeks PR. MAP1B enhances microtubule assembly rates and axon extension rates in developing neurons. Mol Cell Neurosci. 2012;49:110–119. doi: 10.1016/j.mcn.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Viñals J, Kolinski A, Skolnick J. Numerical study of the entropy loss of dimerization and the folding thermodynamics of the GCN4 leucine zipper. Biophys J. 2002;83:2801–2811. doi: 10.1016/S0006-3495(02)75289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J. Promotion of microtubule assembly by oligocations: cooperativity between charged groups. Biochemistry. 1998;37:10722–10729. doi: 10.1021/bi980400n. [DOI] [PubMed] [Google Scholar]
- Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. 2008;135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shen QT, Oristian DS, Lu CP, Zheng Q, Wang HW, Fuchs E. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3beta. Cell. 2011;144:341–352. doi: 10.1016/j.cell.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bauer C, Strasser G, Wollman R, Julien JP, Fuchs E. Integrators of the cytoskeleton that stabilize microtubules. Cell. 1999;98:229–238. doi: 10.1016/s0092-8674(00)81017-x. [DOI] [PubMed] [Google Scholar]