Abstract
Hypoxia mimic nickel(II) is a human respiratory carcinogen with a suspected epigenetic mode of action. We examined whether Ni(II) elicits a toxicologically significant activation of the tumor suppressor p53, which is typically associated with genotoxic responses. We found that treatments of H460 human lung epithelial cells with NiCl2 caused activating phosphorylation at p53-Ser15, accumulation of p53 protein and depletion of its inhibitor MDM4 (HDMX). Confirming activation of p53, its knockdown suppressed the ability of Ni(II) to upregulate MDM2 and p21 (CDKN1A). Unlike DNA damage, induction of GADD45A by Ni(II) was p53-independent. Ni(II) also increased p53-Ser15 phosphorylation and p21 expression in normal human lung fibroblasts. Although Ni(II)-induced stabilization of HIF-1α occurred earlier, it had no effect on p53 accumulation and Ser15 phosphorylation. Ni(II)-treated H460 cells showed no evidence of necrosis and their apoptosis and clonogenic death were suppressed by p53 knockdown. The apoptotic role of p53 involved a transcription-dependent program triggering the initiator caspase 9 and its downstream executioner caspase 3. Two most prominently upregulated proapoptotic genes by Ni(II) were PUMA and NOXA but only PUMA induction required p53. Knockdown of p53 also led to derepression of antiapoptotic MCL1 in Ni(II)-treated cells. Overall, our results indicate that p53 plays a major role in apoptotic death of human lung cells by Ni(II). Chronic exposure to Ni(II) may promote selection of resistant cells with inactivated p53, providing an explanation for the origin of p53 mutations by this epigenetic carcinogen.
Keywords: nickel, p53, apoptosis, HIF-1, hypoxia, cancer
Introduction
Nickel is an important industrial metal that has long been used in plating and manufacturing of several large-volume alloys and Ni-Cd batteries. A more recent application of Ni is its use in the production of carbon nanotubes, which release Ni(II) ions inside the cells causing toxic effects (Liu et al., 2007). Chronic inhalation of Ni(II) compounds has been associated with the impairment of pulmonary functions and pathological changes in the lung. Occupational exposures to Ni(II)-containing compounds have also been linked to the increased incidence of respiratory cancers (Goodman et al., 2011; Salnikow and Zhitkovich, 2008). The carcinogenic potential of Ni(II) is further supported by its ability to cause tumors in experimental animals via inhalation and other routes of administration. Ni(II) also displayed strong transformation activities in various cellular models (Bierdermann and Landolph, 1987; Maehle et al., 1992; Miura et al., 1989). However, unlike most other human carcinogens, Ni(II) showed almost no mutagenic activity in the standard test systems (Costa et al., 2005; Salnikow and Zhitkovich, 2008) although it has been found to promote the formation of pre-mutagenic 8-oxo-dG (Huang et al., 1995; Qu et al., 2001). The apparent lack of mutagenicity along with evidence of chromatin remodeling and gene silencing point to the importance of nongenotoxic mechanisms in Ni(II) carcinogenicity (Lee et al., 1995; Costa et al., 2005; Salnikow and Zhitkovich, 2008).
Treatments of cells with Ni(II) led to the activation of several stress-sensitive signaling processes and transcription factors, such as ATF-1, NF-κB, NFAT and others (Ding et al., 2006; Gao et al., 2010; Huang et al., 2006; Salnikow et al., 1997). Surprisingly, Ni(II) was also a very potent inducer of a gene expression program driven by the hypoxia-responsive transcriptional factor HIF-1 (Salnikow et al., 2000; Salnikow et al., 2003). Similar to hypoxia, upregulation of hypoxia-sensitive genes by Ni(II) resulted from stabilization of HIF-1α protein. Blockage of proteasomal destruction of HIF-1α in Ni(II)-treated cells was attributed to inhibition of HIF-1α-targeting prolyl hydroxylases through Fe(II) displacement (Davidson et al., 2006) and a depletion of their cofactor ascorbate (Salnikow et al., 2004). Accumulation of HIF-1α in hypoxic cells is responsible for stabilization of the p53 transcriptional factor (An et al., 1998; Chen et al., 2003). Human lung cancers frequently harbor mutations in the p53 gene (Pfeifer and Besaratinia, 2009) and Ni(II)-transformed human cells also contained mutated p53 (Maehle et al., 1992). Thus, p53 is expected to play a significant role in Ni(II) carcinogenicity. Published data on the ability of Ni(II) to upregulate p53 are contradictory (Ding et al., 2009; Huang et al., 2006; Salnikow et al., 1999), which could be related to the use of different cellular models and biochemical readouts of p53 activation. No studies have yet examined whether p53 is involved in the manifestation of toxicological responses to Ni(II), such as apoptosis or other forms of cytotoxicity.
In this work, we conducted a detailed examination of p53 activation by Ni(II) in human lung cells that have normal p53 responses. We found that Ni(II) ions induced a robust upregulation of the p53 pathway, which was a major contributor to apoptosis and clonogenic death. HIF-1α was completely dispensable for the p53 stabilization by Ni(II), pointing to a major difference in stress signaling for this hypoxia mimic versus hypoxia. Our findings suggest that p53 inactivation in carcinogenesis by nonmutagenic Ni(II) could result from a selective outgrowth of apoptosis-resistant cells containing mutated p53.
Materials and Methods
Chemicals
NiCl2•6H2O (BioReagent grade) was obtained from Sigma-Aldrich (St. Louis, Missouri, USA). Freshly prepared stock solutions of NiCl2 were always used. All other salts and buffers were also purchased from Sigma. Z-VAD-fmk was from BD Biosciences (San Jose, California, USA).
Cells and treatments
H460 cells were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA) and maintained in RPMI 1640 medium supplemented with 10% serum and pennicilin/streptomycin. IMR90 fibroblasts were obtained from ATCC and propagated in DMEM medium containing 15% serum. H460 cells were grown in the humidified atmosphere containing 95% air and 5% CO2 while IMR90 fibroblasts were kept at 5% O2 and 5% CO2. Cells were treated with NiCl2 in complete growth media.
Knockdowns with shRNA and siRNA
A stable knockdown of p53 expression in H460 cells was created by infection with an shRNA-expressing pSUPER-RETRO vector. Empty and scrambled shRNA-coding vectors were obtained from Oligoengine (Seattle, Washington, USA). The supplier's recommended conditions were followed for the insertion of the desired oligonucleotides. The targeting sequences was GACTCCAGTGGTAATCTAC. Retrovirus packaging, infection and purmycin selection conditions have been described previously (Reynolds et al., 2004). A loss of vector expression in the infected populations was prevented by a continous presence of 1.5 µg/mL puromycin in the growth media. HIF-1α was downregulated by a transfection of H460 cells with 50 nM ON-TARGETplus SMARTpool siRNA from Thermo Scientific (Waltham, Massachusetts, USA). H460 cells were transfected with 50 nM siRNA using Lipofectamine RNAimax from Invitrogen (Grand Island, New York, USA) one day after seeding. Ni(II) treatments were done 24 hr after the transfection.
Western blotting
Soluble protein extracts were prepared as described earlier (Reynolds and Zhitkovich, 2007). Proteins were separated by a standard SDS-PAGE and electrotransferred to ImmunoBlot PVDF membrane (Bio-Rad, Hercules, California, USA). Primary antibodies: PARP (9542), pS15-p53 (9284), hexokinase II (2106), cleaved caspase 3 (9661) and caspase 9 (9502) were from Cell Signaling Technology (Danvers, Massachusetts, USA), p21 (SX118) and HIF-1α (610958) were from BD Biosciences (San Jose, California, USA), MDM4 (A300-287A) from Bethyl Laboratories (Montgomery, Texas, USA), p53 (DO-1) from Santa Cruz (Santa Cruz, California, USA), tubulin (T6557) from Sigma (St. Louis, Missouri, USA).
Cell death
The FITC-Annexin V staining kit from BD Biosciences (San Jose, California, USA) was used for a simutaneous detection of apoptotic and necrotic cells. Attached and floating cells were combined and washed twice with PBS and a binding buffer. Cells were then stained with FITC-Annexin V and 7-aminoactinomycin D (7-AAD) for 15 min at room temperature followed by immediate FACS analysis (FACSCalibur, BD Biosciences). Leakage of cellular lactate dehydrogenase (LDH) was measured by a kit from Cayman Chemical (Ann Arbor, Michigan, USA).
RT-qPCR
RNA was extracted with the TRIzol reagent from Invitrogen (Grand Island, New York, USA), purified with the RNeasy mini kit from Qiagen (Valencia, California, USA) and treated with DNase I. RNA samples were assessed for purity by a Nanodrop spectrophotometer and stored at −80°C. cDNA was produced by the RT First Strand kit according to manufacturer’s instructions (SABiosciences-Qiagen, Valencia, California, USA). All PCR primers were obtained from SABiosciences and amplification reactions were carried out using the ABI 7900HT Real-Time PCR system (Applied Biosystems, Carlsbad, California, USA). Four housekeeping genes (B2M, HPRT1, RPL13A, ACTB) were used for normalization purposes. Differences in gene expression were calculated by the 2−DDCt method.
Clonogenic survival
Cells (400–800 per 60-mm dish) were seeded in triplicates per dose and allowed to attach overnight. The medium was replaced and cells were treated with Ni(II) for 24 hr. At 7–8 days post-exposure, colonies were fixed with methanol and stained with a commercial Giemsa solution (Sigma-Aldrich, St. Louis, Missouri, USA).
Results
p53 activation by Ni(II) ions
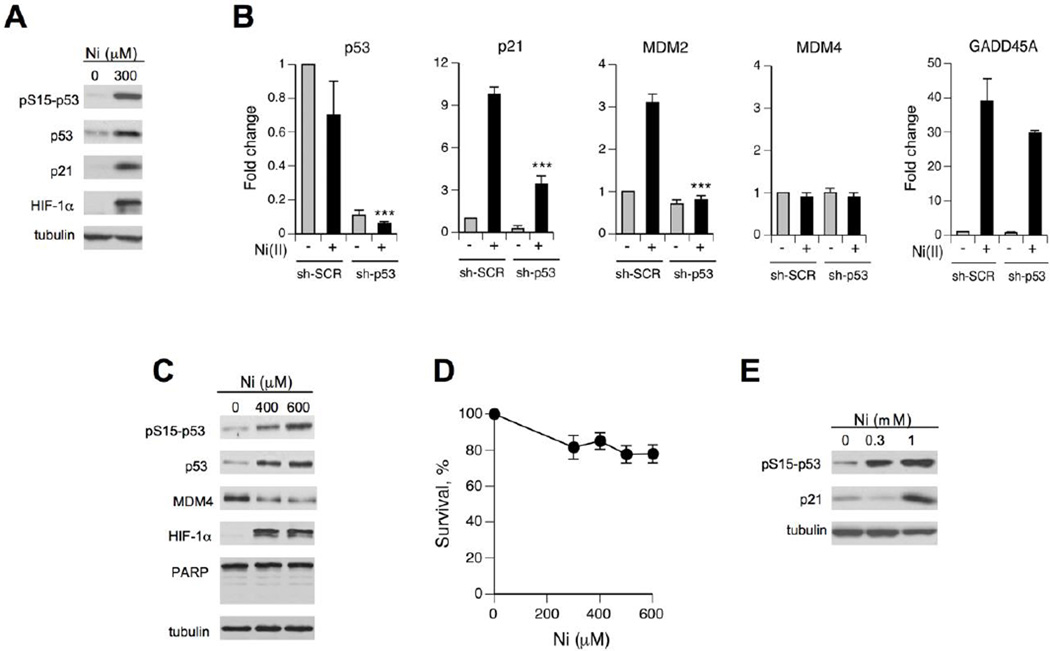
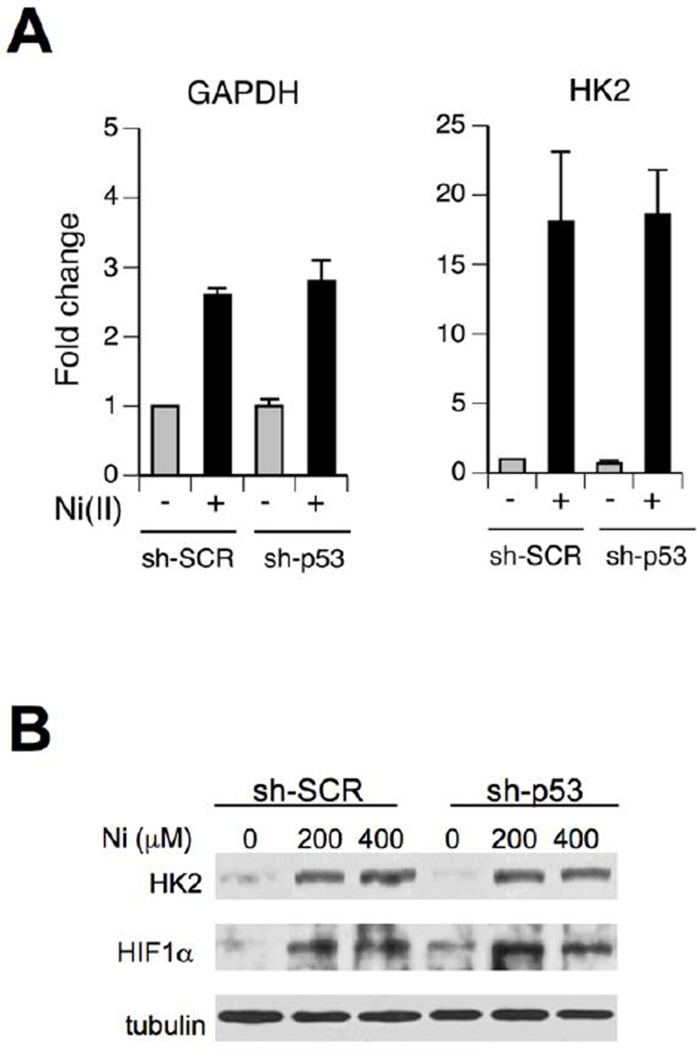
Many common cell lines, including nontumorigenic immortalized lines, are p53-deficient genetically or functionally, which can partially explain contradictory findings in the previous studies on the ability of Ni2+ ions to activate p53 (Ding et al., 2009; Huang et al., 2006; Salnikow et al., 1999). We have chosen H460 human lung epithelial cells as our primary cellular model because this cell line displayed normal p53 responses to different stressors (Reynolds and Zhitkovich, 2007; Wong et al., 2012; Zhang et al., 2006). H460 are also sensitive to Ni(II)-induced apoptosis (Patel et al., 2012; Pietruska et al., 2011). We first examined biochemical parameters related to p53 activation after treatments of H460 cells with Ni(II) for 24 hr, as this metal typically exhibits delayed stress responses. Ni(II)-exposed H460 cells showed a very strong accumulation of HIF-1α protein, verifying Ni(II) uptake and confirming the intactness of this important stress response in the selected cell model (Fig. 1A). Our assessement of p53 included measurements of its Ser15 phosphorylation, protein levels and induction of its target gene the CDK inhibitor p21 (CDKN1A). Ser15 phosphorylation is important for dissociation of the E3 ligase MDM2 and accumulation of p53 via stabilization (Kruse and Gu, 2009; Toledo and Wahl, 2006). We found that Ni(II) was a potent activator of the p53 pathway as evidenced by clear increases for all three measured parameters (Fig. 1A). Next, we examined mRNA levels for several p53-associated proteins and their dependence on the presence of p53. Consistent with the stabilization mechanism for the observed p53 protein accumulation, Ni(II)-treated cells showed no increase in the amount of the p53 mRNA (Fig. 1B, left panel). The same panel demonstrates a very efficient, >10-fold reduction in the p53 mRNA concentration in H460 cells stably expressing p53-targeting shRNA. Expression of two well-known target genes for transactivation by p53, p21 and MDM2, was increased by Ni(II) in a p53-dependent manner (Fig. 1B). GADD45A, another commonly induced gene by p53, showed a very strong upregulation by Ni(II) but this response was similar in cells with normal and shRNA-depleted p53 levels (Fig. 1B).
Figure 1. Activation of the p53 pathway by Ni(II) in human lung cells.
(A) Western blots of protein extracts from H460 cells treated with Ni(II) for 24 hr. (B) RT-qPCR analysis of mRNA levels in H460 cells expressing nonspecific (sh-SCR) and p53-targeting (sh-p53) shRNA. Cells were treated with 400 µM Ni(II) for 24 hr. Data are means±SD for three independent RNA samples (***- p<0.0001 relative to sh-SCR+Ni). (C) Western blots for H460 cells treated with Ni(II) for 6 hr. (D) Clonogenic survival of H460 cells treated with Ni(II) for 6 hr (means±SD, n=3). (E) Phosphorylation of p53 at Ser15 and p21 expression in IMR90 fibroblasts treated with Ni(II) for 24 hr.
Activation of p53 by prolonged treatments with Ni(II) can potentially result from a nonspecific cytotoxicity-induced stress, which would not be very significant biologically. To test a potential involvement of cell death processes, we treated H460 cells only for 6 hr and examined p53-related stress signaling. We found these short treatments with 0–600 µM Ni(II) were sufficient to strongly increase protein levels of HIF-1α (biomarker of Ni2+ uptake) but they produced no immediate cell death, as evidenced by a complete absence of the early apoptotic marker, PARP cleavage (Fig. 1C). A long-term viability of cells treated with 0–600 µM Ni(II) for 6 hr was also very high (Fig. 1D). Despite a lack of cytotoxicity, these short Ni(II) treatments clearly increased p53 protein levels and its Ser15 phosphorylation (Fig. 1C). Ni(II)-treated cells also showed a depletion of MDM4 (MDMX) protein, which acts as an inhibitor of transactivation activity of p53. MDM4 mRNA levels were unchanged by Ni(II) irrespective of p53 status (Fig. 1B, middle panel). Downregulation of MDM4 after genotoxic stress is caused by its increased proteolysis (Toledo and Wahl, 2006; Wong et al., 2012). Finally, we determined that activation of p53 was not specific to H460 cells, as normal human lung fibroblasts also showed strongly increased Ser15-p53 phosphorylation and p21 induction by Ni(II) (Fig. 1E).
Role of p53 in Ni(II)-induced cell death
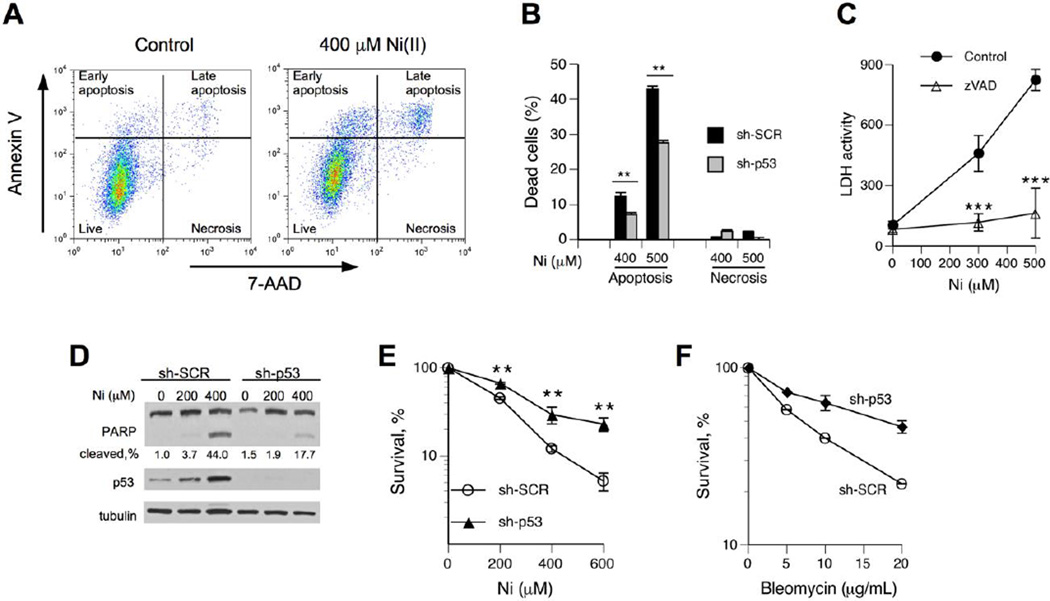
Biochemical and gene expression evidence of p53 activation raised a question about its potential involvement in the engagement of cell death programs in Ni(II)-treated cells. It is known that the biological effects of transcriptionally active p53 can be suppressed by other branches of stress signaling (Beckerman et al., 2009; Gottifredi et al., 2001). Our findings on p53-independence of GADD45A induction by Ni(II) also pointed to a more limited spectrum of gene upregulation compared to the canonical p53 response to DNA damage by ionizing radiation (Toledo and Wahl, 2006). The role of p53 in cell fate decisions can involve activation of either apoptosis (Toledo and Wahl, 2006) or a recently discovered form of necrosis triggered by opening of the mitochondrial permeability transition pore (Vaseva et al., 2012). To simultaneously assess both necrosis and apoptosis, we performed FACS analysis of intact H460 cells stained with Annexin V and 7-AAD. Fig. 2A shows typical FACS profiles of control and Ni(II)-treated cells co-stained for apoptotic (Annexin V) and necrotic (7-AAD) markers. Apoptosis was a principal form of cell death by Ni(II), as evidenced by only marginal amounts of Annexin V-negative/7-AAD-positive cells (Fig. 2A,B). Our assignment of double-positive cells (top-right quadrant) in Ni(II)-treated groups as late apoptotic cells was based on three considerations: (i) presence of early apoptotic cells but no early necrotic cells, (ii) a visible track of cells from live to the top-left quadrant (early apoptosis) to the top-right quadrant, and (iii) the top-right quadrant cells had a significantly lower forward scattering, which is indicative of apoptotic cell shrinkage. To further investigate a mode of cell death by Ni(II). we examined a leakage of cellular LDH, which occurs in all forms of necrosis but also in late apoptosis. We found a dose-dependent increase in extracellular LDH but this response was completely suppressed by the addition of the pancaspase inhibitor z-VAD-fmk (Fig. 2C), confirming the absence of necrotic cell death by Ni(II). Cells with stable knockdown of p53 showed significantly lower levels of apoptotis by Ni(II) (Fig. 2B). Ni(II) treatments also produced much lower amounts of caspase-mediated PARP cleavage in p53-depleted cells (Fig. 2D), further supporting the pro-apoptotic role of p53. The loss of p53 also resulted in a significantly higher long-term viability of Ni(II)-treated cells (Fig. 2E), indicating that a suppression of p53-dependent apoptosis did not result in a compensatory upregulation of alternative forms of cell death. The survival advantage of Ni(II)-treated cells with p53 deficiency was comparable to that of cells treated with the radiomimetic bleomycin (Fig. 2F).
Figure 2. Role of p53 in Ni(II)-induced apoptosis.
(A) Representative FACS profiles of control and Ni(II)-treated H460 cells stained with Annexin V and 7-AAD. Attached and floating cells were collected immediately after 24 hr exposure to Ni(II). (B) Ni(II)-induced apoptosis and necrosis in H460 cells with nonspecific (sh-SCR) and p53-targeting shRNA (sh-p53). Cells were stained with Annexin V and 7-AAD and analyzed by FACS. Background-subtracted values are shown (means±SD, n=3, **- p<0.01 relative to sh-SCR). (C) LDH leakage from H460 cells treated with Ni(II) for 24 hr in the presence or absence of 50 µM z-VAD-fmk (means±SD, n=4, ***- p<0.001 relative to samples without z-VAD-fmk). (D) PARP cleavage in H460 cells following 24 hr exposure to Ni(II). (E) Clonogenic survival of H460 cells expressing nonspecific and p53-targeting shRNA. Cells were treated with Ni(II) for 24 hr. Data are means±SD from three clonogenic experiments with 3 dishes per dose (**- p<0.01 relative to sh-SCR). (F) Clonogenic survival of control and p53-depleted H460 cells treated with bleomycin for 3 hr (means±SD for 3 dishes).
Mechanism of p53-induced apoptosis
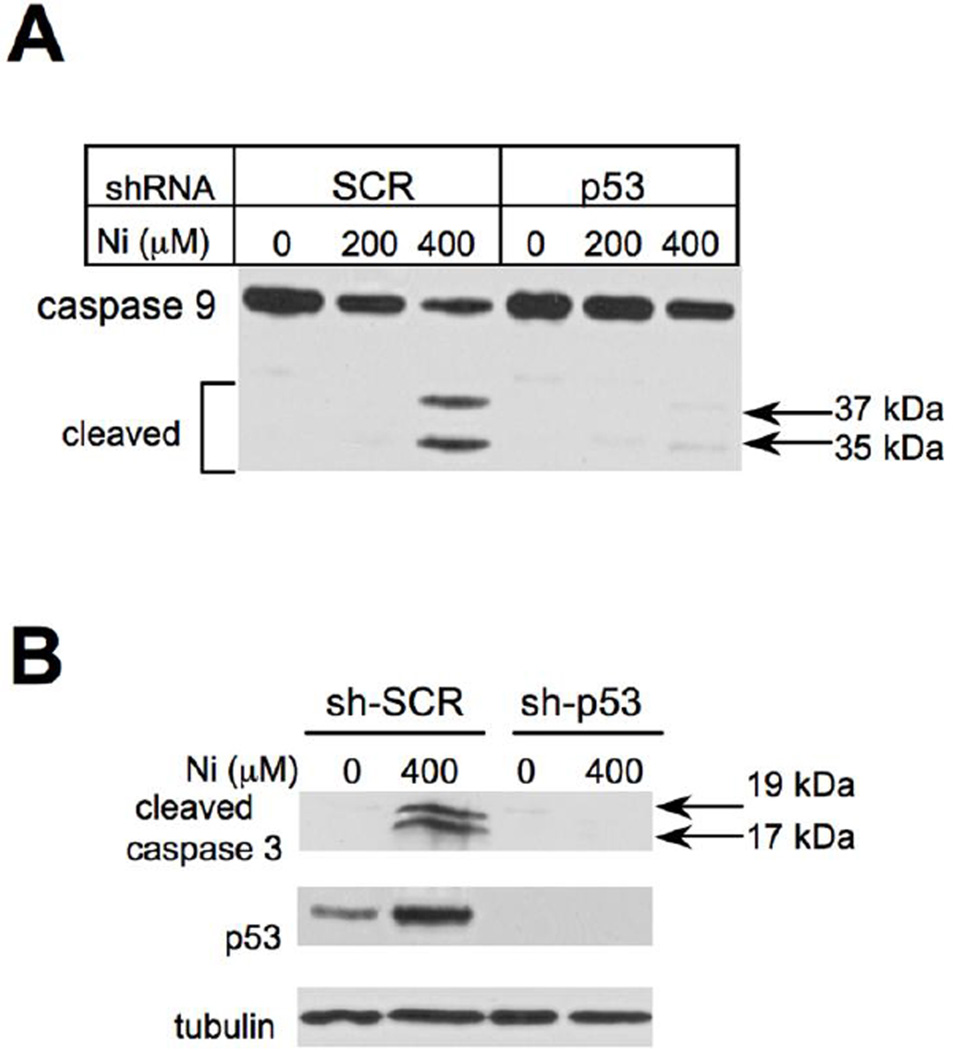
The involvement of p53 in apoptosis can occur via transcription-dependent and -independent mechanisms (Green and Kroemer, 2009; Mihara et al., 2003), which could trigger a mitochondrial apoptotic pathway targeting the initiator caspase 9. Activating autocleavage of procaspase 9 at Asp315 and Asp330 generates p35 and p37 active subunits, respectively (Li et al., 1997; Zou et al., 1999). We found that Ni(II) treatments of cells with normal levels of p53 produced both forms of active caspase 9 (Fig. 3A), with a dose-dependence similar to that of PARP cleavage (Fig. 2C). Caspase 9 activation showed a strong dependence on the presence of p53, as evidenced by a barely detectable presence of active 35 and 37 kDa caspase 9 forms in p53 knockdown cells (Fig. 3A). Suppression of caspase 9 processing by p53 depletion also eliminated activating cleavage of its downstream executioner caspase 3 (Fig. 3B), which plays a major role in PARP cleavage and apoptotic death in general (Gray et al., 2012).
Figure 3. Effect of p53 knockdown on activation of caspases 9 and 3.
(A) Western blot of H460 lysates with antibodies against a full-length caspase 9. Cells were treated with Ni(II) for 24 hr. (B) Levels of cleaved caspase 3 in control and p53-depleted H460 cells following 24 hr exposure to Ni(II).
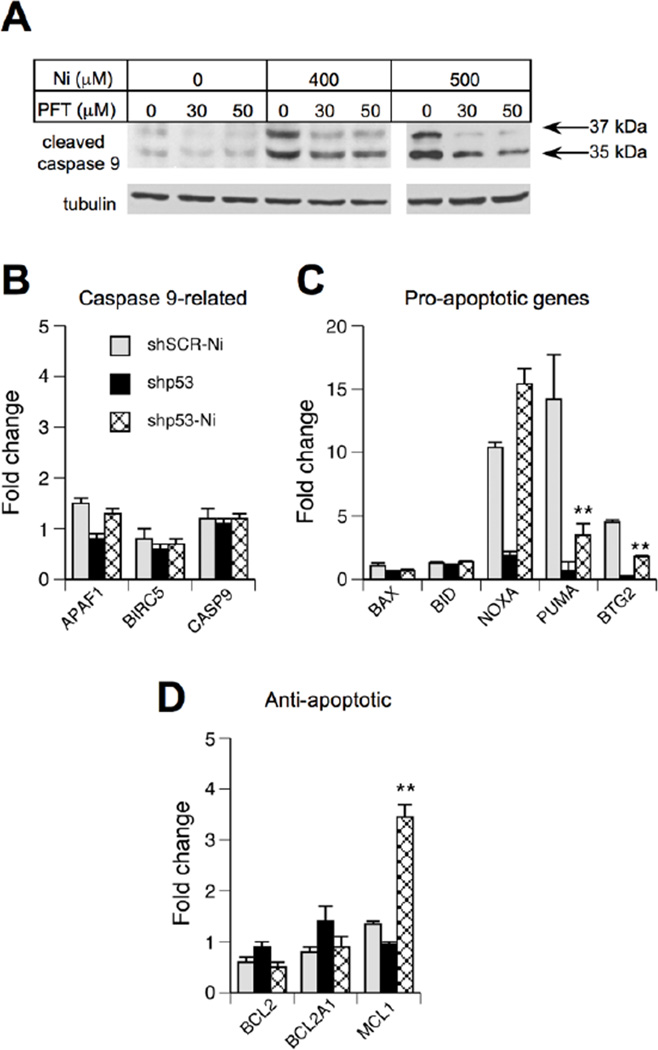
To assess a potential involvement of the transcription-dependent apoptotic mechanism, we first examined the effects of the p53 inhibitor, pifithrin-α (Komarov et al., 1999). We found that the addition of pifithrin-α completely abrogated the formation of the 37 kDA form of caspase 9 and strongly diminished the levels of the 35 kDa product in Ni(II)-treated cells (Fig. 4A). Next, we examined expression of 11 apoptosis-related genes by RT-qPCR. We found that Ni(II) did not cause any significant changes in mRNA levels for proteins that form complexes with caspase 9, such as CASP9 itself, its adaptor protein APAF1 or the inhibitory protein BIRC5 (Fig. 4B). Among the main pro-apoptotic genes that are known to be p53-responsive, expression of PUMA (BBC3) and NOXA (PMAIP1) showed the strongest increases following Ni(II) exposure but only PUMA induction was suppressed by p53 knockdown (Fig. 4C). Another gene with the p53-dependent upregulation by Ni(II) was BTG2, which promotes apoptotic mitochondrial depolarization (Hong et al., 2005). Among anti-apoptotic genes, BCL2 and BCL2A1 showed no signifcant changes in response to Ni(II), wheareas MCL1 levels were moderately higher in Ni(II)-treated shp53 cells (Fig. 4D). Taken together, suppression of caspase 9 cleavage by pifithrin-α and upregulation of pro-apoptotic PUMA and BTG2 on mRNA level support the importance of the transcriptional mechanism in the induction of apoptosis by p53.
Figure 4. Role of the transcriptional mechanism in p53-mediated apoptosis.
(A) Inhibition of caspase 9 autocleavage in Ni(II)-treated H460 cells by pifithrin-α (PFT). Cells were treated with PFT and Ni(II) for 24 hr. (B–D) Expression of apoptosis-related genes in control and p53-depleted H460 cells following 24 hr exposure to 400 µM Ni(II). Data are means±SD for three independent RNA samples (**-p<0.01 relative to sh-SCR+Ni).
Independence of HIF-1α and p53 responses to Ni(II)
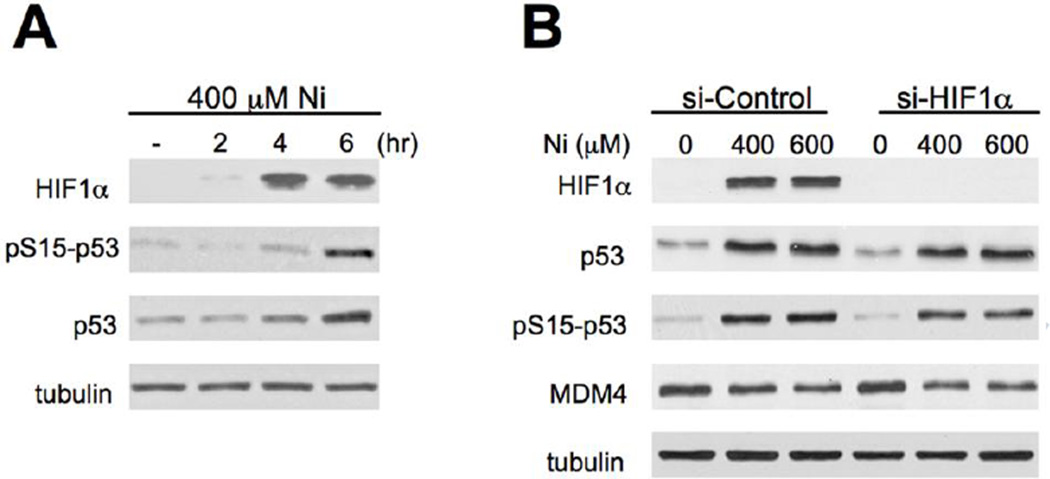
Activation of both HIF-1α and p53 pathways by Ni(II) raises a question whether these two stress responses are interconnected. We found that p53 knockdown had no effect on Ni(II)-induced upregulation of mRNA levels for GAPDH and HK2 (Fig. 5A), which are known HIF-1α target genes in hypoxic and Ni(II)-treated cells (Salnikow et al. 2003). Upregulation of HK2 (hexokinase II) on protein level and HIF-1α protein stabilization by Ni(II) were also unchanged by shRNA-mediated depletion of p53 (Fig. 5B). Consistent with the p53-independence of HIF-1α responses, time-course studies showed that HIF-1α stabilization preceeded accumulation of p53 and its Ser15 phosphorylation in Ni(II)-treated cells (Fig. 6A). Despite its earlier upregulation, HIF-1α was dispensable for p53 activation by Ni(II), as evidenced by normal p53 stabilization, Ser15-p53 phosphorylation and MDM4 depletion in cells with HIF-1α knockdown by siRNA (Fig. 6B).
Figure 5. HIF-1α responses in p53-depleted cells.
(A) GAPDH and HK2 mRNA levels in H460 cells expressing nonspecific (sh-SCR) and p53-targeting shRNA (sh-p53). Cells were treated with 400 µM Ni(II) for 24 hr. Data are means±SD for three independent RNA samples. (B) Western blot of cell lysates collected following 24-hr long Ni(II) exposures.
Figure 6. Activation of p53 in cells with depleted HIF-1α.
(A) Western blot of H460 cells treated with Ni(II) for 0–6 hr. (B) Activation of p53 in H460 cells with normal and siRNA-depleted expression of HIF-1α. Cells were treated with Ni(II) for 6 hr.
Discussion
Biological significance of p53 activation by Ni(II)
Our examination of Ni(II)-treated H460 human lung epithelial cells and normal human lung fibroblasts found several lines of methologically diverse evidence supporting activation of the p53 pathway. The biochemical set of evidence includes elevated Ser15-p53 phosphorylation, accumulation of p53 protein and diminished levels of its inhibitor MDM4. Ni(II) also caused a strong upregulation of the p53 target genes, such as the E3 ubiquitin ligase MDM2, the CDK inhibitor p21 (CDKN1A) and the proapoptotic factor PUMA (BBC3). Importantly, the induction of these genes was suppressed by p53 knockdown, providing a genetic confirmation for the presence of the transcriptionally active p53 in Ni(II)-treated cells. The activated p53 was also important for manifestation of Ni(II)-induced cytotoxic effects, such as apoptosis and clonogenic death. Although p53 activates a necrotic cell death in response to H2O2 and ischemia (Vaseva et al., 2012), we found no detectable necrosis in H460 cells treated with the hypoxia mimic Ni(II). Ni(II) has been reported to elevate cellular levels of H2O2 (Huang et al., 2006), however, it appears that these increases are insifficiently high to induce necrosis.
The p53-mediated apoptosis involved a transcription-dependent apoptotic program resulting in activation of the initiator caspase 9 and the executioner caspase 3. Caspase 9 is activated by pro-apoptotic factors released from mitochondria by pores-forming oligomers of conformationally altered BAX and BAK (Li et al., 1997; Wei et al., 2001; Zou et al., 1999). Upregulation of PUMA expression probably played a major role in p53-mediated apoptosis by Ni(II), as this BH3 domain-containing protein alone can cause apoptosis via a conformational activation of BAX and BAK (Garrison et al., 2012; Jeffers et al., 2003; Ren et al., 2010). The p53-mediated induction of BTG2 by Ni(II) was more moderate than that for PUMA but it can help promote mitochondrial depolarization (Hong et al., 2005) and release of caspase 9-activating mitochondrial proteins. Unlike ionizing radiation (Oda et al., 2000), a strong induction of another BH3-only member NOXA was p53-independent for nongenotoxic Ni(II) and it appeared to have only little if any direct effect on caspase 9 activation. Similarly to other members of the “sensitizers” group of proapoptotic BH3 proteins, NOXA alone is unable to induce BAX/BAK-dependent apoptosis (Green and Kroemer, 2009), which explains a strong antiapoptotic effect of p53 depletion (no PUMA induction) despite the remaining high levels of NOXA. The main role of NOXA in apoptosis involves release of proapoptotic proteins from sequestration by the antiapoptotic protein MCL1 (Green and Kroemer, 2009; Kim et al., 2009). The observed upregulation of MCL1 in p53-depleted cells would help neutralize pro-apoptotic functions of NOXA. In p53-proficient human lung cells, Ni(II)-induced apoptosis most likely involves cooperative effects of both PUMA and NOXA, with the p53-dependent increase in PUMA providing the apoptotic trigger. Ni(II)-treated rat pancreatic β-cells also showed cleavage of caspases 9 and 3, which was dependent on JNK activity (Wu et al., 2011). However, it is unclear whether Ni(II) and/or JNK affected Puma or Noxa levels in this cellular model, as this study examined only Bak and Bid expression and detected very modest ~1.5-fold increases for both genes by 2.2 mM Ni(II). We found no changes in BID expression in H460 cells treated with 0.4 mM Ni(II), a dose that caused >10-fold induction of PUMA and NOXA.
Previous studies have reported contradictory findings on the ability of Ni(II) to activate p53. Salnikow et al. (1999) have found increased levels of p53 protein in Ni(II)-treated human A549 and MCF7 cells although the toxieologieal role of this response was not established. Two subsequent studies were unable to detect p53 activation by Ni(II) (Ding et al., 2009; Huang et al., 2006). We confirmed the earlier observation on p53 stabilization (Salnikow et al., 1999) and provided genetic and biochemical evidence for transeriptional activity of p53 and its role in apoptotic responses to Ni(II). Our RT-qPCR studies showed that p53 upregulated only a subset of its target genes, which would make it difficult to detect p53 activation using reporter constructs and this can explain negative findings by Huang et al. (2006). The negative study by Ding et al. (2009) used SV40-transformed BEAS-2B cells in which p53 is inactivated by binding with SV40 large T-antigen, making p53 completely unresponsive in this cell line.
p53 activation is independent of HIF-1α
Upregulation of HIF-1α protein and its transeriptional network is a prominent cellular response to the hypoxia-mimicking Ni(II) ions (Costa et al., 2005; Salnikow et al., 2003; Salnikow and Zhitkovich, 2008). A rapid and strong stabilization of HIF-1α followed by increased expression of its target genes was also found in our experiments with Ni(II)-treated H460 human lung cells. Hypoxia-induced accumulation of p53 has been shown to be dependent on the presence of HIF-1α (An et al., 1998). Stabilization of p53 protein in hypoxic cells has been attributed to binding of its E3 ligase MDM2 by HIF-1α, which inactivates p53 ubiquitylation (Chen et al., 2003). We found that a loss of HIF-1α did not prevent p53 stabilization by Ni(II), pointing to a mechanistically distinct mode of p53 activation by this hypoxia mimic. The postulated inhibition of MDM2 by HIF-1α binding (Chen et al., 2003) also cannot explain other p53-stimulating responses, such as Ser15-p53 phosphorylation and lower MDM4 levels in Ni(II)-treated cells. We noticed that the microarray gene expression data showed a similar induction of the p53 target gene p21 in Ni(II)-treated wild-type and Hifla-null mouse fibroblasts (Salnikow et al., 2003), further supporting our conclusion on the HIF-1α independence of p53 activation by Ni(II).
Implications for Ni-induced carcinogenesis
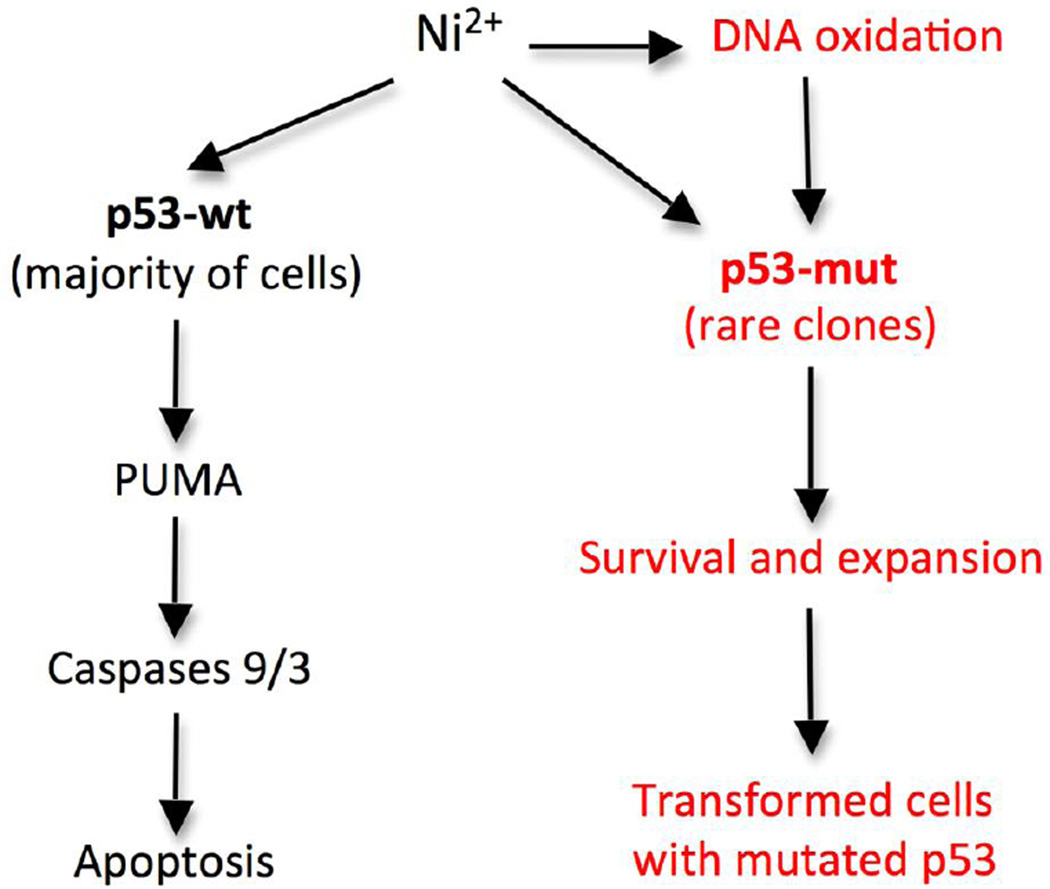
Chronic exposures to Ni-containing compounds cause lung cancers (Goodman et al., 2011; Salnikow and Zhitkovich, 2008). A high percentage of human lung tumors contain mutations in p53 gene (Pfeifer and Besaratinia, 2009), which was also mutationally inactivated during Ni(II)-induced transformation of human cells (Maehle et al., 1992). Ni(II) is a potent carcinogen but its mutagenicity is low in both bacterial and mammalian cells (Costa et al., 2005; Salnikow and Zhitkovich, 2008), which leads to a question how p53 mutations can arise in Ni(II)-dependent carcinogenesis. Our findings on the importance of p53 in Ni(II)-induced cell death suggest that chronic exposures to cytotoxic doses of Ni(II) ions could select for resistant lung cells with the pre-existing p53 mutations. Malignant transformation by Ni(II) is associated with a sustained presence of high cellular concentrations of Ni(II) ions (Costa and Mollenhauer, 1980; Goodman et al., 2011; Ke et al., 2007; Munoz and Costa, 2012), which is consistent with the proposed cytotoxicity-based selection mechanism. The formation of pre-mutagenic DNA lesions (Huang et al., 1995; Qu et al., 2001) and the proposed selection mechanism are not necessarily mutually exclusive causes of Ni(II)-transformed cells with mutated p53, as both processes are expected to act cooperatively by increasing the initial number of the mutated clones and promoting their expansion, respectively (Fig. 7).
Figure 7. Major steps in p53-mediated apoptosis by Ni(II) and a selection model for p53 mutations in Ni(II)-induced tumors.
PUMA is the main pro-apoptotic factor upregulated by Ni(II) via p53 activation. Apoptotic resistance of Ni(II)-treated cells lacking functional p53 results from their induction of antiapoptotic MLCl and the inability to transactivate PUMA. Both endogenous and Ni(II)-promoted mutations can serve a source of resistant cells with inactivated p53 (p53-mut).
Highlights.
Ni(II) is a strong activator of the transcription factor p53.
Apoptosis is a principal form of death by Ni(II) in human lung epithelial cells.
Ni(II)-activated p53 triggers a caspase 9/3-mediated apoptotic program.
NOXA and PUMA are two main proapoptotic genes induced by Ni(II).
HIF-1α and p53 are independent stress responses to hypoxia-mimicking Ni(II).
Acknowledgements
This work was supported by grants ES008786 and ES013660 from the National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, Prives C. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann KA, Landolph JR. Induction of anchorage independence in human diploid foreskin fibroblasts by carcinogenic metal salts. Cancer Res. 1987;47:3815–3823. [PubMed] [Google Scholar]
- Chen D, Li M, Luo J, Gu W. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J. Biol. Chem. 2003;278:13595–13598. doi: 10.1074/jbc.C200694200. [DOI] [PubMed] [Google Scholar]
- Costa M, Davidson TL, Chen H, Ke Q, Zhang P, Yan Y, Huang C, Kluz T. Nickel carcinogenesis: epigenetics and hypoxia signaling. Mutat. Res. 2005;592:79–88. doi: 10.1016/j.mrfmmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Costa M, Mollenhauer HH. Carcinogenic activity of particulate nickel compounds is proportional to their cellular uptake. Science. 1980;209:515–517. doi: 10.1126/science.7394519. [DOI] [PubMed] [Google Scholar]
- Davidson TL, Chen H, Di Toro DM, D'Angelo G, Costa M. Soluble nickel inhibits HIF-prolyl-hydroxylases creating persistent hypoxic signaling in A549 cells. Mol. Carcinog. 2006;45:479–489. doi: 10.1002/mc.20176. [DOI] [PubMed] [Google Scholar]
- Ding J, Zhang X, Li J, Song L, Ouyang W, Zhang D, Xue C, Costa M, Meléndez JA, Huang C. Nickel compounds render anti-apoptotic effect to human bronchial epithelial Beas-2B cells by induction of cyclooxygenase-2 through an IKKbeta/p65-dependent and IKKalpha- and p50-independent pathway. J. Biol. Chem. 2006;281:39022–39032. doi: 10.1074/jbc.M604798200. [DOI] [PubMed] [Google Scholar]
- Ding J, He G, Gong W, Wen W, Sun W, Ning B, Huang S, Wu K, Huang C, Wu M, Xie W, Wang H. Effects of nickel on cyclin expression, cell cycle progression and cell proliferation in human pulmonary cells. Cancer Epidemiol. Biomarkers Prev. 2009;18:1720–1729. doi: 10.1158/1055-9965.EPI-09-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Brant KA, Ward RM, Cattley RT, Barchowsky A, Fabisiak JP. Multiple protein kinase pathways mediate amplified IL-6 release by human lung fibroblasts co-exposed to nickel and TLR-2 agonist, MALP-2. Toxicol. Appl. Pharmacol. 2010;247:146–157. doi: 10.1016/j.taap.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Phillips DC, Jeffers JR, Chipuk JE, Parsons MJ, Rehg JE, Opferman JT, Green DR, Zambetti GP. Genetically defining the mechanism of Puma- and Bim-induced apoptosis. Cell Death Differ. 2012;19:642–649. doi: 10.1038/cdd.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JE, Prueitt RL, Thakali S, Oller AR. The nickel ion bioavailability model of the carcinogenic potential of nickel-containing substances in the lung. Crit. Rev. Toxicol. 2011;41:142–174. doi: 10.3109/10408444.2010.531460. [DOI] [PubMed] [Google Scholar]
- Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl. Acad. Sci. USA. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2012;142:637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Ryu MS, Lim IK. Phosphorylation of serine 147 of tis21/BTG2/pc3 by p-Erk1/2 induces Pin-1 binding in cytoplasm and cell death. J. Biol. Chem. 2005;280:21256–21263. doi: 10.1074/jbc.M500318200. [DOI] [PubMed] [Google Scholar]
- Huang C, Li J, Costa M, Zhang Z, Leonard SS, Castranova V, Vallyathan V, Ju G, Shi X. Hydrogen peroxide mediates activation of nuclear factor of activated T cells (NFAT) by nickel subsulfide. Cancer Res. 2006;61:8051–8057. [PubMed] [Google Scholar]
- Huang X, Kitahara J, Zhitkovich A, Dowjat K, Costa M. Heterochromatic proteins specifically enhance nickel-induced 8-oxo-dG formation. Carcinogenesis. 1995;16:1753–1759. doi: 10.1093/carcin/16.8.1753. [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- Ke Q, Davidson T, Kluz T, Oller A, Costa M. Fluorescent tracking of nickel ions in human cultured cells. Toxicol. Appl. Pharmacol. 2007;219:18–23. doi: 10.1016/j.taap.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YW, Klein CB, Kargacin B, Salnikow K, Kitahara J, Dowjat K, Zhitkovich A, Christie NT, Costa M. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol. Cell. Biol. 1995;15:2547–2557. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liu X, Gurel V, Morris D, Murray D, Zhitkovich A, Kane AB, Hurt RH. Bioavailability of nickel in single-wall carbon nanotubes. Advanced Materials. 2007;19:2790–2796. [Google Scholar]
- Maehle L, Metcalf RA, Ryberg D, Bennett WP, Harris CC, Haugen A. Altered p53 gene structure and expression in human epithelial cells after exposure to nickel. Cancer Res. 1992;52:218–221. [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Miura T, Patierno SR, Sakuramoto T, Landolph JR. Morphological and neoplastic transformation of C3H/10T1/2 Cl 8 mouse embryo cells by insoluble carcinogenic nickel compounds. Environ. Mol. Mutagen. 1989;14:65–78. doi: 10.1002/em.2850140202. [DOI] [PubMed] [Google Scholar]
- Muñoz A, Costa M. Elucidating the mechanisms of nickel compound uptake: a review of particulate and nano-nickel endocytosis and toxicity. Toxicol. Appl. Pharmacol. 2012;260:1–16. doi: 10.1016/j.taap.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Patel E, Lynch C, Ruff V, Reynolds M. Co-exposure to nickel and cobalt chloride enhances cytotoxicity and oxidative stress in human lung epithelial cells. Toxicol. Appl. Pharmacol. 2012;258:367–375. doi: 10.1016/j.taap.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Besaratinia A. Mutational spectra of human cancer. Hum. Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietruska JR, Liu X, Smith A, McNeil K, Weston P, Zhitkovich A, Hurt R, Kane AB. Bioavailability, intracellular mobilization of nickel, and HIF-1α activation in human lung epithelial cells exposed to metallic nickel and nickel oxide nanoparticles. Toxicol. Sci. 2011;124:138–148. doi: 10.1093/toxsci/kfr206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu W, Kasprzak KS, Kadiiska M, Liu J, Chen H, Maciag A, Mason RP, Waalkes MP. Mechanisms of arsenic-induced cross-tolerance to nickel cytotoxicity, genotoxicity, and apoptosis in rat liver epithelial cells. Toxicol. Sci. 2001;63:189–195. doi: 10.1093/toxsci/63.2.189. [DOI] [PubMed] [Google Scholar]
- Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–3139. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M, Peterson E, Quievryn G, Zhitkovich A. Human nucleotide excision repair efficiently removes DNA phosphate-chromium adducts and protects cells against chromate toxicity. J. Biol. Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- Reynolds M, Zhitkovich A. Cellular vitamin C increases chromate toxicity via a death program requiring mismatch repair but not p53. Carcinogenesis. 2007;28:1613–1620. doi: 10.1093/carcin/bgm031. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Wang S, Costa M. Induction of activating transcription factor 1 by nickel and its role as a negative regulator of thrombospondin I gene expression. Cancer Res. 1997;57:5060–5066. [PubMed] [Google Scholar]
- Salnikow K, An WG, Melillo G, Blagosklonny MV, Costa M. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999;20:1819–1823. doi: 10.1093/carcin/20.9.1819. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Blagosklonny MV, Ryan H, Johnson R, Costa M. Carcinogenic nickel induces genes involved with hypoxic stress. Cancer Res. 2000;60:38–41. [PubMed] [Google Scholar]
- Salnikow K, Davidson T, Zhang Q, Chen LC, Su W, Costa M. The involvement of hypoxia-inducible transcription factor-1-dependent pathway in nickel carcinogenesis. Cancer Res. 2003;63:3524–3530. [PubMed] [Google Scholar]
- Salnikow K, Donald SP, Bruick RK, Zhitkovich A, Phang JM, Kasprzak KS. Depletion of intracellular ascorbate by the carcinogenic metals nickel and cobalt results in the induction of hypoxic stress. J. Biol. Chem. 2004;279:40337–40344. doi: 10.1074/jbc.M403057200. [DOI] [PubMed] [Google Scholar]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic and chromium. Chem. Res. Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VC, Cash HL, Morse JL, Lu S, Zhitkovich A. S-phase sensing of DNA-protein crosslinks triggers TopBpl-independent ATR activation and p53-mediated cell death by formaldehyde. Cell Cycle. 2012;11:2526–2537. doi: 10.4161/cc.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Yang CY, Hung DZ, Su CC, Chen KL, Yen CC, Ho TJ, Su YC, Huang CF, Chen CH, Tsai LM, Chen YW. Nickel(II) induced JNK activation-regulated mitochondria-dependent apoptotic pathway leading to cultured rat pancreatic β-cell death. Toxicology. 2011;289:103–111. doi: 10.1016/j.tox.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]