Abstract
Bacterial surface (S) layers are the outermost proteinaceous cell envelope structures found on members of nearly all taxonomic groups of bacteria and Archaea. They are composed of numerous identical subunits forming a symmetric, porous, lattice-like layer that completely covers the cell surface. The subunits are held together and attached to cell wall carbohydrates by non-covalent interactions, and they spontaneously reassemble in vitro by an entropy-driven process. Due to the low amino acid sequence similarity among S-layer proteins in general, verification of the presence of an S-layer on the bacterial cell surface usually requires electron microscopy. In lactobacilli, S-layer proteins have been detected on many but not all species. Lactobacillus S-layer proteins differ from those of other bacteria in their smaller size and high predicted pI. The positive charge in Lactobacillus S-layer proteins is concentrated in the more conserved cell wall binding domain, which can be either N- or C-terminal depending on the species. The more variable domain is responsible for the self-assembly of the monomers to a periodic structure. The biological functions of Lactobacillus S-layer proteins are poorly understood, but in some species S-layer proteins mediate bacterial adherence to host cells or extracellular matrix proteins or have protective or enzymatic functions. Lactobacillus S-layer proteins show potential for use as antigen carriers in live oral vaccine design because of their adhesive and immunomodulatory properties and the general non-pathogenicity of the species.
Keywords: Surface layer protein, S-layer protein, Lactobacillus, Adhesion
Introduction
Bacterial surface (S) layers are proteinaceous cell envelope structures ubiquitously found in Gram-positive and Gram-negative bacterial species and in Archaea (Sára and Sleytr 2000). When present, they form the outermost layer of the cell, being occasionally covered only by capsules (Fouet et al. 1999). S-layers are composed of numerous identical (glyco)protein subunits, 40–200 kDa in molecular weight, which form a two-dimensional, regular and highly porous array with oblique (p1, p2), square (p4) or hexagonal (p3, p6) symmetry. The subunits are held together and attached to the underlying cell surface by non-covalent interactions and have an intrinsic, entropy-driven tendency to form regular structures either in solution or on a solid support in vitro. The subunit proteins are typically rich in acidic and hydrophobic amino acids but low in sulphur-containing amino acids and have a low predicted overall pI value (Sára and Sleytr 2000). S-layer protein genes are highly expressed. Several S-layer protein genes in the genome of a single strain have been described, but all of the genes are not necessarily expressed at the same time; silent genes, antigenic variation based on S-layer gene expression (reviewed by Boot and Pouwels 1996; Sára and Sleytr 2000; Thompson 2002), alternative expression of S-layer protein genes in or ex vivo (reviewed by Fouet 2009), sequential expression during growth (Mignot et al. 2004) and, rarely, superimposed S-layers (Stewart and Murray 1982; Cerquetti et al. 2000) or S-layers composed of two different S-layer proteins (Rothfuss et al. 2006; Fagan et al. 2009; Goh et al. 2009; Sekot et al. 2012) have been described. Due to the low overall sequence similarity among S-layer protein genes and the lack of a universal signature sequence, confirmation of the presence of an S-layer still relies largely on electron microscopy.
In recent decades, information about the biological functions of S-layer proteins has accumulated, but no common function for all S-layers has emerged. The functions characterized thus far include, e.g., the determination or maintenance of cell shape (Mescher and Strominger 1976; Engelhardt 2007a) and functions as a molecular sieve (Sára and Sleytr 1987; Sára et al. 1990), as a binding site for large molecules (Kay et al. 1985; Phipps and Kay 1988; Matuschek et al. 1994; Egelseer et al. 1995, 1996; Peters et al. 1995), ions (Schultze-Lam et al. 1992; Pollmann et al. 2006; Klingl et al. 2011) or phages (Howard and Tipper 1973; Ishiguro et al. 1984; Fouet 2009) and as a mediator of bacterial adhesion (Doig et al. 1992; Toba et al. 1995; Noonan and Trust 1997; Hynönen et al. 2002; Buck et al. 2005; Sakakibara et al. 2007; Poppinga et al. 2012). In pathogenic bacteria, S-layers may contribute to virulence by several mechanisms, including adhesion, coaggregation (Shimotahira et al. 2013), antigenic variation (Thompson 2002; Spigaglia et al. 2011), protection from complement or from phagocytosis (Doig et al. 1992; Thompson 2002; Shimotahira et al. 2013) or modulation of T-cell or cytokine responses (Wang et al. 2000; Ausiello et al. 2006; Sekot et al. 2011; Settem et al. 2013). Further, S-layer proteins may protect the bacterial cell from various environmental factors such as mechanical and osmotic stresses (Engelhardt 2007a, b), antimicrobial peptides (de la Fuente-Núñez et al. 2012), radiation (Kotiranta et al. 1999), changes in environmental pH (Gilmour et al. 2000), bacteriophages (Howard and Tipper 1973), bacterial or eukaryotic microbial predators (Koval and Hynes 1991; Tarao et al. 2009) or bacteriolytic enzymes (Lortal et al. 1992). Some S-layer proteins have the potential to act as degradative enzymes (Calabi et al. 2001; Ahn et al. 2006; Prado Acosta et al. 2008), and the S-layer protein of a marine Synechococcus strain is involved in motility (Brahamsha 1996; McCarren et al. 2005).
Due to the self-assembly properties and the highly ordered, regular structure down to the nanometer scale, S-layers have a vast application potential in (nano)biotechnology. Applications of S-layers can be roughly divided into two groups. The first comprises applications utilizing (genetically engineered) S-layered bacterial cells, S-layer (fusion) proteins or only the expression and/or secretion signals of S-layer protein genes in various biological systems, including vaccine development, heterologous protein production and surface display. The second group utilizes isolated, usually recombinant, S-layer proteins for non-life (nano) technological applications (see the comprehensive recent reviews by Schuster et al. 2006; Sleytr et al. 2007; Schuster and Sleytr 2009; Ilk et al. 2011; Pum et al. 2013).
Lactic acid bacteria are Gram-positive, non-pathogenic micro-organisms characterized by the production of lactic acid as the main end-product of carbohydrate metabolism. Within lactic acid bacteria, the genus Lactobacillus forms a large, heterogeneous group consisting of non-sporulating, anaerobic or microaerophilic, catalase-negative, fermentative organisms with a low G+C content (32–53 %) and complex nutritional requirements. Lactobacilli have been isolated from various environments, including plants, foodstuffs, silage and sewage, and they have been found in the gastrointestinal and genital tracts of humans and animals, where they form part of the normal flora (Kandler and Weiss 1986; Axelsson 1998; Hayashi et al. 2005; Felis and Dellaglio 2007). Besides having a long history of use in food and feed fermentations, lactic acid bacteria have aroused interest owing to the health beneficial (probiotic) properties of some strains. They have proved promising also as potential vehicles for the delivery of therapeutic and prophylactic molecules, such as vaccine antigens in humans (Seegers 2002; Wells and Mercenier 2008).
Occurrence and general properties of Lactobacillus S-layer proteins
In the genus Lactobacillus, S-layers have been found in several but not all species. Biochemical or functional data have been published about the S-layer proteins of Lactobacillus brevis, Lactobacillus buchneri, Lactobacillus helveticus and Lactobacillus hilgardii and organisms of the former Lactobacillus acidophilus group (Johnson et al. 1980), including L. acidophilus, Lactobacillus amylovorus, Lactobacillus crispatus and Lactobacillus gallinarum. In addition, strains of Lactobacillus amylolyticus, Lactobacillus gigeriorum, Lactobacillus kefiranofaciens, Lactobacillus pasteurii and Lactobacillus ultunensis carry predicted S-layer protein genes in their completely or partially sequenced genomes (see Table 1). Lactobacillus kefir and Lactobacillus parakefir have been shown to possess an S-layer (Garrote et al. 2004), although the genes have not been sequenced. In earlier studies, S-layers have been demonstrated by electron microscopy on Lactobacillus fermentum and Lactobacillus delbrueckii subspecies bulgaricus (Kawata et al. 1974; Masuda and Kawata 1983), but species identification of these strains has subsequently been questioned (Boot et al. 1996c). Supporting this is the fact that no L. fermentum S-layer protein gene sequence can be found in public databases, and the whole genome sequencing of L. delbrueckii subspecies bulgaricus did not reveal any S-layer protein gene (Hao et al. 2011; Makarova et al. 2006); thus, at present these species can be considered as non-S-layer producers. Likewise, in an earlier study, a regular layer was observed on Lactobacillus casei (Barker and Thorne 1970), but according to Boot et al. (1996b), no S-layer protein-encoding gene is present in this species, and the isolate probably would now be reclassified to another species. Moreover, as the demonstration of S-layer proteins on the surface of L. casei, Lactobacillus paracasei subspecies paracasei and Lactobacillus rhamnosus by Zhang et al. (2010b) and Guo et al. (2012) is not yet confirmed, L. casei is also currently considered as a non-S-layer producer.
Table 1.
Lactobacillus strains carrying S-layer protein genes with sequences in public databases
| Strain | Slp referencea | Genome | ||
|---|---|---|---|---|
| Status | Reference | |||
| L. acidophilus | ATCC 4356 | Boot et al. (1995) | ||
| JCM 1038 | AAF65561 | – | ||
| Unspecified | AEW12794 | – | ||
| NCFM | Buck et al. (2005) | Completed | Altermann et al. (2005) | |
| 30SC | Annotation | Completed | Oh et al. (2011) | |
| ATCC 4796 | Annotation | Ongoing | ||
| L. amylolyticus | DSM 11664 | Annotation | Ongoing | |
| L. amylovorus | GRL 1112 | Jakava-Viljanen and Palva (2007) | Completed | Kant et al. (2011a) |
| GRL 1118 | Jakava-Viljanen and Palva (2007) | Completed | Kant et al. (2011b) | |
| GRL 1115 | Jakava-Viljanen and Palva (2007) | Ongoing | ||
| DSM 16698 | Palva et al. unpublished | Ongoing | ||
| L. brevis | ATCC 8287 | Vidgren et al. (1992) | Ongoing | |
| ATCC 14869 | Jakava-Viljanen et al. (2002) | – | ||
| KB290 | BAK78870 | – | ||
| M8 | AFD33419 | – | ||
| ATCC 367 | Åvall-Jääskeläinen et al. (2008) | Completed | Makarova et al. (2006) | |
| DSM 20054 ABBC45 | Annotation | Ongoing | ||
| L. brevis ssp gravesensis | ATCC 27305 | Annotation | Ongoing | |
| L. buchneri | CD034 | Annotation | Completed | Heinl et al. (2012) |
| ATCC 11577 | Annotation | Ongoing | ||
| L. crispatus | JCM 5810 | Sillanpää et al. (2000) | – | |
| LMG 12003 | Sillanpää et al. (2000) | – | ||
| F5.7 | Mota et al. (2006) | – | ||
| ZJ001 | Chen et al. (2009) | – | ||
| K2-4-3 | Hu et al. (2011) | – | ||
| K313 | Sun et al. (2012) | – | ||
| M247 | AJ007839 | – | ||
| MH315 | AB110090, AB11091 | – | ||
| Unspecified | AAY41912 | – | ||
| Unspecified | AAY41916 | – | ||
| ST1 | Hurmalainen et al. 2007 | Completed | Ojala et al. (2010) | |
| 125-2-CHN CTV-05 MV-1A-US MV-3A-US 214–1 JV-V01 | Annotation | Ongoing | ||
| L. gallinarum | D109, D195-2, D256, D109, D255, ATCC 33199, D42 D44-2 | Hagen et al. (2005) | – | |
| DSMZ 10532 | AAS83409 | – | ||
| L. gigeriorum | CRBIP 24.85 | Annotation | Ongoing | |
| L. helveticus /suntoryeus | CNRZ 892 | Callegari et al. (1998) | – | |
| CNRZ 1269 | CAA63409 | – | ||
| ATCC 12046 | Lortal et al. (1992) CAB46984 | – | ||
| GCL 1001b | BAB72065 | – | ||
| JCM 1003b | BAB72066 | – | ||
| JCM 1007 | BAB72067 | – | ||
| JCM 1008 | BAB72068 | – | ||
| IMPC M696 CNRZ 35 IMPC HLM1 IMPC I60 CNRZ 303 ATCC 15009 | Ventura et al. (2000) | – | ||
| K1/R0052 | ADK74769 | – | ||
| Y10 M4 | Cachat and Priest (2005) | – | ||
| Slh02 | AGD98690 | – | ||
| MIMLh5 | Taverniti et al. (2012) | – | ||
| Unspecified | AAL36968 | – | ||
| DPC4571 | Annotation | Completed | Callanan et al. (2008) | |
| H10 | Annotation | Completed | Zhao et al. (2011) | |
| MTCC5463 | Annotation | Completed | Prajapati et al. (2011) | |
| R0052 | Annotation | Completed | Tompkins et al. (2012) | |
| DSM20075 | Annotation | Ongoing | ||
| L. hilgardii | B706 | Dohm et al. (2011) | – | |
| ATCC 8290 | Annotation | Completed | – | |
| L. kefiranofaciens | ZW3 | Annotation | Completed | Wang et al. (2011) |
| L. pasteurii | CRBIP 24.76 | Annotation | Ongoing | |
| L. ultunensis | DSM 16047 | Annotation | Ongoing | |
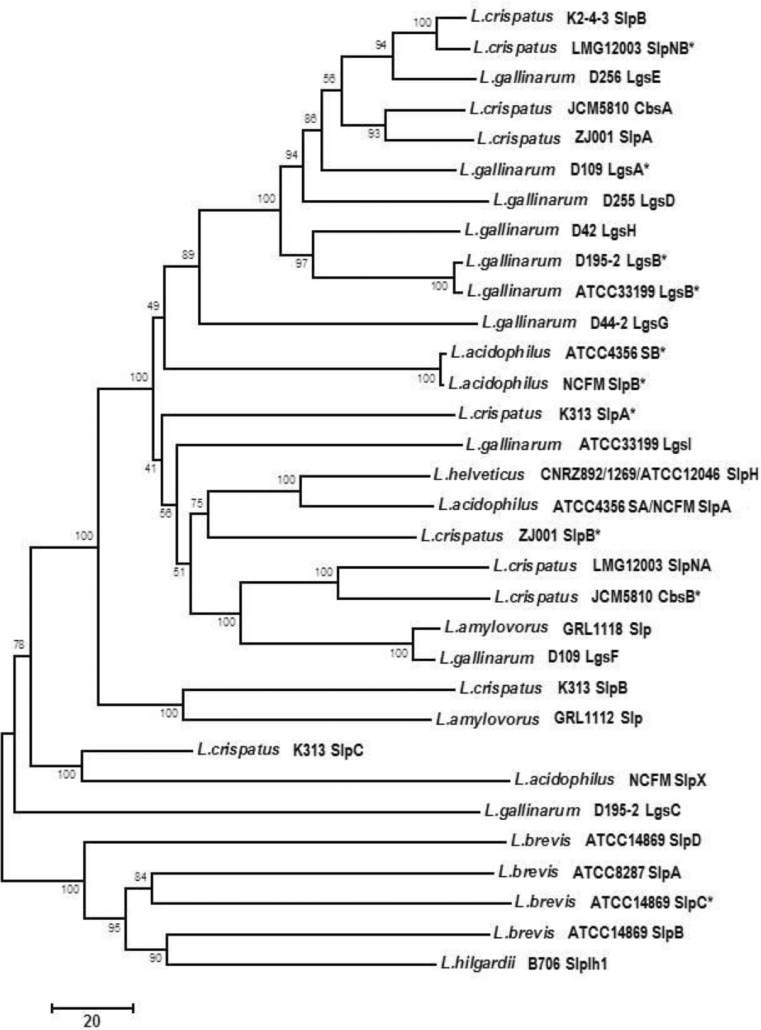
All of the Lactobacillus S-layer proteins characterized thus far are preceded by a 25–32-amino-acid signal peptide, indicating secretion through the general secretory pathway. The deduced amino acid sequences of mature Lactobacillus S-layer proteins vary considerably (Åvall-Jääskeläinen and Palva 2005), and even the S-layer proteins of the same strain, when present, may be markedly different in sequence (Jakava-Viljanen et al. 2002; Hagen et al. 2005). As in the case of S-layers in general, a remarkable similarity between the deduced amino acid sequences can only be found between related species, e.g. between the S-layer proteins of the former L. acidophilus group organisms and some L. helveticus strains (Antikainen et al. 2002; Hagen et al. 2005). Methods for the identification of L. crispatus (Horie et al. 2002b) and L. helveticus (Ventura et al. 2000) based on the presence of the S-layer protein genes have been developed, but the applicability for the latter species has subsequently been questioned due to the observed heterogeneity within L. helveticus S-layer protein gene sequences. Moreover, the relationship between L. helveticus cell surface proteinase and surface layer proteins is currently not clear (Gatti et al. 2005). The comparison of phylogenetic trees based on 22 deduced Lactobacillus S-layer protein (or the S-layer-like Apf protein) sequences and the 16S rRNA gene sequences of the corresponding Lactobacillus species revealed a similar overall clustering of strains (Åvall-Jääskeläinen and Palva 2005). However, when the phylogenetic trees constructed on the basis of the S-layer protein genes of a set of L. acidophilus-related organisms, including strains of the novel Lactobacillus suntoryeus species [later reclassified as L. helveticus (Naser et al. 2006)], were compared with trees constructed on the basis of 16S rRNA or elongation factor Tu (tuf) gene sequences of the same species, the novel strains grouped together in the latter tree, but not in the former tree based on the S-layer protein genes. This indicates a strong selective pressure driving the diversification of S-layer protein genes within at least some L. acidophilus-related organisms as well (Cachat and Priest 2005). Nevertheless, the remarkable similarities on the amino acid level between the S-layer proteins of L. acidophilus-related organisms have led to the proposal of using LC-MS/MS analysis of S-layer proteins for typing strains within this group (Podlesny et al. 2011). A phylogenetic tree based on those Lactobacillus S-layer protein sequences for which gene expression data are currently available is shown in Fig. 1. It clearly separates the S-layer proteins of L. brevis and L. hilgardii from the S-layer proteins of L. helveticus and the L. acidophilus group organisms but indicates great variability within the S-layer proteins of L. acidophilus group as, e.g., the S-layer proteins of different strains of L. gallinarum may be very distantly related (see, e.g., LgsF of L. gallinarum D109).
Fig. 1.
A neighbour-joining phylogenetic tree based on Lactobacillus S-layer protein sequences for which gene expression data are currently available. The scale bar indicates the phylogenetic distances expressed as the number of amino acid substitutions per sequence. Bootstrap values are indicated at the nodes for 500 replicates. The partial S-layer protein sequence of L. helveticus MIMLh5 was excluded from the analysis. Asterisk corresponding gene is known to be silent
The S-layer protein SlpA of L. acidophilus NCFM is identical in sequence with SA of L. acidophilus ATCC 4356, although the strains are clearly distinquishable by pulse field gel electrophoresis of chromosomal DNA (Sanders and Klaenhammer 2001). L. acidophilus NCFM harbours a larger diversity of mobile genetic elements than other probiotic lactic acid bacteria (Altermann et al. 2005). Although the elements do not show similarity to currently known integrative and conjugative elements (Wozniak and Waldor 2010), the possibility of horizontal gene transfer in the acquisition of slpA gene in L. acidophilus cannot be ruled out. A more likely explanation for the presence of an identical slp gene in two genetically different strains is, however, that the strains have a common origin as their genomic organization is predominantly the same (Sanders and Klaenhammer 2001).
S-layer proteins of lactobacilli differ from S-layer proteins in general in their smaller size (25–71 kDa) and high predicted overall pI value (9.4–10.4). The lattice symmetry of Lactobacillus S-layer proteins, when known, is of oblique or hexagonal type (reviewed by Åvall-Jääskeläinen and Palva 2005). A glycan structure of a Lactobacillus S-layer protein has to date been determined only for L. buchneri (Messner et al. 2008), whereas glycosylated S-layer proteins have been described in L. kefir (Mobili et al. 2009a). Secondary structure predictions for S-layer proteins are of limited value thus far as the prediction algorithms are based on the available structures of very dissimilar types of proteins. A prediction performed for the amino acid sequences of the unprocessed forms of six Lactobacillus S-layer proteins suggested on average 14 % α-helices, 39 % extended strands and 47 % random coils in these proteins (Åvall-Jääskeläinen and Palva 2005). Physical measurements revealing secondary structures have been conducted for a few Lactobacillus species. A Fourier transform infrared spectroscopy study performed for the S-layer proteins of L. kefir and L. brevis indicated α-helix contents of 0–21 %, β-sheet contents of 23–50 % and other structure contents, including β-turns and random coils, of 37–63 % in these proteins. For example, the proportions of α-helix, β-sheet, and other structures in SlpA of L. brevis ATCC 8287 were 0, 50 and 50 %, respectively (Mobili et al. 2009b). Atomic force microscopy studies of the S-layer protein CbsA of L. crispatus and its N- and C-terminal fragments suggested the presence of at least four α-helical structures of variable sizes, rather than β-sheets, in the N-terminal part of CbsA (Verbelen et al. 2007). Elucidation of the tertiary structure of S-layer proteins has been hindered by their molecular weights mostly not being in the suitable range (<40 kDa) for nuclear magnetic resonance studies and by their low solubility, i.e., their tendency to form two-dimensional lattices rather than three-dimensional crystals in solution. Consequently, no three-dimensional structures of Lactobacillus S-layer proteins at atomic resolution are currently available.
In addition to the verified S-layer proteins presented earlier, surface proteins described as “S-layer like” have been identified on the surface of several Lactobacillus gasseri and Lactobacillus johnsonii strains. These aggregation promoting factor (Apf) proteins share several characteristics with Lactobacillus S-layer proteins, such as their relative abundance on the cell surface, extractability by lithium chloride (LiCl), amino acid composition, predicted physical properties like high pI and indispensability for growth (Ventura et al. 2002). The Apf proteins of L. gasseri 4B2 are critical for cell shape maintenance, but they are not the sole components responsible for the aggregative phenotype (Jankovic et al. 2003). Despite the similarities with surface layer proteins, the S-layer nature of Apf proteins has not yet been fully confirmed. Each L. gasseri and L. johnsonii strain carries two apf genes in tandem orientation, both of which are expressed at the same time as monocistronic units (Ventura et al. 2002; Jankovic et al. 2003), a feature not very common among Lactobacillus slp genes (see "Expression of Lactobacillus S‐layer protein genes). A three-layered cell envelope structure similar to that observed on S-layered Lactobacillus strains and modifiable by LiCl has been observed in thin-section samples of L. johnsonii cells by electron microscopy (Ventura et al. 2002). However, no micrographs obtained by freeze-fracturing techniques or by other methods revealing the symmetric organization of the proteins have been published, and no reports are available about the recrystallization of the proteins in vitro. Proteins designated as Apf, with C-terminal similarity to Apf proteins of L. gasseri and L. johnsonii, have subsequently been described for several Lactobacillus and other bacterial species (Goh and Klaenhammer 2010), like L. crispatus M247 (Marcotte et al. 2004) and L. acidophilus NCFM (Goh and Klaenhammer 2010). L. acidophilus NCFM has also a true S-layer (Konstantinov et al. 2008), and L. crispatus M247 at least carries an S-layer gene (Table 1). Thus, based on the current data, the designation of L. gasseri and L. johnsonii Apf proteins as belonging to an S-layer like family (Ventura et al. 2002), but not as true S-layer proteins, is justifiable.
Expression of Lactobacillus S-layer protein genes
The very efficient synthesis of S-layer proteins in lactobacilli is achieved by several means: (1) The half lives of the S-layer protein gene transcripts have been determined to be exceptionally long (14 and 15 min for the S-layer protein genes of L. brevis ATCC 8287 and L. acidophilus ATCC 4356, respectively) (Boot et al. 1996b; Kahala et al. 1997). At least in the case of L. acidophilus ATCC 4356 (Narita et al. 2006) and L. crispatus K313 (Sun et al. 2012), this is due to the long 5′ untranslated region (UTR) of the transcript forming a stabilizing secondary structure, as originally suggested by Boot et al. (1996b). However, the 5′ UTR of L. brevis ATCC 8287 slpA transcript is not exceptionally long (Vidgren et al. 1992); (2) A biased codon usage, correlating with efficient gene expression in lactobacilli (Pouwels and Leunissen 1994), has been observed for the S-layer protein genes of L. brevis ATCC 8287 (Vidgren et al. 1992) and L. acidophilus ATCC 4356 (Boot et al. 1995); (3) The promoters of S-layer protein genes are efficient, even to the extent that they have been used in heterologous expression and protein production systems (see "Applications of Lactobacillus S‐layer proteins); (4) The presence of two promoters, which may act to enhance and/or regulate gene expression, has been identified upstream of the slpA gene of L. brevis ATCC 8287 (Vidgren et al. 1992; Hynönen et al. 2010), slpB of L. brevis ATCC 14869 (Jakava-Viljanen et al. 2002) and slpA of L. acidophilus ATCC 4356 (Boot et al. 1996b). In L. brevis ATCC 8287, both slp promoters were shown to be active (Kahala et al. 1997; Hynönen et al. 2010), whereas in L. acidophilus ATCC 4356, only the downstream promoter is functional under the conditions tested (Boot et al. 1996b).
The presence of multiple S-layer protein genes in the same strain is common in lactobacilli (Boot et al. 1996c; Hagen et al. 2005), but so far only the S-layer protein genes slpB and slpD of L. brevis ATCC 14869 (Jakava-Viljanen et al. 2002), slpB and slpC of L. crispatus K313 (Sun et al. 2012) and slpA and slpX of L. acidophilus NCFM (or slpB and slpX of the slpA knock-out mutant of L. acidophilus NCFM) (Goh et al. 2009) have been shown to be expressed simultaneously. Thus, silent S-layer protein genes, under the conditions tested, are common and represented, e.g., by the slpB genes of L. acidophilus ATCC 4356, NCIMB 8607, LMG 11428, LMG 11469 (Boot et al. 1995) and NCFM (Konstantinov et al. 2008), cbsB of L. crispatus JCM 5810 (Sillanpää et al. 2000) and ZJ001 (Chen et al. 2009), SlpNB of L. crispatus LMG 12003 (personal communication; Sillanpää et al. 2000), slpA of L. crispatus K313 (Sun et al. 2012), slpC of L. brevis ATCC 14869 (Jakava-Viljanen et al. 2002), several lgs genes of L. gallinarum (Hagen et al. 2005), and probably also by one of the two S-layer protein genes identified in L. amylovorus by DNA hybridization (Boot et al. 1996c), although the presence of two identical-sized S-layer proteins on the bacterial surface cannot be excluded. According to a preliminary SDS-PAGE analysis of seven porcine L. amylovorus isolates, only one isolate was suggested to express two S-layer protein genes at the same time, while for the remaining strains, only one S-layer protein was present (Jakava-Viljanen and Palva 2007). The genomes of L. gallinarum strains have two genes encoding S-layer proteins: a common one and a strain-specific one, but each strain produces only a single S-layer protein, which is always encoded by the strain-specific gene (Hagen et al. 2005). In the sequenced genomes of L. brevis ATCC 367 (Makarova et al. 2006) and L. buchneri CD034 (Heinl et al. 2012), two or several complete genes, respectively, have been identified by sequence homology (Åvall-Jääskeläinen et al. 2008; Heinl et al. 2012), but the expression of these genes is unknown.
The mechanism of the differential expression of slp genes has been well documented in L. acidophilus 4356, in which an inversion of a chromosomal segment leads to the placement of the silent gene in front of the active slp promoter (Boot et al. 1996a). This event seems to be unfavoured under laboratory conditions as the silent gene is at the expression site only in 0.3 % of the chromosomes of a broth culture of L. acidophilus 4356. No condition favouring the expression of the silent gene has thus far been characterized (Boot et al. 1996a). A similar chromosomal inversion mechanism has subsequently been shown to operate in L. acidophilus NCFM, where the inactivation of the S-layer protein gene slpA by homologous recombination led to the appearance of an alternate S-layer protein, SlpB, in the mutant strain NCK1377-CI (Buck et al. 2005; Konstantinov et al. 2008).
Information about adaptive changes in Lactobacillus S-layer gene expression, not known to involve chromosomal rearrangements, is scarce. In L. brevis ATCC 14869, the differential expression of the slpB and slpD genes is related to the oxygen content of the growth medium and the growth stage: slpB is expressed irrespective of oxygen content and equally in different growth phases, while slpD is predominantly expressed in aerated cultures and mainly in the exponential phase. The onset of slpD expression is most likely mediated by a soluble cytoplasmic factor, and it was surmised to be part of a stress response; a concomitant change in colony morphology, presumably not directly linked to the S-layer protein type, was also observed. Neither the nature/mechanism of action of the soluble regulator nor the reason for the silence of the slpC gene in this strain is known (Jakava-Viljanen et al. 2002). Stress-mediated regulation has been suggested also on other occasions. The expression of the S-layer protein gene of L. acidophilus NCC 2628 was induced when the strain was cultivated under conditions of limited protein supply (Schär-Zammaretti et al. 2005). An effect of bile salts was observed in L. acidophilus ATCC 4356, where concentrations of 0.01–0.05 % were shown to increase slpA gene expression, while the expression was decreased in 0.1 % bile; concomitant changes were observed in colony morphology and cell surface hydrophobicity (Khaleghi et al. 2010). In the same strain, sublethal concentrations of penicillin G were shown to increase the amount of SlpA on the bacterial surface, but the results were not in accordance with slpA gene expression (Khaleghi et al. 2011). The expression of the S-layer protein gene of L. acidophilus NCFM, in contrast, was not significantly induced during the passage through an in vitro gastrointestinal tract model (Weiss and Jespersen 2010). In L. brevis ATCC 8287, a slight repression effect of bile on slpA promoter activity was observed, but neither bile, pancreatin nor an uncommon carbon source had an effect on the amount of SlpA expressed on the bacterial cell surface (Hynönen et al. 2010). Similarly, in L. hilgardii, wine-related stress factors, like the presence of ethanol, copper sulphate or p-coumaric acid, did not affect S-layer protein production measurable by SDS-PAGE (Dohm et al. 2011).
Completely S-layer-negative Lactobacillus mutants are difficult or impossible to create (Palva et al. unpublished data; Boot et al. 1996a; Martinez et al. 2000; Buck et al. 2005), indicating that at least one functional S-layer protein gene is essential for S-layered lactobacilli, and expression of S-layer protein genes thus could be anticipated to be constitutive. However, some of the examples earlier indicate that variation and regulation at the transcriptional and/or translational level also exist. Recently, genes encoding alternative sigma factors have been identified in the sequenced genomes of several Lactobacillus species, and numerous potential transcription factor genes are also present (Azcarate-Peril et al. 2008), indicating potential for the regulated expression of slp genes under special conditions in these organisms. However, currently the transcriptional and translational regulation mechanisms of Lactobacillus S-layer protein genes on a molecular level are almost completely unexplored.
Cell wall binding and self-assembly regions of Lactobacillus S-layer proteins
One or both of the two structural regions generally present in S-layer proteins, i.e., the region involved in the attachment of the S-layer subunit to the cell envelope and the region involved in S-layer assembly, have so far been characterized in the following S-layer proteins: in the SA protein of L. acidophilus ATCC 4356 (Smit et al. 2001), in CbsA of L. crispatus JCM 5810 (Antikainen et al. 2002), in SlpB of L. crispatus strains K313 and K2-4-3 (Hu et al. 2011; Sun et al. 2012), in SlpA of L. crispatus ZJ001 (Chen et al. 2009), in SlpA of L. brevis ATCC 8287 (Åvall-Jääskeläinen et al. 2008) and in the S-layer protein of L. hilgardii B706 (Dohm et al. 2011).
The first five strains listed above belong to the former L. acidophilus group (Johnson et al. 1980), and the amino acid sequences of their S-layer proteins show extensive similarity in the C-terminal region (Hu et al. 2011; Sun et al. 2012). Extending the alignment to the S-layer proteins of organisms of the closely related L. helveticus (Collins et al. 1991; Felis and Dellaglio 2007) also indicates a remarkable conservation of the C-terminal region (Antikainen et al. 2002), suggesting a conserved function for this region. Indeed in SA (Smit et al. 2001), CbsA (Antikainen et al. 2002) and in the SlpB proteins of K313 (Sun et al. 2012) and K2-4-3 (Hu et al. 2011), the C-terminal part of the S-layer protein, approximately 125 amino acids in length, is responsible for binding to the cell envelope. In SA, only one of the two 65-amino-acid repeats of the cell wall binding region is necessary for binding, and an enhancing role for the other repeat has been suggested (Smit and Pouwels 2002). In contrast, in the S-layer proteins of L. brevis ATCC 8287 and L. hilgardii B706, organisms completely unrelated to L. acidophilus, the N-terminal parts of the proteins comprise the cell wall binding region (Åvall-Jääskeläinen et al. 2008; Dohm et al. 2011). Nevertheless, all of the S-layer proteins described earlier have a similar charge distribution with a high predicted pI in the cell wall binding part (Smit et al. 2001; Antikainen et al. 2002; Åvall-Jääskeläinen et al. 2008; Sun et al. 2012; Dohm et al. 2011). Thus, an electrostatic interaction occurring between the cell wall binding regions and the negatively charged cell wall polymers has been proposed (Antikainen et al. 2002).
Lactobacillus S-layer proteins do not possess surface layer homology domains (Lupas et al. 1994), repeated motifs 50–60 amino acids in length, which are known to be involved in the binding of many S-layer proteins to the cell wall, for instance, those of Bacillus anthracis (Mesnage et al. 1999), Geobacillus stearothermophilus PV72/p2 (Ries et al. 1997; Sára et al. 1998), Lysinibacillus sphaericus CCM 2177 (Ilk et al. 1999) and Clostridium thermocellum NCIMB 10682 (Lemaire et al. 1998). Instead two repeated amino acid sequences with homology to the tyrosine/phenylalanine containing carbohydrate-binding motifs of clostridial toxins and streptococcal glucosyltransferases (Wren 1991; von Eichel-Streiber et al. 1992) are present in the cell wall binding regions of the above-mentioned SA, CbsA and SlpA of L. acidophilus, L. crispatus and L. brevis, respectively. These motifs are also found in the C-terminal parts of the silent S-layer protein SB of L. acidophilus ATCC 4356, the S-layer protein of L. helveticus CNRZ 892 and the non-S-layer proteins of lactic acid bacteria known to be associated with the cell envelope (Smit et al. 2001; Åvall-Jääskeläinen et al. 2008). The cell wall receptors of the S-layer proteins of L. acidophilus and L. crispatus have indeed been shown to be carbohydrates: SA of L. acidophilus ATCC 4356 and CbsA and SlpB of L. crispatus JCM 5810 and K313, respectively, interact with teichoic acids (Antikainen et al. 2002; Smit and Pouwels 2002; Sun et al. 2012); CbsA also binds to lipoteichoic acids isolated from Staphylococcus aureus and Streptococcus faecalis, but not to the teichuronic acid/polysaccharide fraction of the cell wall of L. crispatus JCM 5810 (Antikainen et al. 2002). On the contrary, the cell wall components interacting with the S-layer proteins of L. brevis ATCC 8287 and L. hilgardii apparently are non-teichoic acid polysaccharides as trichloroacetic acid (TCA) treatment of the cell walls had no effect on the interaction (Åvall-Jääskeläinen et al. 2008; Dohm et al. 2011); TCA treatment at +4 °C has been reported to selectively remove teichoic acids (Hancock and Poxton 1988). Supporting this is the fact that, in earlier studies, the cell walls of L. brevis and L. buchneri were shown to contain neutral polysaccharides that were suggested to be involved in the anchoring of the S-layer protein via hydrogen bonding (Masuda and Kawata 1980, 1985). In contrast to the well-characterized exopolysaccharides of lactic acid bacteria (De Vuyst and Degeest 1999; Welman and Maddox 2003), the cell wall polysaccharides of lactobacilli other than teichoic acids are poorly known. The detailed structure of a neutral wall polysaccharide of L. casei has been determined (Nagaoka et al. 1990), but no precise structures for such polysaccharides of L. buchneri or L. brevis strains are available.
In SA of L. acidophilus, CbsA of L. crispatus and SlpA of L. crispatus ZJ001 and L. brevis, the more variable part of the protein (N-terminal in the S-layer proteins of L. acidophilus or L. crispatus, C-terminal in SlpA of L. brevis) is responsible for the self-assembly of the S-layer protein monomers to a periodic S-layer lattice, as shown by the mapping of the self-assembly properties of truncated recombinant S-layer proteins by transmission electron microscopy (Sillanpää et al. 2000; Smit et al. 2001, 2002; Antikainen et al. 2002; Åvall-Jääskeläinen et al. 2008; Chen et al. 2009); these regions thus most likely represent the surface-exposed parts of the proteins. SA, CbsA and SlpA of L. brevis, and apparently also the other S-layer proteins described earlier, can thus be viewed as two-domain proteins with a cell wall binding domain and a self-assembly domain facing the extracellular environment, the former being not or less exposed to the environment. In SA, this view is supported by extensive proteolytic and chemical breakdown experiments (Smit et al. 2001); moreover, in the S-layer proteins of L. brevis and L. hilgardii, the C-terminal parts were found to be trypsin resistant (Åvall-Jääskeläinen et al. 2008; Dohm et al. 2011).
More detailed information is available about the structures of the self-assembly domains of SA of L. acidophilus ATCC 4356, CbsA of L. crispatus JCM 5810 and SlpA of L. brevis ATCC 8287. According to insertion and deletion mutagenesis and proteolytic studies of SA, the N-terminal self-assembly domain is probably organized into two subdomains of approximately 12 and 18 kDa, linked by a surface-exposed loop. The very N-terminus of SA is not critical for crystallization and is probably buried inside the domain or facing the cell wall or S-layer pore. Conserved regions and regions predicted to form secondary structures in SA are necessary for the formation of a regular lattice (Smit et al. 2002). The lack of necessity of the very N-terminal end and the importance of the conserved regions for self-assembly have also been demonstrated for CbsA, where the conserved, valine-rich flanking regions of the self-assembly domain are especially important for the formation of the S-layer lattice and may have a role in directing the formation of a regular polymer; changes in the morphology of the self-assembly products of CbsA fragments were seen accompanying a mutation of even a single residue in these conserved border regions as well as with the stepwise truncation of the self-assembly region. The C-terminal cell wall binding domain has a stabilizing role in the recrystallization of CbsA monomers by allowing a more efficient sheet formation (Antikainen et al. 2002). The locations of a set of defined amino acids in SlpA of L. brevis have been mapped by cysteine-scanning mutagenesis combined with sulfhydryl modification. The analysis, based on measuring the surface accessibilities of the residues when the protein is in a monomeric or self-assembled form, grouped the residues according to their locations within the polymerized S-layer structure: to those located in the interior of the subunit, to those on the outer surface of the polymerized protein layer, to those on the inner surface of the layer and to those likely located in the subunit–subunit interface and pore or inner surface of the layer. The results confirmed the two-domain structure of SlpA and revealed several sites of high surface accessibility (Vilen et al. 2009).
Functions of Lactobacillus S-layer proteins
Adhesive functions
The most often proposed function for Lactobacillus S-layers is the mediation of bacterial adherence to various targets. In a number of studies, the loss of the S-layer protein from the bacterial surface by chemical means (Kos et al. 2003; Garrote et al. 2004; Frece et al. 2005; Chen et al. 2007; Jakava-Viljanen and Palva 2007; Tallon et al. 2007) or the covering of the layer by other molecules during prolonged cultivation (Schneitz et al. 1993) has been shown to decrease adhesion to different targets, but the role of the S-layer protein in adherence in these studies has not been directly demonstrated. The haemagglutinating activity of L. acidophilus JCM 1034 and the mucin binding activities of related strains were shown to be linked to their S-layer proteins, although the involvement of other guanidine hydrochloride-extractable components of the cell wall in this lectin-like activity could not be excluded, and/or the effect of aggregation of the S-layer proteins possibly causing unspecific effects could not be completely ruled out (Yamada et al. 1994; Takahashi et al. 1996). Likewise, in the study of Golowczyc et al. (2009), where the carbohydrate-dependent co-aggregation of L. kefir with yeast or red blood cells was suggested to be S-layer-mediated, conclusions were drawn from the effects of LiCl and SDS treatments of L. kefir cells, and the solubility of the S-layer proteins in the LiCl extracts of L. kefir used in the aggregation assays was not demonstrated. Also, in the study of Uchida et al. (2006), which showed an interaction between the S-layer protein of L. brevis OLL 2772 and human blood group A antigen by a surface plasmon resonance assay, a dialysed guanidine hydrochloride extract of bacterial cells was used as an analyte, leaving the effects of the levels of purity and solubility of the protein debatable.
The role of a Lactobacillus S-layer protein in bacterial adherence has been unequivocally shown in a few instances, where recombinant S-layer proteins (Toba et al. 1995; Sun et al. 2012), S-layer-negative mutants (Konstantinov et al. 2008), highly purified monomeric proteins (de Leeuw et al. 2006) or a surface display system for the S-layer protein (Hynönen et al. 2002) was used.
Recombinant forms of CbsA of L. crispatus JCM 5810 (Toba et al. 1995; Sillanpää et al. 2000) and SlpB of L. crispatus K313 (Sun et al. 2012) both bind collagen types I and IV. In contrast, the recombinant form of the non-expressed SlpB protein of L. crispatus JCM 5810, which showed 43 % sequence identity to CbsA at the amino acid level, does not bind these collagens (Sillanpää et al. 2000). L. crispatus JCM 5810 cells also bind to collagen-rich regions of chicken colon in vitro, while guanidine hydrochloride-treated cells are unable to bind, suggesting biological relevance for the observed collagen binding of CbsA (Sillanpää et al. 2000). The N-terminal amino acid residues at position 31–274 of mature CbsA are needed for collagen binding, and mostly the same residues (32–271) are needed for the reassembly of CbsA monomers to an S-layer, suggesting the dependence of collagen binding on the periodic structure (Sillanpää et al. 2000). The display of CbsA on the surface of a non-S-layered L. casei strain through a PrtP cell wall anchor rendered the recombinant cells able to bind collagens, although the anchoring system probably does not allow the monomers to form a true S-layer (Martinez et al. 2000). While the sequence similarity between CbsA and the S-layer protein SlpB of L. crispatus K313 is restricted to the C-terminal cell wall binding region, the N-terminal part of SlpB also binds collagen. More than 341 N-terminal amino acid residues are needed for binding (Sun et al. 2012), but no data are available if the collagen binding and polymerization require the same amino acid residues as demonstrated for CbsA. The recombinant protein comprising the N-terminal part of the S-layer protein of L. crispatus strain ZJ001, in turn, binds to detached HeLa cells (Chen et al. 2009).
A further well-characterized adhesive Lactobacillus S-layer protein is SlpA on L. acidophilus NCFM cells, which binds to the dendritic cell-specific ICAM-3-grabbing nonintegrin (DC-SIGN) receptor on human immature dendritic cells, leading to cytokine production and modulation of the immune response. The slpA knock-out mutant expressing SlpB and SlpX is significantly reduced in binding to DC-SIGN, and the interaction leads to the induction of different cytokines (Konstantinov et al. 2008). Initially, a role for SlpA of L. acidophilus NCFM was demonstrated in binding to Caco-2 cells as the binding of the knock-out mutant of the gene in locus LBA 1377 was decreased by 84 % compared with the wild type (Buck et al. 2005). However, the gene in locus LBA 1377 was subsequently annotated as a putative mucus binding protein, and SlpA was localized in locus LBA 0169. Nevertheless, SlpA encoded by the gene in LBA 0169 has later been detected on the surface of several Caco-2 cell binding L. acidophilus isolates (Ashida et al. 2011). It is identical in sequence with the SA protein of L. acidophilus ATCC 4356, suggesting that these strains might have similar adhesive and immunomodulatory properties as well as surface layer-associated murein hydrolase activity (see “Protective, enzymatic and other functions”).
Finally, the S-layer protein SlpA of L. brevis ATCC 8287 mediates the binding of the bacterial cells to several human epithelial cell lines and fibronectin, as revealed by expressing fragments of slpA in a surface display system based on the H7 flagella of Escherichia coli. Eighty-one amino acids from the N-terminal part of SlpA were sufficient to confer binding to epithelial cells (Hynönen et al. 2002). The binding functions of SlpA were verified using a non-adhesive Lactococcus strain, in which a nicin-inducible surface display system with a cell wall-anchoring peptide from lactococcal AcmA was used to display the binding region of SlpA on the cell surface (Åvall-Jääskeläinen et al. 2003). Khang et al. (2009), in turn, used purified SlpA– green fluorescent protein (GFP) fusion proteins to show the binding of SlpA to undifferentiated human HT-29 cells, although more attention could have been focused on controlling the specificity of the interaction in this study. The binding of SlpA to extracellular matrix proteins has been further confirmed by de Leeuw et al. (2006), who demonstrated a direct interaction between the chromatographically purified, monomeric form of SlpA and soluble fibronectin or laminin by surface plasmon resonance assays. The binding mechanisms to fibronectin and laminin were found to be different and proposed to be mediated by different regions of SlpA.
In addition to the above-mentioned examples of specific binding, the S-layers of lactobacilli may have a non-specific enhancing effect on binding to surfaces, like those encountered in the gastrointestinal or urogenital tract, as they are generally hydrophobic and may thus enhance adhesion to hydrophobic surfaces (van der Mei et al. 2003). This effect is, however, dependent on the ionic strength of the environment (Vadillo-Rodríguez et al. 2005). Some Lactobacillus S-layers, but not all, have even been found to change their surface hydrophobicity in response to environmental ionic strength, thus possibly offering different binding capacities. In the case of the SA protein of L. acidophilus ATCC 4356, the decrease in hydrophobicity associated with higher environmental ionic strength is hypothesized to be due to the shrinkage of the S-layer and the consequent partial exposure of the inner, more hydrophilic C-terminal domain (Vadillo-Rodriguez et al. 2004).
Protective, enzymatic and other functions
To date, a couple of functions other than adhesion have been shown or proposed for Lactobacillus S-layer proteins. The presence of the S-layer protein decreases the susceptibility of L. helveticus ATCC 12046 to mutanolysin (Lortal et al. 1992), the susceptibility of L. acidophilus M92 to gastric and pancreatic juice (Frece et al. 2005) and the susceptibility of L. hilgardii wine isolate B706 to wine-related conditions like the presence of copper sulphate or tannic acid (Dohm et al. 2011). On the other hand, the S-layer proteins of brewery isolates of L. brevis were deduced not to act as barriers for the hop bittering substance isohumulone (Yasui et al. 1995). The auxiliary S-layer component SlpX of L. acidophilus NCFM probably affects the permeability of the S-layer as the slpX-negative mutant is more susceptible to SDS and more resistant to bile than the wild type (Goh et al. 2009). The C-terminal part of the S-layer protein SA of L. acidophilus ATCC 4356 has been shown to have murein hydrolase (endopeptidase) activity against the cell wall of, e.g., Salmonella enterica (Prado Acosta et al. 2008), but the biological relevance of this finding was not investigated. A role as a phage receptor has been suggested for the S-layer protein of L. helveticus CNRZ 892 (Callegari et al. 1998).
Applications of Lactobacillus S-layer proteins
Vaccine development
During the recent years, the number of applications developed or suggested for Lactobacillus S-layer proteins has gradually increased. One of the fields currently extensively studied is the construction of S-layer fusion proteins for use in immunization in man or animals. Especially the development of live Lactobacillus strains carrying S-layers composed of hybrid proteins on their surface is of interest as such strains have potential for use as live mucosal vaccines. Several findings support this approach: (1) The non-pathogenicity of lactobacilli and their ability to survive the passage through the gastrointestinal tract enables a simple, safe and efficient route of oral antigen delivery; (2) A clear relationship exists between antigen expression levels and immune response (Grangette et al. 2001; Seegers 2002), and surface display with an S-layer protein as a carrier results in the simultaneous expression of the foreign peptide as hundreds of thousands of regularly arranged copies on the cell; (3) Lactobacillus cells as well as surface layer arrays have intrinsic adjuvant properties (Smith et al. 1993; Miettinen et al. 1996; Maassen et al. 2000; Seegers 2002; Beganović et al. 2011), and the simultaneous display of immunomodulating molecules in the S-layer could further enhance or direct the immune response; (4) As antigen carrier systems can be significantly improved by the co-display of adhesins (Cano et al. 1999; Liljeqvist et al. 1999), the various binding functions described earlier might prove advantageous in the targeted delivery of antigenic molecules. For instance, the identification of the S-layer protein of L. acidophilus NCFM as the binding ligand for the dendritic cell-specific antigen DC-SIGN (Konstantinov et al. 2008) makes this probiotic strain or its S-layer an attractive tool for oral vaccine design. So far, only a system utilizing L. acidophilus NCFM cells, not yet its S-layer, as a carrier for an antigen with a small dendritic cell-targeting peptide has been developed (Mohamadzadeh et al. 2009).
The development of Lactobacillus vaccine carriers based on hybrid S-layers is at an early stage. Small model peptides have been displayed in each monomer of the S-layer of L. brevis ATCC 8287 (Åvall-Jääskeläinen et al. 2002) and L. acidophilus ATCC 4356 (Smit et al. 2002) by chromosomal integration based on homologous recombination. Similarly, surface display of GFP in the S-layer proteins on chicken Lactobacillus isolates has been achieved by utilizing the gene fragment encompassing the expression and secretion signals and the region encoding the cell wall binding domain of the S-layer protein of L. crispatus (Mota et al. 2006). As a prerequisite for hybrid S-layer-based vaccine development, a systematic mapping of surface-accessible amino acids has been performed for the S-layer protein of L. brevis ATCC 8287 (Vilen et al. 2009). Apart from hybrid S-layer proteins, non-adhesive antigen delivery vehicles like lactococci have been rendered adhesive by the surface display of adhesive S-layer proteins or S-layer-derived peptides such as those of L. crispatus JCM 5810 (Martinez et al. 2000) or L. brevis ATCC 8287 (Åvall-Jääskeläinen et al. 2003). Preliminary experiments have also been performed in the field of passive immunization by utilizing the epithelial cell binding S-layer protein of L. brevis KCTC 3102 (ATCC 8287) as a purified, immunoglobulin binding fusion protein to target antibodies to the intestinal surfaces of calves in order to prevent neonatal diarrhoea (Khang et al. 2009). In this small field study, a higher recovery of calves from diarrhoea was obtained by administering antiviral and antibacterial antibodies in combination with the fusion protein than by administering the antibodies alone, although the mechanism of protection remained speculative.
Applications based on anti-adhesive and anti-infectious effects
Another potential application is the use of S-layers or S-layered lactobacilli as anti-adhesive agents or as other therapeutic or preventative measures against infectious diseases. In many studies, however, the anti-adhesive effects observed for S-layer proteins against different pathogens, as described, e.g., for the S-layer proteins of L. crispatus (Horie et al. 2002a; Chen et al. 2007), L. helveticus (Sherman et al. 2005; Johnson-Henry et al. 2007) and L. kefir (Golowczyc et al. 2007), cannot unequivocally be attributed to the surface layer proteins per se, as the dialysed extracts used in the inhibition studies apparently contained also other LiCl or guanidium hydrochloride-extractable cell surface components as well as aggregates of the S-layer protein, and thus the specificities of the inhibitions were compromised. The same holds true for the study of Martínez et al. (2012), which demonstrated the inhibition of JUNV infection by the surface protein extract of L. acidophilus ATCC 4356 in a DC-SIGN expressing cell culture model, although the interaction between SlpA of ATCC 4356/NCFM and DC-SIGN has previously been demonstrated (Konstantinov et al. 2008). Similarly, the results of the study of Carasi et al. (2011), which showed the potential of L. kefir S-layer proteins for decreasing the cytopathic effect of Clostridium difficile culture supernatants or toxins to Vero cells, were obtained using unpurified LiCl extracts of L. kefir cells and thus cannot be considered as fully conclusive.
The identification of the S-layer protein of L. acidophilus NCFM as the binding ligand for the dendritic cell-specific antigen DC-SIGN and the different cytokine responses elicited by SlpA and the alternative S-layer protein SlpB (Konstantinov et al. 2008) have raised interest in studying the contribution of the S-layer protein of NCFM to its probiotic action. There is an association between DC-SIGN polymorphisms and allergic sensitization, and the colonization of 1-month-old infants by L. acidophilus slightly decreases the risk of allergic dermatitis (Penders et al. 2010), but still the role of the SlpA–DC-SIGN interaction in immunological tolerance and its biological significance is far from clear. On the other hand, in the cellular mechanisms of inflammatory bowel disease, lipoteichoic acids of L. acidophilus NCFM seem to have a major role, as pre-treatment of mice by LTA-negative L. acidophilus NCFM ameliorated dextran sulphate sodium-induced inflammatory colitis (Mohamadzadeh et al. 2011). Interestingly, the presence of SlpA on an slpB - slpX - mutant actually increases the pro-inflammatory action of LTA compared with the LTA-expressing parental strain (Zadeh et al. 2012). Thus, a role for SlpB and SlpX in regulating LTA-induced inflammation has been suggested (Lightfoot and Mohamadzadeh 2013). A mutant lacking SlpB and SlpX but carrying SlpA also tends to be cleared from the mouse gastrointestinal tract more rapidly than the wild type, but the mechanism is not known (Zadeh et al. 2012). Some indications about the contribution of the S-layer protein of L. acidophilus ATCC 4356/NCFM to probiotic action were obtained in the study of Li et al. (2011b), in which the chromatographically purified S-layer protein SlpA was shown to counteract a Salmonella-induced transepithelial electric resistance decrease and IL-8 secretion as well as to inhibit Salmonella-induced F-actin rearrangements and JNK and p38 activation in Caco-2 cells. In another study, the same protein was shown to activate the ERK1/2 signaling pathway and to inhibit caspase-3 activity in Salmonella-infected Caco-2 cells, thereby decreasing Salmonella-induced Caco-2 cell apoptosis and cell damage (Li et al. 2011a). Interestingly, apart from probiotics, even the S-layer protein of a dairy strain of L. helveticus was found to reduce NF-κB activation in Caco-2 cells while triggering the expression of TLR2-mediated pro-inflammatory factors in human and mouse macrophages, thus showing stimulatory effects on innate immunity. In this study, special effort was exerted to demonstrate the purity of the S-layer preparation used (Taverniti et al. 2012).
A novel application has been suggested for the S-layer protein SA of L. acidophilus ATCC 4356. The murein hydrolase activity of the C-terminal part of SA, shown by the degradation of cell wall preparations of Gram-negative pathogens (Prado Acosta et al. 2008), acts synergistically with the well-documented antibacterial agent, nisin, against bacterial pathogens. The combination of these two inhibits the growth of both Gram-positive and Gram-negative pathogens, as exemplified by Salmonella enterica, Staphylococcus aureus and Bacillus cereus, through a mechanism that involves the dissipation of the transmembrane proton motive force (Prado-Acosta et al. 2010).
Chemical conjugates and liposomes
Some biochemical and physical studies of isolated Lactobacillus S-layer proteins aiming at biotechnological or clinical applications have also been initiated. Small molecular probes like biotin or fluorescein isothiocyanate have been conjugated to purified S-layer proteins of L. brevis using amine-based coupling chemistry. The S-layer protein bioconjugates formed, purified by affinity chromatography, were capable of self-assembling into regular layers, where the surface coverage of the conjugated molecules is homogeneous and the density controllable. The method offers a way to display several different and high molecular weight molecules at an interface (Sampathkumar and Gilchrist 2004). Further, positively charged liposomes have been coated by the S-layer proteins of L. brevis and L. kefir (Hollmann et al. 2007, 2010a). Importantly for future vaccine applications, the S-layer proteins markedly increased the stability of the liposomes under unfavourable conditions, e.g. at low pH, at high temperature or in the presence of bile salts or pancreatic extract, as measured by the release of a fluorescent marker compound, and the effect could be further enhanced by cross-linking the proteins with glutaraldehyde (Hollmann et al. 2007). The stabilizing effect was shown to be based on the neutralization of the charge repulsion between stearylamine molecules in the liposome, leading to increased acyl chain packing and membrane rigidity. The glycosylated S-layer protein of L. kefir had higher affinity to the liposomes than the non-glycosylated one of L. brevis (Hollmann et al. 2010a), but no striking differences were found between the liposome-stabilizing effects of the two proteins (Hollmann et al. 2007). The kinetics of the interaction between the S-layer protein and a lipid monolayer was found to be dependent on the composition of the membrane and could be modulated by components that modify the hydration state of the lipid interface (Hollmann et al. 2010b).
Expression/secretion signals in heterologous gene expression
The expression and/or secretion signals of Lactobacillus S-layer protein genes have also been utilized in biotechnological applications (Savijoki et al. 1997; Kahala and Palva 1999; Novotny et al. 2005; Lizier et al. 2010; Zhang et al. 2010a). The region encompassing the double promoter and the ribosome-binding sequence up to the start of the slpA gene of L. brevis ATCC 8287 (Kahala and Palva 1999), or the region containing additionally the slpA signal peptide gene sequence (Savijoki et al. 1997), have been used in Lactobacillus and Lactococcus hosts for intracellular or extracellular protein production. Using slpA expression and secretion signals, secretion levels of beta-lactamase up to 80 mg/l have been achieved. Differences exist between the recognition efficiency of the signals in different hosts; high-level extracellular protein production with slpA signals was achieved in Lactococcus lactis and Lactobacillus plantarum and moderate production in L. gasseri, while in L. casei the expression signals were not recognized (Savijoki et al. 1997). On the other hand, the promoter region of L. acidophilus ATCC 4356 S-layer protein gene was highly active in L. casei (Boot et al. 1996b) but functioned poorly in L. reuteri (Lizier et al. 2010). The transcriptional activity in heterologous hosts could be significantly improved or decreased by the modification of native slp gene promoter, and both strain- and context-dependent effects of the introduced sequences have been detected (McCracken and Timms 1999). Adding merely the signal peptide encoding sequence of slpA from L. brevis ATCC 8287 upstream of the 5′ end of the human interferon alpha gene increased the secretion efficiency of interferon alpha in L. lactis threefold compared to the signal peptide encoding sequence of lactococcal Usp45, but the total interferon production was lower in the strain with the slpA signal peptide encoding sequence (Zhang et al. 2010a). The recent development of a counterselective gene replacement system for the chromosomal integration of genes in L. acidophilus (Goh et al. 2009) has enabled protein production using chromosomally located S-layer protein promoters in lactobacilli. The promoter of the S-layer protein gene slpA of L. acidophilus NCFM was found to direct the expression of the reporter gene gusA3, leading to a higher expression level than that obtained from a plasmid, when the reporter gene was placed between the stop codon and the transcriptional terminator of slpA (Douglas and Klaenhammer 2011).
Concluding remarks
Present knowledge about Lactobacillus S-layer proteins supports the view of Gram-positive S-layer proteins as two-domain entities, where one domain is responsible for cell wall binding and the other for the self-assembly of the regular surface layer. The common theme of carbohydrates as the binding sites for S-layer proteins in the cell walls of Gram-positive bacteria is also supported, although the detailed anchoring molecules and mechanisms vary among different lactobacilli. Biophysical methods are increasingly utilized in the structural studies of S-layers, and together with computer modelling-based methods they will probably allow for high-resolution structures of Lactobacillus S-layer proteins, which currently are scarce owing to difficulties in obtaining high-quality crystals for X-ray crystallography.
As food-grade and potentially probiotic organisms, lactobacilli are excellent candidates for health-related applications like live oral vaccines, where their ability to survive in the gastrointestinal tract could be utilized and their S-layer proteins could be used as carriers of antigens or other medically important molecules, possibly in combination with immunostimulatory or adhesive molecules. In this approach, the polymeric nature and inherent adjuvant properties of S-layers are apparently an advantage. Further, immobilization of recombinant S-layer proteins combined with the display of foreign molecules in the S-layer forms the basis for the development of different solid-phase reagents, such as biocatalysts, diagnostic devices, biosensors and biosorbents, where the typical positive charge of the cell wall-binding domain of Lactobacillus S-layer proteins could augment the immobilization. While most of the biotechnological applications of S-layer proteins so far have been designed for the S-layer proteins of thermophilic bacilli, the increasing knowledge about the structure and biology of Lactobacillus S-layer proteins, as well as the developing tools to genetically manipulate these organisms, will pave the way to applications utilizing the S-layer proteins of these beneficial and easily cultivable bacteria.
Acknowledgments
This work was performed in the Centre of Excellence on Microbial Food Safety Research, Academy of Finland.
References
- Ahn JS, Chandramohan L, Liou LE, Bayles KW. Characterization of CidR-mediated regulation in Bacillus anthracis reveals a previously undetected role of S-layer proteins as murein hydrolases. Mol Microbiol. 2006;62:1158–1169. doi: 10.1111/j.1365-2958.2006.05433.x. [DOI] [PubMed] [Google Scholar]
- Altermann E, Russell WM, Azcarate-Peril A, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. PNAS. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antikainen J, Anton L, Sillanpää J, Korhonen TK. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol. 2002;2:381–394. doi: 10.1046/j.1365-2958.2002.03180.x. [DOI] [PubMed] [Google Scholar]
- Ashida N, Yanagihara S, Shinoda T, Yamamoto N. Characterization of adhesive molecule with affinity to Caco-2 cells in Lactobacillus acidophilus by proteome analysis. J Biosci Bioeng. 2011;112:333–337. doi: 10.1016/j.jbiosc.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Ausiello CM, Cerquetti M, Fedele G, Spensieri F, Palazzo R, Nasso M, Frezza S, Mastrantonio P. Surface layer proteins from Clostridium difficile induce inflammatory and regulatory cytokines in human monocytes and dendritic cells. Microbes Infect. 2006;8:2640–2646. doi: 10.1016/j.micinf.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Åvall-Jääskeläinen S, Palva A. Lactobacillus surface layers and their applications. FEMS Microbiol Rev. 2005;29:511–529. doi: 10.1016/j.fmrre.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Åvall-Jääskeläinen S, Kylä-Nikkilä K, Kahala M, Miikkulainen-Lahti T, Palva A. Surface display of foreign epitopes on the Lactobacillus brevis S-layer. Appl Environ Microbiol. 2002;68:5943–5951. doi: 10.1128/AEM.68.12.5943-5951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åvall-Jääskeläinen S, Lindholm A, Palva A. Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl Environ Microbiol. 2003;69:2230–2236. doi: 10.1128/AEM.69.4.2230-2236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åvall-Jääskeläinen S, Hynönen U, Ilk N, Pum D, Sleytr UB, Palva A. Identification and characterization of domains responsible for self-assembly and cell wall binding of the surface layer protein of Lactobacillus brevis ATCC 8287. BMC Microbiol. 2008;8:165. doi: 10.1186/1471-2180-8-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson L (1998) Lactic acid bacteria: Classification and physiology. Lactic acid bacteria: microbiology and functional aspects In: Salminen S, von Wright A (eds), pp. 1–73. Marcel Dekker, New York.
- Azcarate-Peril MA, Altermann E, Goh YJ, Sanozky-Dawes RB, Pfeiler EA, O'Flaherty S, Buck BL, Dobson A, Duong T, Miller MJ, Barrangou R, Klaenhammer TR. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl Environ Microbiol. 2008;74:4610–4625. doi: 10.1128/AEM.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DC, Thorne KJ. Spheroplasts of Lactobacillus casei and the cellular distribution of bactoprenol. J Cell Sci. 1970;7:755–785. doi: 10.1242/jcs.7.3.755. [DOI] [PubMed] [Google Scholar]
- Beganović J, Frece J, Kos B, Leboš Pavunc A, Habjanič K, Sušković J. Functionality of the S-layer protein from the probiotic strain Lactobacillus helveticus M92. Antonie Van Leeuwenhoek. 2011;100:43–53. doi: 10.1007/s10482-011-9563-4. [DOI] [PubMed] [Google Scholar]
- Boot HJ, Pouwels PH. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol Microbiol. 1996;21:1117–1123. doi: 10.1046/j.1365-2958.1996.711442.x. [DOI] [PubMed] [Google Scholar]
- Boot HJ, Kolen CP, Pouwels PH. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J Bacteriol. 1995;177:7222–7230. doi: 10.1128/jb.177.24.7222-7230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot HJ, Kolen CP, Pouwels PH. Interchange of the active and silent S-layer protein genes of Lactobacillus acidophilus by inversion of the chromosomal slp segment. Mol Microbiol. 1996;21:799–809. doi: 10.1046/j.1365-2958.1996.401406.x. [DOI] [PubMed] [Google Scholar]
- Boot HJ, Kolen CP, Andreadaki FJ, Leer RJ, Pouwels PH. The Lactobacillus acidophilus S-layer protein gene expression site comprises two consensus promoter sequences, one of which directs transcription of stable mRNA. J Bacteriol. 1996;178:5388–5394. doi: 10.1128/jb.178.18.5388-5394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot HJ, Kolen CP, Pot B, Kersters K, Pouwels PH. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology. 1996;142:2375–2384. doi: 10.1099/00221287-142-9-2375. [DOI] [PubMed] [Google Scholar]
- Brahamsha B. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc Natl Acad Sci U S A. 1996;93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck BL, Altermann E, Svingerud T, Klaenhammer TR. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2005;71:8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat E, Priest FG. Lactobacillus suntoryeus sp. nov., isolated from malt whisky distilleries. Int J Syst Evol Microbiol. 2005;55:31–34. doi: 10.1099/ijs.0.63266-0. [DOI] [PubMed] [Google Scholar]
- Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, Dell A, Dougan G, Fairweather N. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol Microbiol. 2001;40:1187–1199. doi: 10.1046/j.1365-2958.2001.02461.x. [DOI] [PubMed] [Google Scholar]
- Callanan M, Kaleta P, O'Callaghan J, O'Sullivan O, Jordan K, McAuliffe O, Sangrador-Vegas A, Slattery L, Fitzgerald GF, Beresford T, Ross RP. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol. 2008;190:727–735. doi: 10.1128/JB.01295-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari ML, Riboli B, Sanders JW, Cocconcelli PS, Kok J, Venema G, Morelli L. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology. 1998;144(Pt 3):719–726. doi: 10.1099/00221287-144-3-719. [DOI] [PubMed] [Google Scholar]
- Cano F, Liljeqvist S, Nguyen TN, Samuelson P, Bonnefoy JY, Ståhl S, Robert A. A surface-displayed cholera toxin B peptide improves antibody responses using food-grade staphylococci for mucosal subunit vaccine delivery. FEMS Immunol Med Microbiol. 1999;25:289–298. doi: 10.1111/j.1574-695X.1999.tb01354.x. [DOI] [PubMed] [Google Scholar]
- Carasi P, Trejo FM, Pérez PF, De Antoni GL, Serradell Mde L. Surface proteins from Lactobacillus kefir antagonize in vitro cytotoxic effect of Clostridium difficile toxins. Anaerobe. 2011;18:135–142. doi: 10.1016/j.anaerobe.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Cerquetti M, Molinari A, Sebastianelli A, Diociaiuti M, Petruzzelli R, Capo C, Mastrantonio P. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb Pathog. 2000;28:363–372. doi: 10.1006/mpat.2000.0356. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu J, Shuai J, Chen J, Zhang Z, Fang W. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157:H7 and Salmonella typhimurium. Int J Food Microbiol. 2007;115:307–312. doi: 10.1016/j.ijfoodmicro.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Chen X, Chen Y, Li X, Chen N, Fang W. Characterization of surface layer proteins in Lactobacillus crispatus isolate ZJ001. J Microbiol Biotechnol. 2009;19:1176–1183. [PubMed] [Google Scholar]
- Collins MD, Rodrigues U, Ash C, Aguirre M, Farrow JAE, Martinez-Murcia A, Phillips BA, Williams AM, Wallbanks S. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett. 1991;77:5–12. doi: 10.1111/j.1574-6968.1991.tb04313.x. [DOI] [Google Scholar]
- de la Fuente-Núñez C, Mertens J, Smit J, Hancock RE. The bacterial surface layer provides protection against antimicrobial peptides. Appl Environ Microbiol. 2012;78:5452–5456. doi: 10.1128/AEM.01493-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw E, Li X, Lu W. Binding characteristics of the Lactobacillus brevis ATCC 8287 surface layer to extracellular matrix proteins. FEMS Microbiol Lett. 2006;260:210–215. doi: 10.1111/j.1574-6968.2006.00313.x. [DOI] [PubMed] [Google Scholar]
- De Vuyst L, Degeest B. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev. 1999;23:153–177. doi: 10.1111/j.1574-6976.1999.tb00395.x. [DOI] [PubMed] [Google Scholar]
- Dohm N, Petri A, Schlander M, Schlott B, König H, Claus H. Molecular and biochemical properties of the S-layer protein from the wine bacterium Lactobacillus hilgardii B706. Arch Microbiol. 2011;193:251–261. doi: 10.1007/s00203-010-0670-9. [DOI] [PubMed] [Google Scholar]
- Doig P, Emödy L, Trust TJ. Binding of laminin and fibronectin by the trypsin-resistant major structural domain of the crystalline virulence surface array protein of Aeromonas salmonicida. J Biol Chem. 1992;267:43–49. [PubMed] [Google Scholar]
- Douglas GL, Klaenhammer TR. Directed chromosomal integration and expression of the reporter gene gusA3 in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2011;77:7365–7371. doi: 10.1128/AEM.06028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelseer E, Schocher I, Sára M, Sleytr UB. The S-layer from Bacillus stearothermophilus DSM 2358 functions as an adhesion site for a high-molecular-weight amylase. J Bacteriol. 1995;177:1444–1451. doi: 10.1128/jb.177.6.1444-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelseer EM, Schocher I, Sleytr UB, Sára M. Evidence that an N-terminal S-layer protein fragment triggers the release of a cell-associated high-molecular-weight amylase in Bacillus stearothermophilus ATCC 12980. J Bacteriol. 1996;178:5602–5609. doi: 10.1128/jb.178.19.5602-5609.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt H. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J Struct Biol. 2007;160:115–124. doi: 10.1016/j.jsb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Engelhardt H. Mechanism of osmoprotection by archaeal S-layers: a theoretical study. J Struct Biol. 2007;160:190–199. doi: 10.1016/j.jsb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Fagan RP, Albesa-Jove D, Qazi O, Svergun DI, Brown KA, Fairweather NF. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol Microbiol. 2009;71:1308–1322. doi: 10.1111/j.1365-2958.2009.06603.x. [DOI] [PubMed] [Google Scholar]
- Felis GE, Dellaglio F. Taxonomy of Lactobacilli and Bifidobacteria. Curr Issues Intest Microbiol. 2007;8:44–61. [PubMed] [Google Scholar]
- Fouet A. The surface of Bacillus anthracis. Mol Aspects Med. 2009;30:374–385. doi: 10.1016/j.mam.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Fouet A, Mesnage S, Tosi-Couture E, Gounon P, Mock M. Bacillus anthracis surface: capsule and S-layer. J Appl Microbiol. 1999;87:251–255. doi: 10.1046/j.1365-2672.1999.00882.x. [DOI] [PubMed] [Google Scholar]
- Frece J, Kos B, Svetec IK, Zgaga Z, Mrsa V, Suskovic J. Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J Appl Microbiol. 2005;98:285–292. doi: 10.1111/j.1365-2672.2004.02473.x. [DOI] [PubMed] [Google Scholar]
- Garrote GL, Delfederico L, Bibiloni R, Abraham AG, Perez PF, Semorile L, De Antoni GL. Lactobacilli isolated from kefir grains: evidence of the presence of S-layer proteins. J Dairy Res. 2004;71:222–230. doi: 10.1017/S0022029904000160. [DOI] [PubMed] [Google Scholar]
- Gatti M, Rossetti L, Fornasari ME, Lazzi C, Giraffa G, Neviani E. Heterogeneity of putative surface layer proteins in Lactobacillus helveticus. Appl Environ Microbiol. 2005;71:7582–7588. doi: 10.1128/AEM.71.11.7582-7588.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour R, Messner P, Guffanti AA, Kent R, Scheberl A, Kendrick N, Krulwich TA. Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J Bacteriol. 2000;182:5969–5981. doi: 10.1128/JB.182.21.5969-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh YJ, Klaenhammer TR. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2010;76:5005–5012. doi: 10.1128/AEM.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh YJ, Azcarate-Peril MA, O'Flaherty S, Durmaz E, Valence F, Jardin J, Lortal S, Klaenhammer TR. Development and application of a upp-based counterselective gene replacement system for study of the S-layer protein SlpX in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2009;75:3093–3105. doi: 10.1128/AEM.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golowczyc MA, Mobili P, Garrote GL, Abraham AG, De Antoni GL. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar Enteritidis. Int J Food Microbiol. 2007;118:264–273. doi: 10.1016/j.ijfoodmicro.2007.07.042. [DOI] [PubMed] [Google Scholar]
- Golowczyc MA, Mobili P, Garrote GL, de Los Angeles Serradell M, Abraham AG, De Antoni GL. Interaction between Lactobacillus kefir and Saccharomyces lipolytica isolated from kefir grains: evidence for lectin-like activity of bacterial surface proteins. J Dairy Res. 2009;76:111–116. doi: 10.1017/S0022029908003749. [DOI] [PubMed] [Google Scholar]
- Grangette C, Müller-Alouf H, Goudercourt D, Geoffroy MC, Turneer M, Mercenier A. Mucosal immune responses and protection against tetanus toxin after intranasal immunization with recombinant Lactobacillus plantarum. Infect Immun. 2001;69:1547–1553. doi: 10.1128/IAI.69.3.1547-1553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CF, Zhang LW, Han X, Yi HX, Li JY, Tuo YF, Zhang YC, Du M, Shan YJ, Yang L. Screening for cholesterol-lowering probiotic based on deoxycholic acid removal pathway and studying its functional mechanisms in vitro. Anaerobe. 2012;18:516–522. doi: 10.1016/j.anaerobe.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Hagen KE, Guan LL, Tannock GW, Korver DR, Allison GE. Detection, characterization, and in vitro and in vivo expression of genes encoding S-proteins in Lactobacillus gallinarum strains isolated from chicken crops. Appl Environ Microbiol. 2005;71:6633–6643. doi: 10.1128/AEM.71.11.6633-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock IC, Poxton IR. Bacterial cell surface techniques. New York: Wiley; 1988. [Google Scholar]
- Hao P, Zheng H, Yu Y, Ding G, Gu W, Chen S, Yu Z, Ren S, Oda M, Konno T, Wang S, Li X, Ji ZS, Zhao G. Complete sequencing and pan-genomic analysis of Lactobacillus delbrueckii subsp. bulgaricus reveal its genetic basis for industrial yogurt production. PLoS One. 2011;17:e15964. doi: 10.1371/journal.pone.0015964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005;54:1093–1101. doi: 10.1099/jmm.0.45935-0. [DOI] [PubMed] [Google Scholar]
- Heinl S, Wibberg D, Eikmeyer F, Szczepanowski R, Blom J, Linke B, Goesmann A, Grabherr R, Schwab H, Pühler A, Schlüter A. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J Biotechnol. 2012;161:153–166. doi: 10.1016/j.jbiotec.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Hollmann A, Delfederico L, Glikmann G, De Antoni G, Semorile L, Disalvo EA. Characterization of liposomes coated with S-layer proteins from lactobacilli. Biochim Biophys Acta. 2007;1768:393–400. doi: 10.1016/j.bbamem.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Hollmann A, Delfederico L, De Antoni G, Semorile L, Disalvo EA. Interaction of bacterial surface layer proteins with lipid membranes: synergysm between surface charge density and chain packing. Colloids Surf B: Biointerfaces. 2010;79:191–197. doi: 10.1016/j.colsurfb.2010.03.046. [DOI] [PubMed] [Google Scholar]
- Hollmann A, Delfederico L, De Antoni G, Semorile L, Disalvo EA. Relaxation processes in the adsorption of surface layer proteins to lipid membranes. J Phys Chem B. 2010;114:16618–16624. doi: 10.1021/jp107062e. [DOI] [PubMed] [Google Scholar]
- Horie M, Ishiyama A, Fujihira-Ueki Y, Sillanpää J, Korhonen TK, Toba T. Inhibition of the adherence of Escherichia coli strains to basement membrane by Lactobacillus crispatus expressing an S-layer. J Appl Microbiol. 2002;92:396–403. doi: 10.1046/j.1365-2672.2002.01539.x. [DOI] [PubMed] [Google Scholar]
- Horie M, Kajikawa HS, Toba T. Identification of Lactobacillus crispatus by polymerase chain reaction targeting S-layer protein gene. Lett Appl Microbiol. 2002;35:57–61. doi: 10.1046/j.1472-765X.2002.01141.x. [DOI] [PubMed] [Google Scholar]
- Howard L, Tipper DJ. A polypeptide bacteriophage receptor: modified cell wall protein subunits in bacteriophage-resistant mutants of Bacillus sphaericus strain P-1. J Bacteriol. 1973;113:1491–1504. doi: 10.1128/jb.113.3.1491-1504.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Kong J, Sun Z, Han L, Kong W, Yang P. Heterologous protein display on the cell surface of lactic acid bacteria mediated by the S-layer protein. Microb Cell Fact. 2011;10:86. doi: 10.1186/1475-2859-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurmalainen V, Edelman S, Antikainen J, Baumann M, Lähteenmäki K, Korhonen TK. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology. 2007;153:1112–1122. doi: 10.1099/mic.0.2006/000901-0. [DOI] [PubMed] [Google Scholar]
- Hynönen U, Westerlund-Wikström B, Palva A, Korhonen TK. Identification by flagellum display of an epithelial cell- and fibronectin-binding function in the SlpA surface protein of Lactobacillus brevis. J Bacteriol. 2002;184:3360–3367. doi: 10.1128/JB.184.12.3360-3367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynönen U, Åvall-Jääskeläinen S, Palva A. Characterization and separate activities of the two promoters of the Lactobacillus brevis S-layer protein gene. Appl Microbiol Biotechnol. 2010;87:657–668. doi: 10.1007/s00253-010-2500-z. [DOI] [PubMed] [Google Scholar]
- Ilk N, Kosma P, Puchberger M, Egelseer EM, Mayer HF, Sleytr UB, Sára M. Structural and functional analyses of the secondary cell wall polymer of Bacillus sphaericus CCM 2177 that serves as an S-layer specific anchor. J Bacteriol. 1999;181:7643–7646. doi: 10.1128/jb.181.24.7643-7646.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilk N, Egelseer EM, Sleytr UB. S-layer fusion proteins—construction principles and applications. Curr Opin Biotechnol. 2011;22:824–831. doi: 10.1016/j.copbio.2011.05.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro EE, Ainsworth T, Harkness RE, Kay WW, Trust TJ. A temperate bacteriophage specific for strains of Aeromonas salmonicida possessing A-layer, a cell surface virulence factor. Curr Microbiol. 1984;10:199–202. doi: 10.1007/BF01627255. [DOI] [Google Scholar]
- Jakava-Viljanen M, Palva A. Isolation of surface (S) layer protein carrying Lactobacillus species from porcine intestine and faeces and characterization of their adhesion properties to different host tissues. Vet Microbiol. 2007;124:264–273. doi: 10.1016/j.vetmic.2007.04.029. [DOI] [PubMed] [Google Scholar]
- Jakava-Viljanen M, Åvall-Jääskeläinen S, Messner P, Sleytr UB, Palva A. Isolation of three new surface layer protein genes (slp) from Lactobacillus brevis ATCC 14869 and characterization of the change in their expression under aerated and anaerobic conditions. J Bacteriol. 2002;184:6786–6795. doi: 10.1128/JB.184.24.6786-6795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic I, Ventura M, Meylan V, Rouvet M, Elli M, Zink R. Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillus gasseri 4B2. J Bacteriol. 2003;185:3288–3296. doi: 10.1128/JB.185.11.3288-3296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Phelps CF, Cummins CS, London J, Gasser F. Taxonomy of the Lactobacillus acidophilus group. Int J Syst Bacteriol. 1980;30:53–68. doi: 10.1099/00207713-30-1-53. [DOI] [Google Scholar]
- Johnson-Henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 2007;9:356–367. doi: 10.1111/j.1462-5822.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- Kahala M, Palva A. The expression signals of the Lactobacillus brevis slpA gene direct efficient heterologous protein production in lactic acid bacteria. Appl Microbiol Biotechnol. 1999;51:71–78. doi: 10.1007/s002530051365. [DOI] [PubMed] [Google Scholar]
- Kahala M, Savijoki K, Palva A. In vivo expression of the Lactobacillus brevis S-layer gene. J Bacteriol. 1997;179:284–286. doi: 10.1128/jb.179.1.284-286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler O, Weiss N. In: Regular, nonsporing Gram-positive rods. Bergey's manual of systematic bacteriology. Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Baltimore: Williams and Wilkins; 1986. pp. 1208–1234. [Google Scholar]
- Kant R, Paulin L, Alatalo E, de Vos WM, Palva A. Genome sequence of Lactobacillus amylovorus GRL1112. J Bacteriol. 2011;193:789–790. doi: 10.1128/JB.01365-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant R, Paulin L, Alatalo E, de Vos WM, Palva A. Genome sequence of Lactobacillus amylovorus GRL1118, isolated from pig ileum. J Bacteriol. 2011;193:3147–3148. doi: 10.1128/JB.00423-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T, Masuda K, Yoshino K, Fujimoto M. Regular array in the cell wall of Lactobacillus fermenti as revealed by freeze-etching and negative staining. Jpn J Microbiol. 1974;18:469–476. doi: 10.1111/j.1348-0421.1974.tb00836.x. [DOI] [PubMed] [Google Scholar]
- Kay WW, Phipps BM, Ishiguro EE, Trust TJ. Porphyrin binding by the surface array virulence protein of Aeromonas salmonicida. J Bacteriol. 1985;164:1332–1336. doi: 10.1128/jb.164.3.1332-1336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghi M, Kermanshahi RK, Yaghoobi MM, Zarkesh-Esfahani SH, Baghizadeh A. Assessment of bile salt effects on S-layer production, slp gene expression and some physicochemical properties of Lactobacillus acidophilus ATCC 4356. J Microbiol Biotechnol. 2010;20:749–756. [PubMed] [Google Scholar]
- Khaleghi M, Kasra Kermanshahi R, Zarkesh-Esfahani SH. Effects of penicillin G on morphology and certain physiological parameters of Lactobacillus acidophilus ATCC 4356. J Microbiol Biotechnol. 2011;21:822–829. doi: 10.4014/jmb.1012.12020. [DOI] [PubMed] [Google Scholar]
- Khang YH, Park HY, Jeong YS, Kim JA, Kim YH. Recombinant S-layer proteins of Lactobacillus brevis mediating antibody adhesion to calf intestine alleviated neonatal diarrhea syndrome. J Microbiol Biotechnol. 2009;19:511–519. doi: 10.4014/jmb.0805.325. [DOI] [PubMed] [Google Scholar]
- Klingl A, Moissl-Eichinger C, Wanner G, Zweck J, Huber H, Thomm M, Rachel R. Analysis of the surface proteins of Acidithiobacillus ferrooxidans strain SP5/1 and the new, pyrite-oxidizing Acidithiobacillus isolate HV2/2, and their possible involvement in pyrite oxidation. Arch Microbiol. 2011;193:867–882. doi: 10.1007/s00203-011-0720-y. [DOI] [PubMed] [Google Scholar]
- Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, van Kooyk Y. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A. 2008;105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 2003;94:981–987. doi: 10.1046/j.1365-2672.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- Kotiranta AK, Ito H, Haapasalo MP, Lounatmaa K. Radiation sensitivity of Bacillus cereus with and without a crystalline surface protein layer. FEMS Microbiol Lett. 1999;179:275–280. doi: 10.1111/j.1574-6968.1999.tb08738.x. [DOI] [PubMed] [Google Scholar]
- Koval SF, Hynes SH. Effect of paracrystalline protein surface layers on predation by Bdellovibrio bacteriovorus. J Bacteriol. 1991;173:2244–2249. doi: 10.1128/jb.173.7.2244-2249.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire M, Miras I, Gounon P, Beguin P. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology. 1998;144(Pt 1):211–217. doi: 10.1099/00221287-144-1-211. [DOI] [PubMed] [Google Scholar]
- Li P, Yin Y, Yu Q, Yang Q. Lactobacillus acidophilus S-layer protein-mediated inhibition of Salmonella-induced apoptosis in Caco-2 cells. Biochem Biophys Res Commun. 2011;409:142–147. doi: 10.1016/j.bbrc.2011.04.131. [DOI] [PubMed] [Google Scholar]
- Li P, Yu Q, Ye X, Wang Z, Yang Q. Lactobacillus S-layer protein inhibition of Salmonella-induced reorganization of the cytoskeleton and activation of MAPK signalling pathways in Caco-2 cells. Microbiology. 2011;157(Pt 9):2639–2646. doi: 10.1099/mic.0.049148-0. [DOI] [PubMed] [Google Scholar]
- Lightfoot YL, Mohamadzadeh M. Tailoring gut immune responses with lipoteichoic acid-deficient Lactobacillus acidophilus. Front Immunol. 2013;4:25. doi: 10.3389/fimmu.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeqvist S, Cano F, Nguyen TN, Uhlen M, Robert A, Ståhl S. Surface display of functional fibronectin-binding domains on Staphylococcus carnosus. FEBS Lett. 1999;446:299–304. doi: 10.1016/S0014-5793(99)00232-X. [DOI] [PubMed] [Google Scholar]
- Lizier M, Sarra PG, Cauda R, Lucchini F. Comparison of expression vectors in Lactobacillus reuteri strains. FEMS Microbiol Lett. 2010;308:8–15. doi: 10.1111/j.1574-6968.2010.01978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortal S, van Heijenoort J, Gruber K, Sleytr UB. S-layer of Lactobacillus helveticus ATCC 12046: isolation, chemical characterization and reformation after extraction with lithium chloride. J Gen Microbiol. 1992;138:611–618. doi: 10.1099/00221287-138-3-611. [DOI] [Google Scholar]
- Lupas A, Engelhardt H, Peters J, Santarius U, Volker S, Baumeister W. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol. 1994;176:1224–1233. doi: 10.1128/jb.176.5.1224-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maassen CB, van Holten-Neelen C, Balk F, den Bak-Glashouwer MJ, Leer RJ, Laman JD, Boersma WJ, Claassen E. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine. 2000;18:2613–2623. doi: 10.1016/S0264-410X(99)00378-3. [DOI] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte H, Ferrari S, Cesena C, Hammarström L, Morelli L, Pozzi G, Oggioni MR. The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J Appl Microbiol. 2004;97:749–756. doi: 10.1111/j.1365-2672.2004.02364.x. [DOI] [PubMed] [Google Scholar]
- Martinez B, Sillanpää J, Smit E, Korhonen TK, Pouwels PH. Expression of cbsA encoding the collagen-binding S-protein of Lactobacillus crispatus JCM5810 in Lactobacillus casei ATCC 393(T) J Bacteriol. 2000;182:6857–6861. doi: 10.1128/JB.182.23.6857-6861.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez MG, Prado Acosta M, Candurra NA, Ruzal SM. S-layer proteins of Lactobacillus acidophilus inhibits JUNV infection. Biochem Biophys Res Commun. 2012;422:590–595. doi: 10.1016/j.bbrc.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Kawata T. Reassembly of the regularly arranged subunits in the cell wall of Lactobacillus brevis and their reattachment to cell walls. Microbiol Immunol. 1980;24:299–308. doi: 10.1111/j.1348-0421.1980.tb02833.x. [DOI] [PubMed] [Google Scholar]
- Masuda K, Kawata T. Distribution and chemical characterization of regular arrays in the cell walls of strains of the genus Lactobacillus. FEMS Microbiol Lett. 1983;20:145–150. doi: 10.1111/j.1574-6968.1983.tb00105.x. [DOI] [Google Scholar]
- Masuda K, Kawata T. Reassembly of a regularly arranged protein in the cell wall of Lactobacillus buchneri and its reattachment to cell walls: chemical modification studies. Microbiol Immunol. 1985;29:927–938. [PubMed] [Google Scholar]
- Matuschek M, Burchhardt G, Sahm K, Bahl H. Pullulanase of Thermoanaerobacterium thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene, composite structure of the enzyme, and a common model for its attachment to the cell surface. J Bacteriol. 1994;176:3295–3302. doi: 10.1128/jb.176.11.3295-3302.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren J, Heuser J, Roth R, Yamada N, Martone M, Brahamsha B. Inactivation of swmA results in the loss of an outer cell layer in a swimming Synechococcus strain. J Bacteriol. 2005;187:224–230. doi: 10.1128/JB.187.1.224-230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A, Timms P. Efficiency of transcription from promoter sequence variants in Lactobacillus is both strain and context dependent. J Bacteriol. 1999;181:6569–6572. doi: 10.1128/jb.181.20.6569-6572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mescher MF, Strominger JL. Structural (shape-maintaining) role of the cell surface glycoprotein of Halobacterium salinarium. Proc Natl Acad Sci U S A. 1976;73:2687–2691. doi: 10.1073/pnas.73.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Tosi-Couture E, Fouet A. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol Microbiol. 1999;31:927–936. doi: 10.1046/j.1365-2958.1999.01232.x. [DOI] [PubMed] [Google Scholar]
- Messner P, Steiner K, Zarschler K, Schäffer C. S-layer nanoglycobiology of bacteria. Carbohydr Res. 2008;343:1934–1951. doi: 10.1016/j.carres.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen M, Vuopio-Varkila J, Varkila K. Production of human tumor necrosis factor alpha, interleukin-6, and interleukin-10 is induced by lactic acid bacteria. Infect Immun. 1996;64:5403–5405. doi: 10.1128/iai.64.12.5403-5405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Couture-Tosi E, Mesnage S, Mock M, Fouet A. In vivo Bacillus anthracis gene expression requires PagR as an intermediate effector of the AtxA signalling cascade. Int J Med Microbiol. 2004;293:619–624. doi: 10.1078/1438-4221-00306. [DOI] [PubMed] [Google Scholar]
- Mobili P, Serradell Mde L, Trejo SA, Aviles Puigvert FX, Abraham AG, De Antoni GL. Heterogeneity of S-layer proteins from aggregating and non-aggregating Lactobacillus kefir strains. Antonie Van Leeuwenhoek. 2009;95:363–372. doi: 10.1007/s10482-009-9322-y. [DOI] [PubMed] [Google Scholar]
- Mobili P, Londero A, Maria TMR, Eusébio MES, De Antoni GL, Fausto R, Gómez-Zavaglia A. Characterization of S-layer proteins of Lactobacillus by FTIR spectroscopy and differential scanning calorimetry. Vib Spectrosc. 2009;50:68–77. doi: 10.1016/j.vibspec.2008.07.016. [DOI] [Google Scholar]
- Mohamadzadeh M, Duong T, Sandwick SJ, Hoover T, Klaenhammer TR. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proc Natl Acad Sci USA. 2009;106:4331–4336. doi: 10.1073/pnas.0900029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, Bere P, Sarraj B, Khan MW, Pakanati KC, Ansari MJ, O'Flaherty S, Barrett T, Klaenhammer TR. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota RM, Moreira JL, Souza MR, Horta MF, Teixeira SM, Neumann E, Nicoli JR, Nunes AC. Genetic transformation of novel isolates of chicken Lactobacillus bearing probiotic features for expression of heterologous proteins: a tool to develop live oral vaccines. BMC Biotechnol. 2006;6:2. doi: 10.1186/1472-6750-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka M, Muto M, Nomoto K, Matuzaki T, Watanabe T, Yokokura T. Structure of polysaccharide–peptidoglycan complex from the cell wall of Lactobacillus casei YIT9018. J Biochem. 1990;108:568–571. doi: 10.1093/oxfordjournals.jbchem.a123243. [DOI] [PubMed] [Google Scholar]
- Narita J, Ishida S, Okano K, Kimura S, Fukuda H, Kondo A. Improvement of protein production in lactic acid bacteria using 5′-untranslated leader sequence of slpA from Lactobacillus acidophilus. Improvement in protein production using UTLS. Appl Microbiol Biotechnol. 2006;73:366–373. doi: 10.1007/s00253-006-0477-4. [DOI] [PubMed] [Google Scholar]
- Naser SM, Hagen KE, Vancanneyt M, Cleenwerck I, Swings J, Tompkins TA. Lactobacillus suntoryeus Cachat and Priest 2005 is a later synonym of Lactobacillus helveticus (Orla-Jensen 1919) Bergey et al. 1925 (Approved Lists 1980) Int J Syst Evol Microbiol. 2006;56(Pt 2):355–360. doi: 10.1099/ijs.0.64001-0. [DOI] [PubMed] [Google Scholar]
- Noonan B, Trust TJ. The synthesis, secretion and role in virulence of the paracrystalline surface protein layers of Aeromonas salmonicida and A. hydrophila. FEMS Microbiol Lett. 1997;154:1–7. doi: 10.1111/j.1574-6968.1997.tb12616.x. [DOI] [PubMed] [Google Scholar]
- Novotny R, Scheberl A, Giry-Laterriere M, Messner P, Schaffer C. Gene cloning, functional expression and secretion of the S-layer protein SgsE from Geobacillus stearothermophilus NRS 2004/3a in Lactococcus lactis. FEMS Microbiol Lett. 2005;242:27–35. doi: 10.1016/j.femsle.2004.10.036. [DOI] [PubMed] [Google Scholar]
- Oh S, Roh H, Ko HJ, Kim S, Kim KH, Lee SE, Chang IS, Kim S, Choi IG. Complete genome sequencing of Lactobacillus acidophilus 30SC, isolated from swine intestine. J Bacteriol. 2011;193:2882–2883. doi: 10.1128/JB.00343-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala T, Kuparinen V, Koskinen JP, Alatalo E, Holm L, Auvinen P, Edelman S, Westerlund-Wikström B, Korhonen TK, Paulin L, Kankainen M. Genome sequence of Lactobacillus crispatus ST1. J Bacteriol. 2010;192:3547–3548. doi: 10.1128/JB.00399-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, Mommers M, Stobberingh EE, Dompeling E, Reijmerink NE, van den Brandt PA, Kerkhof M, Koppelman GH, Postma DS. Intestinal lactobacilli and the DC-SIGN gene for their recognition by dendritic cells play a role in the aetiology of allergic manifestations. Microbiology. 2010;156:3298–3305. doi: 10.1099/mic.0.042069-0. [DOI] [PubMed] [Google Scholar]
- Peters J, Nitsch M, Kuhlmorgen B, Golbik R, Lupas A, Kellermann J, Engelhardt H, Pfander JP, Müller S, Goldie K. Tetrabrachion: a filamentous archaebacterial surface protein assembly of unusual structure and extreme stability. J Mol Biol. 1995;245:385–401. doi: 10.1006/jmbi.1994.0032. [DOI] [PubMed] [Google Scholar]
- Phipps BM, Kay WW. Immunoglobulin binding by the regular surface array of Aeromonas salmonicida. J Biol Chem. 1988;263:9298–9303. [PubMed] [Google Scholar]
- Podlesny M, Jarocki P, Komon E, Glibowska A, Targonski Z. LC-MS/MS analysis of surface layer proteins as a useful method for the identification of lactobacilli from the Lactobacillus acidophilus group. J Microbiol Biotechnol. 2011;21:421–429. [PubMed] [Google Scholar]
- Pollmann K, Raff J, Merroun M, Fahmy K, Selenska-Pobell S. Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol Adv. 2006;24:58–68. doi: 10.1016/j.biotechadv.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Poppinga L, Janesch B, Fünfhaus A, Sekot G, Garcia-Gonzalez E, Hertlein G, Hedtke K, Schäffer C, Genersch E. Identification and functional analysis of the S-layer protein SplA of Paenibacillus larvae, the causative agent of American foulbrood of honey bees. PLoS Pathog. 2012;8:e1002716. doi: 10.1371/journal.ppat.1002716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels PH, Leunissen JA. Divergence in codon usage of Lactobacillus species. Nucleic Acids Res. 1994;22:929–936. doi: 10.1093/nar/22.6.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado Acosta M, Mercedes Palomino M, Allievi MC, Sanchez Rivas C, Ruzal SM. Murein hydrolase activity in the surface layer of Lactobacillus acidophilus ATCC 4356. Appl Environ Microbiol. 2008;74:7824–7827. doi: 10.1128/AEM.01712-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Acosta M, Ruzal SM, Allievi MC, Palomino MM, Sanchez Rivas C. Synergistic effects of the Lactobacillus acidophilus surface layer and nisin on bacterial growth. Appl Environ Microbiol. 2010;76:974–977. doi: 10.1128/AEM.01427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati JB, Khedkar CD, Chitra J, Suja S, Mishra V, Sreeja V, Patel RK, Ahir VB, Bhatt VD, Sajnani MR, Jakhesara SJ, Koringa PG, Joshi CG. Whole-genome shotgun sequencing of an Indian-origin Lactobacillus helveticus strain, MTCC 5463, with probiotic potential. J Bacteriol. 2011;193:4282–4283. doi: 10.1128/JB.05449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pum D, Toca-Herrera JL, Sleytr UB. S-layer protein self-assembly. Int J Mol Sci. 2013;14:2484–2501. doi: 10.3390/ijms14022484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries W, Hotzy C, Schocher I, Sleytr UB, Sára M. Evidence that the N-terminal part of the S-layer protein from Bacillus stearothermophilus PV72/p2 recognizes a secondary cell wall polymer. J Bacteriol. 1997;179:3892–3898. doi: 10.1128/jb.179.12.3892-3898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfuss H, Lara JC, Schmid AK, Lidström ME. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology. 2006;152:2779–2787. doi: 10.1099/mic.0.28971-0. [DOI] [PubMed] [Google Scholar]
- Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153:866–876. doi: 10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- Sampathkumar P, Gilchrist ML. Synthesis and characterization of bioconjugates of S-layer proteins. Bioconjug Chem. 2004;15:685–693. doi: 10.1021/bc034204r. [DOI] [PubMed] [Google Scholar]
- Sanders ME, Klaenhammer TR. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J Dairy Sci. 2001;84:319–331. doi: 10.3168/jds.S0022-0302(01)74481-5. [DOI] [PubMed] [Google Scholar]
- Sára M, Sleytr UB. Molecular sieving through S-layers of Bacillus stearothermophilus strains. J Bacteriol. 1987;169:4092–4098. doi: 10.1128/jb.169.9.4092-4098.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sára M, Sleytr UB. S-layer proteins. J Bacteriol. 2000;182:859–868. doi: 10.1128/JB.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sára M, Moser-Tjoer L, Kainz O, Sleytr UB. Characterization of S-layers from mesophilic Bacillaceae and studies on their protective role towards muramidases. Arch Microbiol. 1990;153:2096–2114. doi: 10.1007/BF00249069. [DOI] [Google Scholar]
- Sára M, Dekitsch C, Mayer HF, Egelseer EM, Sleytr UB. Influence of the secondary cell wall polymer on the reassembly, recrystallization, and stability properties of the S-layer protein from Bacillus stearothermophilus PV72/p2. J Bacteriol. 1998;180:4146–4153. doi: 10.1128/jb.180.16.4146-4153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savijoki K, Kahala M, Palva A. High level heterologous protein production in Lactococcus and Lactobacillus using a new secretion system based on the Lactobacillus brevis S-layer signals. Genetics. 1997;186:255–262. doi: 10.1016/s0378-1119(96)00717-2. [DOI] [PubMed] [Google Scholar]
- Schär-Zammaretti P, Dillmann ML, D'Amico N, Affolter M, Ubbink J. Influence of fermentation medium composition on physicochemical surface properties of Lactobacillus acidophilus. Appl Environ Microbiol. 2005;71:8165–8173. doi: 10.1128/AEM.71.12.8165-8173.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz C, Nuotio L, Lounatma K. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface layer (S-layer) J Appl Bacteriol. 1993;74:290–294. doi: 10.1111/j.1365-2672.1993.tb03028.x. [DOI] [PubMed] [Google Scholar]
- Schultze-Lam S, Harauz G, Beveridge TJ. Participation of a cyanobacterial S layer in fine-grain mineral formation. J Bacteriol. 1992;174:7971–7981. doi: 10.1128/jb.174.24.7971-7981.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B, Sleytr UB. Composite S-layer lipid structures. J Struct Biol. 2009;168:207–216. doi: 10.1016/j.jsb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B, Pum D, Sára M, Sleytr UB. S-layer proteins as key components of a versatile molecular construction kit for biomedical nanotechnology. Mini-Rev Med Chem. 2006;6:909–920. doi: 10.2174/138955706777935026. [DOI] [PubMed] [Google Scholar]
- Seegers JF. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 2002;20:508–515. doi: 10.1016/S0167-7799(02)02075-9. [DOI] [PubMed] [Google Scholar]
- Sekot G, Posch G, Messner P, Matejka M, Rausch‐Fan X, Andrukhov O, Schäffer C (2011) Potential of the Tannerella forsythia S‐layer to delay the immune response. J Dent Res 90:109–14. [DOI] [PMC free article] [PubMed]
- Settem RP, Honma K, Nakajima T, Phansopa C, Roy S, Stafford GP, Sharma A. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 2013;6:415–426. doi: 10.1038/mi.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PM, Johnson-Henry KC, Yeung HP, Ngo PS, Goulet J, Tompkins TA. Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect Immun. 2005;73:5183–5188. doi: 10.1128/IAI.73.8.5183-5188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotahira N, Oogai Y, Kawada-Matsuo M, Yamada S, Fukutsuji K, Nagano K, Yoshimura F, Noguchi K, Komatsuzawa H (2013) The S-layer of Tannerella forsythia contributes to serum resistance and oral bacterial co-aggregation. Infect Immun 81:1198-206 [DOI] [PMC free article] [PubMed]
- Sillanpää J, Martínez B, Antikainen J, Toba T, Kalkkinen N, Tankka S, Lounatmaa K, Keränen J, Höök M, Westerlund-Wikström B, Pouwels PH, Korhonen TK. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J Bacteriol. 2000;182:6440–6450. doi: 10.1128/JB.182.22.6440-6450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol Lett. 2007;267:131–144. doi: 10.1111/j.1574-6968.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- Smit E, Pouwels PH. One repeat of the cell wall binding domain is sufficient for anchoring the Lactobacillus acidophilus surface layer protein. J Bacteriol. 2002;184:4617–4619. doi: 10.1128/JB.184.16.4617-4619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit E, Oling F, Demel R, Martinez B, Pouwels PH. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterization of domains responsible for S-protein assembly and cell wall binding. J Mol Biol. 2001;305:245–257. doi: 10.1006/jmbi.2000.4258. [DOI] [PubMed] [Google Scholar]
- Smit E, Jager D, Martinez B, Tielen FJ, Pouwels PH. Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein–protein interaction of two subdomains. J Mol Biol. 2002;324:953–964. doi: 10.1016/S0022-2836(02)01135-X. [DOI] [PubMed] [Google Scholar]
- Smith RH, Messner P, Lamontagne LR, Sleytr UB, Unger FM. Induction of T-cell immunity to oligosaccharide antigens immobilized on crystalline bacterial surface layers (S-layers) Vaccine. 1993;11:919–924. doi: 10.1016/0264-410X(93)90378-B. [DOI] [PubMed] [Google Scholar]
- Spigaglia P, Galeotti CL, Barbanti F, Scarselli M, Van Broeck J, Mastrantonio P. The LMW surface-layer proteins of Clostridium difficile PCR ribotypes 027 and 001 share common immunogenic properties. J Med Microbiol. 2011;60(Pt 8):1168–1173. doi: 10.1099/jmm.0.029710-0. [DOI] [PubMed] [Google Scholar]
- Stewart M, Murray RGE. Structure of the regular surface layer of Aquaspirillum serpens MW5. J Bacteriol. 1982;150:348–357. doi: 10.1128/jb.150.1.348-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Kong J, Hu S, Kong W, Lu W, Liu W (2012) Characterization of a S-layer protein from Lactobacillus crispatus K313 and the domains responsible for binding to cell wall and adherence to collagen. Appl Microbiol Biotechnol(Apr 18) [DOI] [PubMed]
- Takahashi N, Saito T, Ohwada S, Ota H, Hashiba H, Itoh T. A new screening method for the selection of Lactobacillus acidophilus group lactic acid bacteria with high adhesion to human colonic mucosa. Biosci Biotechnol Biochem. 1996;60:1434–1438. doi: 10.1271/bbb.60.1434. [DOI] [PubMed] [Google Scholar]
- Tallon R, Arias S, Bressollier P, Urdaci MC. Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J Appl Microbiol. 2007;102:442–451. doi: 10.1111/j.1365-2672.2006.03086.x. [DOI] [PubMed] [Google Scholar]
- Tarao M, Jezbera J, Hahn MW. Involvement of cell surface structures in size-independent grazing resistance of freshwater Actinobacteria. Appl Environ Microbiol. 2009;75:4720–4726. doi: 10.1128/AEM.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, Cordova ZM, Junttila I, Hämäläinen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl Environ Microbiol. 2012;79:1221–1231. doi: 10.1128/AEM.03056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SA. Campylobacter surface-layers (S-layers) and immune evasion. Ann Periodontol. 2002;7:43–53. doi: 10.1902/annals.2002.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba T, Virkola R, Westerlund B, Bjorkman Y, Sillanpää J, Vartio T, Kalkkinen N, Korhonen TK. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl Environ Microbiol. 1995;61:2467–2471. doi: 10.1128/aem.61.7.2467-2471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins TA, Barreau G, Broadbent JR. Complete genome sequence of Lactobacillus helveticus R0052, a commercial probiotic strain. J Bacteriol. 2012;194:6349. doi: 10.1128/JB.01638-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H, Kinoshita H, Kawai Y, Kitazawa H, Miura K, Shiiba K, Horii A, Kimura K, Taketomo N, Oda M, Yajima T, Saito T. Lactobacilli binding human A-antigen expressed in intestinal mucosa. Res Microbiol. 2006;157:659–665. doi: 10.1016/j.resmic.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Vadillo-Rodriguez V, Busscher HJ, Norde W, de Vries J, van der Mei HC. Dynamic cell surface hydrophobicity of Lactobacillus strains with and without surface layer proteins. J Bacteriol. 2004;186:6647–6650. doi: 10.1128/JB.186.19.6647-6650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadillo-Rodríguez V, Busscher HJ, van der Mei HC, de Vries J, Norde W. Role of Lactobacillus cell surface hydrophobicity as probed by AFM in adhesion to surfaces at low and high ionic strength. Colloids Surf B: Biointerfaces. 2005;41:33–41. doi: 10.1016/j.colsurfb.2004.10.028. [DOI] [PubMed] [Google Scholar]
- van der Mei HC, van de Belt-Gritter B, Pouwels PH, Martinez B, Busscher HJ. Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids Surf B: Biointerfaces. 2003;28:127–134. doi: 10.1016/S0927-7765(02)00144-3. [DOI] [Google Scholar]
- Ventura M, Callegari ML, Morelli L. S-layer gene as a molecular marker for identification of Lactobacillus helveticus. FEMS Microbiol Lett. 2000;189:275–279. doi: 10.1111/j.1574-6968.2000.tb09243.x. [DOI] [PubMed] [Google Scholar]
- Ventura M, Jankovic I, Walker DC, Pridmore RD, Zink R. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl Environ Microbiol. 2002;68:6172–6181. doi: 10.1128/AEM.68.12.6172-6181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbelen C, Antikainen J, Korhonen TK, Dufrene YF. Exploring the molecular forces within and between CbsA S-layer proteins using single molecule force spectroscopy. Ultramicroscopy. 2007;107:1004–1011. doi: 10.1016/j.ultramic.2007.02.040. [DOI] [PubMed] [Google Scholar]
- Vidgren G, Palva I, Pakkanen R, Lounatmaa K, Palva A. S-layer protein gene of Lactobacillus brevis: cloning by polymerase chain reaction and determination of the nucleotide sequence. J Bacteriol. 1992;174:7419–7427. doi: 10.1128/jb.174.22.7419-7427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilen H, Hynönen U, Badelt-Lichtblau H, Ilk N, Jääskeläinen P, Torkkeli M, Palva A. Surface location of individual residues of SlpA provides insight into the Lactobacillus brevis S-layer. J Bacteriol. 2009;191:3339–3349. doi: 10.1128/JB.01782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eichel-Streiber C, Sauerborn M, Kuramitsu HK. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J Bacteriol. 1992;174:6707–6710. doi: 10.1128/jb.174.20.6707-6710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Kraig E, Kolodrubetz D. Use of defined mutants to assess the role of the Campylobacter rectus S-layer in bacterium–epithelial cell interactions. Infect Immun. 2000;68:1465–1473. doi: 10.1128/IAI.68.3.1465-1473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Ahmed Z, Bai X, Wang J. Complete genome sequence of Lactobacillus kefiranofaciens ZW3. J Bacteriol. 2011;193:4280–4281. doi: 10.1128/JB.05306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G, Jespersen L. Transcriptional analysis of genes associated with stress and adhesion in Lactobacillus acidophilus NCFM during the passage through an in vitro gastrointestinal tract model. J Mol Microbiol Biotechnol. 2010;18:206–214. doi: 10.1159/000316421. [DOI] [PubMed] [Google Scholar]
- Wells JM, Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat Rev Microbiol. 2008;6:349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welman AD, Maddox IS. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. Trends Biotechnol. 2003;21:269–274. doi: 10.1016/S0167-7799(03)00107-0. [DOI] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- Wren BW. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Yamada M, Saito T, Toba T, Kitazawa H, Uemura J, Itoh T. Hemagglutination activity of Lactobacillus acidophilus group lactic acid bacteria. Biosci Biotechnol Biochem. 1994;58:910–915. doi: 10.1271/bbb.58.910. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Shinoda T, Takano T. Molecular cloning and sequence analysis of a gene encoding an extracellular proteinase from Lactobacillus helveticus CP790. Biosci Biotechnol Biochem. 2000;64:1217–1222. doi: 10.1271/bbb.64.1217. [DOI] [PubMed] [Google Scholar]
- Yasui T, Yoda K, Kamiya T. Analysis of S-layer proteins of Lactobacillus brevis. FEMS Microbiol Lett. 1995;133:181–186. doi: 10.1111/j.1574-6968.1995.tb07881.x. [DOI] [PubMed] [Google Scholar]
- Zadeh M, Khan MW, Goh YJ, Selle K, Owen JL, Klaenhammer T, Mohamadzadeh M (2012) Induction of intestinal pro-inflammatory immune responses by lipoteichoic acid. J Inflamm (Lond) 9:7 [DOI] [PMC free article] [PubMed]
- Zhang Q, Zhong J, Liang X, Liu W, Huan L. Improvement of human interferon alpha secretion by Lactococcus lactis. Biotechnol Lett. 2010;32:1271–1277. doi: 10.1007/s10529-010-0285-x. [DOI] [PubMed] [Google Scholar]
- Zhang YC, Zhang LW, Tuo YF, Guo CF, Yi HX, Li JY, Han X, Du M. Inhibition of Shigella sonnei adherence to HT-29 cells by lactobacilli from Chinese fermented food and preliminary characterization of S-layer protein involvement. Res Microbiol. 2010;161:667–672. doi: 10.1016/j.resmic.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Zhao W, Chen Y, Sun Z, Wang J, Zhou Z, Sun T, Wang L, Chen W, Zhang H. Complete genome sequence of Lactobacillus helveticus H10. J Bacteriol. 2011;193:2666–2667. doi: 10.1128/JB.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]