Abstract
Post-translational modification of proteins with ubiquitin is mediated by dynamic multi-enzyme machinery (E1, E2, E3). E3 ubiquitin ligases play a key role acting as both scaffolds to bring reactants together and activators to catalyze ubiquitin (Ub) transfer from E2~Ub conjugates to substrates. Our recent studies provided insights into the mechanism of the activation event; binding of an E3 to an E2~Ub conjugate was found to affect the motions of E2~Ub and allosterically stimulate Ub transfer. This proposed mechanism implies that the dynamics of the conjugate, which has been shown to occupy a wide range of E2~Ub orientations, will be altered significantly upon binding of E3. To directly assess the effect of E3 binding on E2~Ub dynamics, we undertook an in-depth comparative analysis of 15N NMR relaxation of UbcH5c~Ub in the absence and presence of the E3 ligase, E4B. Challenges encountered in deciphering inter-domain motions for this ternary complex are discussed along with the limitations of the current approaches. Notably, although a reduction in inter-domain dynamics of UbcH5c~Ub is observed upon binding to E4B, Ub retains an extensive degree of flexibility. These results provide strong support for our dynamic model of a significant orientational bias of Ub towards a more closed conformation in the E3/E2~Ub complex.
Post-translational modification of proteins by addition of one or more ubiquitin molecules is a key mechanism used to target proteins for degradation in the proteasome, as well as mediate intracellular signaling. The process of adding ubiquitin to a substrate requires the sequential activity of E1, E2, and E3 enzymes. The E1 activating enzyme uses ATP to drive formation of a covalent bond to the C-terminus of ubiquitin (Ub). The Ub molecule is then transferred to the E2 conjugating enzyme, which carries the activated Ub to the E3 ligating enzyme. In the case of RING and U-box type E3 ubiquitin ligases, the E3 acts as a scaffold to bring together the substrate and the E2~Ub conjugate for transfer of the Ub to the substrate (1). While these basic steps are known, many questions remain unanswered regarding the mechanism of Ub transfer to the substrate.
The current model of E3 function involves not only co-localization of E2~Ub and substrate, but also a mysterious “activation” of the E2~Ub conjugate. This activation event is reflected in the vastly increased rate of E2~Ub hydrolysis upon interaction of the E2 with the U-box (or RING) domain of E3 ligases, relative to the rate of E2~Ub hydrolysis free in solution (2). Models to explain E2~Ub activation have been proposed in which binding to the E3 induces a transition from a highly dynamic state to a “closed” Ub orientation (3-6). Recent x-ray crystal structures revealed Ub in the closed conformation (7, 8), but NMR studies have shown that binding of the conjugate to the E3 causes an alteration in the distribution of inter-domain orientations (9). In our NMR-based model, Ub prefers “closed” conformations, but is not held in a fixed orientation.
While our previous studies support a slowing of Ub motions in the ternary (E4BU/UbcH5c~Ub) complex, they did not establish unambiguously that binding of the E3 causes a shift in inter-domain flexibility that would be required for preferential occupancy of closed conformations. Here we describe an in-depth heteronuclear NMR relaxation analysis to monitor the changes in inter-domain flexibility of UbcH5c~Ub induced by the binding of the U-box domain of E4B, to obtain a more complete and quantitative analysis of the effects of E3 binding to the E2~Ub conjugate.
Heteronuclear NMR relaxation is a powerful method to directly characterize molecular motions. The most commonly used approach for the study of proteins involves measuring 15N relaxation parameters to define the tensor describing rotational diffusion. A ‘model-free7#x2019; analysis is often applied that partitions overall tumbling of the molecule from internal motions, therefore allowing insights into relative flexibility of specific residues of regions of the protein. The majority of NMR relaxation analyses have been carried out on globular single domain proteins, but in the last few years a number of more complex systems have been studied. In two domain systems, an apparent diffusion tensor can be directly extracted from NMR relaxation data. However, the tensor represents a combination of global tumbling and any inter-domain motion (10). A limited number of two-domain systems have been studied (11-14), but to our knowledge there is no study reported for a three component system. Here we present the results for, and discuss the challenges to, using extended model-free and other approaches to quantify changes in Ub flexibility upon binding of an E2~Ub conjugate to an E3.
Materials and Methods
Protein expression and purification
Plasmid constructs, expression, and purification of E1, Ub, UbcH5c S22R/C85S (E2), E4BU, and production of UbcH5c-O-Ub were previously described (9). All proteins were expressed in BL21 DE3 E. coli in either rich LB media or minimal media with 15NH4Cl as the sole nitrogen source. Proteins were stored in a 25 mM sodium phosphate pH 7.0, 150 mM NaCl buffer.
Ub hydrolysis rates
Samples of 200 μM UbcH5c-O-Ub were incubated in a range of buffer and pH conditions with 150 mM NaCl at 20 and 25 °C, with or without E4BU added to a ratio of E3:E2 at 1:1 or 3:1 as noted. To monitor differences in the rates of hydrolysis, small aliquots were incubated for up to 10 days. Samples were extracted for analysis by SDS-PAGE at regular intervals
NMR data collection
Data were collected at 20 °C on Bruker Avance III spectrometers operating at 1H frequencies of 600, 800 or 900 MHz. Heteronuclear 15N-R1, -R2 and NOE parameters were measured using standard pulse sequences provided by the manufacturer [hsqct1etf3gpsi3d.2 (R1), hsqct2etf3gpsi3d (R2), and hsqcnoef3gpsi (NOE)] (10). Protein solutions contained 200 μM 15N-enriched UbcH5c-O-Ub in either the pH 7 buffer described above or 30 mM sodium citrate at pH 5.75, 150 mM NaCl, and 8% D2O, in the absence or presence of 600 μM unlabeled E4BU. Data for the WT E4BU/UbcH5c-O-Ub complex were collected on a set of samples prepared from a common stock, with each sample mixed just prior to use. A new sample was used every 1.5 days.
The recovery delays for 15N-R1 experiments were 7.5 s, 9 s and 12 s (600, 800 and 900 MHz, respectively). Time delays for the R1 measurements were 0.1, 0.4, 0.8, 1.2, 1.8, 2.5, and 4 s at 600 MHz, and 0.1, 0.4, 0.8, 1.2, 2, 3, and 5 s at 800 and 900 MHz. Time delays for R2 measurements were 17, 35, 52, 69, 104, and 173 ms. A three second recovery delay and three second saturation period was used for measuring the {1H}-15N NOE. The data were processed in Topspin (Bruker) and analyzed in Sparky (15). The NOE was calculated as the ratio of intensities Isat/Iref, and the NOE error was calculated as (NOE err/NOE)2= [(Iref err/Iref)2+(Isat err/Isat)2], where the error of each intensity measurement is the rmsd noise of each plane.
NMR data analysis
15N-1H chemical shift assignments at pH 5.75 were transferred from those made at pH 7 reported previously (6). Diffusion tensors for each domain were determined using all available data for each condition. Residues with NOE < 0.7 or for which fewer than three parameters could be measured were removed from analysis, as well as residues with R2/R1 ratios greater than two standard deviations above the mean for that domain. The remaining residues were each fit to a local isotropic correlation time (τiso) using relax (16, 17). These values were then input into the quadric diffusion program (AG Palmer, based on Lee 1997 (18)) to fit global diffusion tensors using a structural model oriented to the inertial mass tensor. The best fit was determined to be isotropic for all domains using an F-statistic. Further model-free analysis with the relax and freemodel programs are described in the Results.
NMR data simulations
Predicted relaxation parameters were calculated with both HYDRONMR (19) and relax. Structural models were based on PDB ID 1FXT, 2GMI, 2KJH, 2OXQ, and 3A33 (3, 20-23). HYDRONMR was used largely to predict the expected overall molecular tumbling parameters of the structural models at multiple fields. The relax function ‘create_ri’ was used to back-calculate 15N NMR relaxation parameters for the final results of extended model-free analysis.
Results
The U-box E3 ligase E4B was selected for this analysis because of its unusual ability to function as a monomer (24, 25); the E4B U-box domain alone is sufficient to bind and activate the UbH5c~Ub conjugate, which greatly facilitates the analysis. Unfortunately, the UbcH5c~Ub conjugate when in the presence of an E3 ligase has a very short lifetime on the time scale of NMR relaxation measurements. To alleviate this problem, we turned to an E2~Ub analog (UbcH5c-O-Ub), in which the E2 Cys residue that conjugates Ub is mutated to Ser (C85S). Changing to an ester conjugate from a thioester conjugate drastically reduces the intrinsic reactivity of the bond linking the E2 to Ub. A second mutation (S22R) was incorporated in UbcH5c to inhibit binding to the “backside” of the E2, which has been shown to cause large increases in NMR linewidths (26). UbcH5c-C85S,S22R readily forms UbcH5c-O-Ub conjugates and was used in the previously reported study of the dynamics of UbcH5c~Ub (6).
Optimization of conditions to maximize the lifetime of UbcH5c~Ub in the E4BU ternary complex
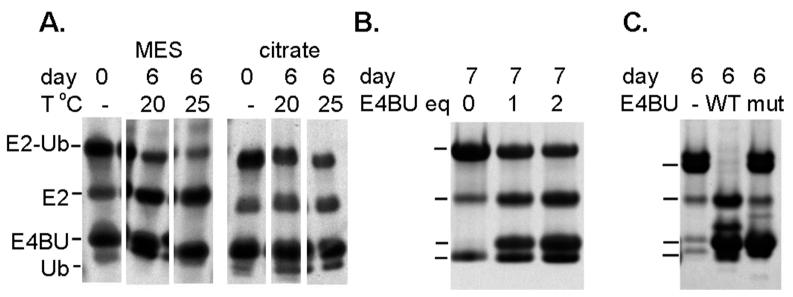
Initial attempts to characterize the ternary E4BU/UbcH5c-O-Ub complex were inhibited by the short lifetime of the conjugate, even in this ester analog, and its relatively low affinity for E4BU (Kd~ 100 μM (9)). The free UbcH5c-O-Ub conjugate generated by direct mixing of E1, E2, Ub, and ATP has been shown to be stable against hydrolysis over several days. However, upon addition of E4BU to UbcH5c-O-Ub, there is a dramatic increase in Ub hydrolysis with the lifetime reduced to hours (9), which is insufficient for detailed relaxation studies. We hypothesized that the reaction is likely base-catalyzed and would be slowed by reducing the pH. A test of the stability of the conjugate at a range of pH values showed that the conjugate lifetime was lengthened considerably under acidic conditions. The conjugate showed signs of aggregation at pH < 5.5 when concentrated to ~200 μM as required for the NMR studies, so further analyses were performed at pH 5.75. With the goal of further increasing the lifetime of the ternary complex, we also tested the effect of buffer identity and temperature on the rate of Ub hydrolysis and found a significant slowing of the rate in citrate buffer at 20 °C (Figure 1A). We note that these changes in solution conditions had a significant effect on the chemical shifts of UbcH5c-O-Ub with chemical shift perturbations up to ~0.2 ppm. However, as described below the overall tumbling of the particle and binding of E4BU were not altered.
Figure 1.
Ub hydrolysis rates are affected by A) buffer and temperature B) addition of E4BU. Complexes were 1:1 in panel A, and as noted in B.
High concentrations of E3 expedite Ub hydrolysis, so a third factor to optimize was the concentration of E3 in the ternary complex (Figure 1B). At a UbcH5c-O-Ub concentration of 200 μM, the chemical shift changes in the conjugate appeared to be complete with a 3-fold excess of unlabeled E4BU as no further changes were observed upon addition of more E4BU. Additionally, the UbcH5c-O-Ub residues most affected by E4BU are those in the expected binding interface (Figure S1). Based on the experimental estimate for the Kd of ~100 μM, this condition corresponds to a fractional saturation of ~80% of the conjugate. This ratio was selected to minimize hydrolysis of the conjugate over the course of the experiment, and by adjusting the experimental conditions we were able to stabilize the E4BU/UbcH5c-O-Ub ternary complex to the point where it remained essentially intact for 1.5 days. These experimental conditions provide a conservative model for the fully saturated system as any observed differences from the free conjugate will be less than if the measurements were made under conditions of 100% saturation.
UbcH5c-O-Ub is flexible with and without E4BU
NMR relaxation parameters are sensitive to a wide range of molecular motions from vibrations of the N-H bond vector, to global rotational diffusion, to conformational exchange (10). NMR relaxation analysis therefore enables detection of fast picosecond motions, rotational tumbling, and larger-scale μs-ms domain movements in proteins. The overall motion of the molecule is characterized by a tensor describing rotational diffusion. For spherical, globular molecules, the diffusion tensor is symmetric and the overall motion can be represented by a global rotational correlation time τc, which directly reflects the size and shape of the tumbling particle. In systems with two flexibly tethered domains, the rotational diffusion of a domain will reflect a combination of overall tumbling and inter-domain motion. Thus, analysis of the data for each domain provides an “apparent” correlation time. The motion can be difficult to model if substantial changes in shape arise in the molecule as a consequence of large changes in inter-domain orientation (27). In such a situation, analysis of the apparent diffusion of each domain enables differentiation of two domains as rigidly or flexibly attached, but the difference in correlation times cannot be directly related to the extent of the inter-domain of flexibility.
To investigate the effect of E3 binding on the inter-domain dynamics of an E2~Ub conjugate, 15N NMR relaxation parameters were collected for UbcH5c-O-Ub without and with E4BU. These included 15N-R1 and NOE parameters at 600 and 900 MHz and 15N-R2 parameters at 600 MHz. Plots of the relaxation parameters reveal clear differences between the E2 and Ub components of the conjugate, indicating that they tumble independently (Figure S2). UbcH5c contains 147 residues and Ub only 76, thus if they are not rigidly oriented we can expect Ub to tumble faster and exhibit relaxation properties consistent with that faster motion. Inspection of the data revealed consistent trends in all of the data sets, except for the R2 values obtained for Ub in the ternary complex. These R2 values were far smaller than anticipated and nearly matched values for free Ub. Since the chemical shifts of free Ub are the same as Ub in the conjugate and cannot be distinguished for the most part, we concluded that a small amount of free Ub must be present in solution. Free Ub has a substantially slower rate of transverse relaxation, so even a small of amount of free protein will completely dominate the decay curve in the measurement of the R2. The reason there is no effect from the free Ub on R1 measurements is that due to the way in which the parameter is measured the decay curve from the larger particle will remain dominant.
Apparent domain-specific diffusion tensors can still be calculated in the absence of the R2 data from 15N R1 and NOE values measured at multiple fields (10) and given the problems with R2 this approach was used here. First, we established that the data were best fit to isotropic diffusion; the more complicated axially symmetric models of Ub diffusion previously reported (9) were determined to be statistically no better than the isotropic model. Consequently, the motion of the domains can be expressed in terms of τc (Table 1). The UbcH5c-O-Ub conjugate has previously been shown to exist as two flexibly linked domains (6). Because they are tethered together, both domains tumble more slowly than the isolated E2 and Ub molecules, and since the tether is flexible the larger E2 domain tumbles more slowly than Ub (6).
Table 1.
Apparent correlation times (τc) for UbcH5c-O-Ub free and in complex with E4BU.
| UbcH5c τc (ns) | Ub τc (ns) | |
|---|---|---|
| UbcH5c-O-Ub | 17.7 +/− 0.1 | 13.0 +/− 0.1 |
| UbcH5c-O-Ub/E4BU | 22.0 +/− 0.2 | 14.3 +/− 0.2 |
| UbcH5c-O-Ub/E4BU | 21.8 +/− 0.2 | 14.8 +/− 0.1 |
| R1143A |
The apparent correlation times for the UbcH5c and Ub domains of the free conjugate are 17.7 and 13 ns, respectively, in agreement with the τc calculated from R2/R1 values reported previously for the conjugate under different buffer and temperature conditions (16.9 ns, 12.4 ns respectively) (6). The presence of E4BU results in slower tumbling of both domains consistent with formation of a ternary complex, as reflected in increased apparent τc values of 22 ns for UbcH5c and 14.3 ns for Ub (Table 1). Notably, the correlation time for Ub is still significantly smaller than that of E2, indicating that Ub remains flexibly linked to UbcH5c in the ternary E4BU/UbcH5c-O-Ub complex.
To obtain deeper insights into the data, we used HYDRONMR to predict relaxation parameters for a range of static E2~Ub and ternary complex structural models. While static models will not accurately predict results for flexibly-tethered systems, they can yield useful insights into the possible distributions of extended and compact states. The models used here were derived from published structures of U-box/E2 and E2-Ub complexes with different relative orientations of Ub (see Methods). Global correlation times were calculated and compared to experimental results for the isolated proteins, the conjugate, and the ternary complexes. Predicted correlation times ranged from 16-21 ns depending on the orientation for E2~Ub conjugates and 27-33 ns for the ternary complex. For UbcH5c in the conjugate, the experimental value is near the low end of the predicted range, indicating that either a compact static conformation is preferred or the system is dynamic with variable inter-domain orientations. [The term inter-domain is used here because when bound together, the E2 and Ub become two domains of a single E2~Ub molecule so technically they are domains.] In the ternary complex, UbcH5c tumbles faster than predicted even from the most compact structural model, an effect that can be attributed to E2~Ub flexibility. The experimental data for Ub follow a similar trend except that the tumbling in both the conjugate and the ternary complex is faster than predicted for any of the static binary complexes. As for UbcH5c, this finding is attributed to E2~Ub flexibility.
Model-free analysis with WT E4BU
Domain-based analysis of apparent rotational diffusion does not specifically parse overall tumbling of the molecule from inter-domain motion. This distinction is necessary for our goal of understanding how the relative flexibility of Ub with respect to UbcH5c is altered by binding to E4BU. We therefore turned to model-free analysis of the NMR relaxation data to investigate relative Ub flexibility in the ternary complex. A number of strategies were tried for fitting the data, starting with the basic model-free approach with a simple two state model of closed and open conformation. While these analyses returned reasonable isotropic correlation times and order parameters, the overall fit to the data was poor. In the end, we found that the extended model-free formalism, which allows separation of fast and slow motions, provided the best fit to the data for this system. This strategy has been used to successfully describe and quantify inter-domain flexibility for other systems (28–30). Our approach involved fitting the relaxation data to a multi-component motional model that includes an overall rotational correlation time τc plus residue-specific order parameters S2 composed of S2f and τf for the fast internal motions of bond vectors, and S2s and τs to represent the slow inter-domain motion. While this analysis can be quite powerful, there are many assumptions about the timescales of motion that can become problematic for detailed mechanistic studies of complex systems, as will be discussed below.
The first step for extended model-free analysis is to define the global τc of the complex. In cases where one domain dominates and is much larger than the second, the global tumbling of the complex is essentially the same as the apparent tumbling of the larger domain (14). Here, UbcH5c is only roughly twice the mass of Ub, so we did not expect global tumbling of the conjugate to be necessarily dominated by the E2 and therefore global tumbling would be significantly slower than the apparent diffusion tensor of the domain. To test this possibility, we searched for the best global τc value using two different programs to fit our data to the extended model-free formalism: freemodel and relax (14, 16, 17). In both cases, a rough grid search was performed to fit the data with a global τc ranging from the apparent τc of UbcH5c up to ~2-times larger than this value. The quality of each fit was assessed by an overall χ2 measure of error, and comparing average χ2 per residue (Figure 2). Remarkably, for both programs the best fit of the data for the isolated conjugate and the ternary complex occurs when the global τc is set to the apparent τc of UbcH5c (17.7 ns for the conjugate and 22 ns for the ternary complex). Testing lower τc values resulted in unrealistically high values of S2, while higher τc values resulted in poorer fits and very low S2 values. We therefore conclude that even though the E2 is only twice the mass of Ub, and rotational diffusion scales with the hydrated radius of the particle, the global tumbling of the complex is best fit by the isotropic τc of the E2 domain.
Figure 2.
Best fit of global correlation time is the apparent value for UbcH5c (E2). The average Χ2 error per residue is plotted for extended model-free fitting to a range of correlation times beginning at the correlation time of UbcH5c for that complex. Searching higher correlation times clearly results in poorer fits.
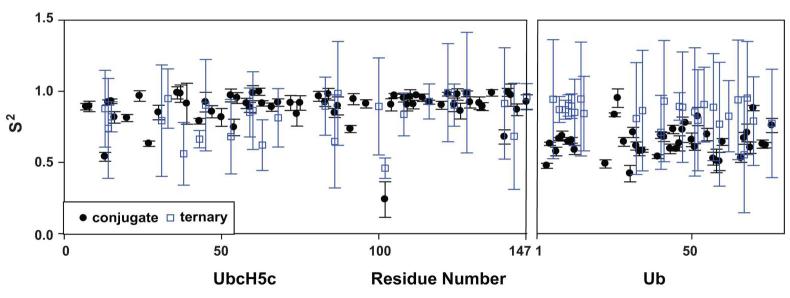
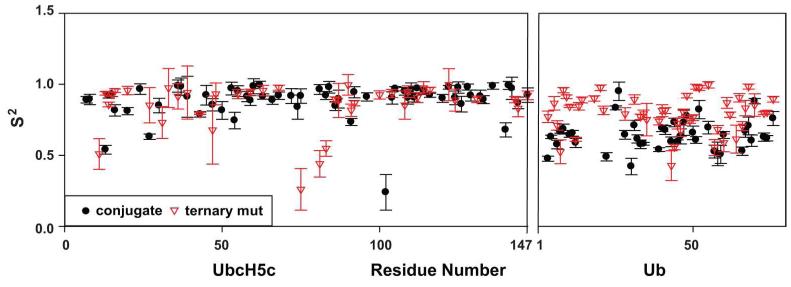
Once the global τc was set, we could analyze the internal dynamic parameters of the extended model-free formalism. This involved fitting S2 and S2f as variables, which allows the S2s values to be derived from the other two. For UbcH5c, the average S2 values are relatively high for both the conjugate and ternary complexes (Figure 3 and Table 2). Average S2s values are also quite high (0.92 and 0.91, respectively), indicating little if any inter-domain motion is observed for UbcH5c. The high values of S2s and absence of inter-domain motion are expected in this case, since the best fit for the global τc was obtained when it coincided with the apparent τc of this domain.
Figure 3.
Ub appears to become less flexible in the presence of E4BU. The determined extended model-free values for S2 are plotted for the conjugate E2-Ub with τc at 17.7 ns and the ternary complex with τc at 22 ns.
Table 2.
Average values for final fits to extended model-free using τc= 17.7 ns for the conjugate and 21.8 ns for ternary complexes. Averages are based on all residues (#aa) for which reasonable values were fit (ie S2 not 0 or 1) using all data available. All of the final analyses were based on the results obtained for the E4B mutant; the results with WT E4B are included for comparison.
| E2 | Ub | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| #aa | S2 | S2f | S2s | τs ns | #aa | S2 | S2f | S2s | τs ns | |
| UbcH5c-O-Ub | 58 | 0.89 +/− 0.05 |
0.95 +/− 0.04 |
0.92 +/− 0.03 |
0.71 +/− 1.31 |
31 | 0.65 +/− 0.03 |
0.83 +/− 0.03 |
0.78 +/− 0.03 |
3.2 +/− 0.72 |
|
UbcH5c-O-Ub/
E4BU R1143A |
41 | 0.85 +/− 0.07 |
0.89 +/− 0.06 |
0.92 +/− 0.06 |
0.49 +/− 0.72 |
46 | 0.79 +/− 0.03 |
0.92 +/− 0.03 |
0.85 +/− 0.02 |
1.7 +/− 0.23 |
|
UbcH5c-0-Ub/
WT E4BU |
26 |
0.80 +/−
0.25 |
0.88 +/−
0.21 |
0.91 +/−
0.23 |
0.49 +/−
1.19 |
24 |
0.85
+/− 0.30 |
0.95 +/−
0.23 |
0.85 +/−
0.28 |
2.4 +/−
1.47 |
The results for Ub were very different. The order parameters are lower in general and there is a dramatic shift between the conjugate and the ternary complex, with S2 increasing from 0.65 to 0.85 when E4BU is bound. This change was also reflected in S2s, which increases from 0.78 in the conjugate to 0.85 in the ternary complex. Thus, the data for Ub report a significant loss of inter-domain motion upon binding of UbcH5c-O-Ub to E4BU.
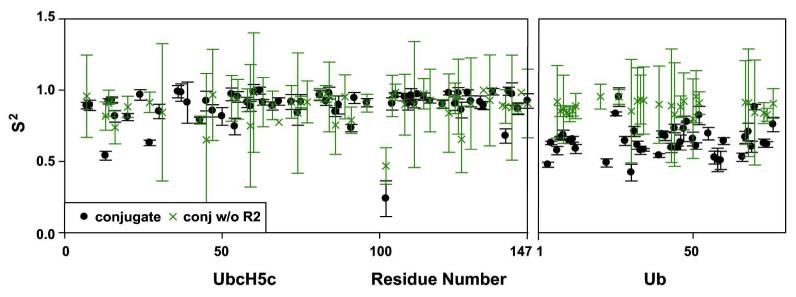
Although the trends from this analysis are clear, the overall tendency toward high values of the order parameters for Ub appears inconsistent with the τc values that indicate Ub retains substantial inter-domain flexibility in the ternary complex. To determine if the discrepancies are due to the lack of accurate R2 data for Ub in the ternary complex (see above), we compared the analysis of the data for the conjugate using only the 15N R1 and NOE data with the analysis that includes 15N R2 values (Figure 4). In making this comparison, we find very little difference in the order parameters for the E2 domain. In contrast, the order parameters for Ub returned from the analysis when no R2 data are used are much higher than those obtained when R2 is included. It therefore appears that the extended model-free analysis requires R2 values to generate reliable order parameters in cases of slow inter-domain motion. Among the standard R1, R2 and NOE parameters, only R2 reports on motional frequencies in the range of the J(0) term of the spectral density function. Although, R1 correctly reports on the apparent rotational tumbling and the dependency on J(0) cannot directly explain the relatively high order parameters observed for Ub, the R2 parameter does appear to be required to quantify the slow inter-domain motions of flexibly tethered domains.
Figure 4.
15N R2 values are critical for observing inter-domain flexibility in UbcH5c-O-Ub. Relaxation data were fit to an extended model-free formalism with and without R2 values and the resulting S2 values are shown.
Analysis of inter-domain dynamics using a mutant E4BU reveals that binding to an E3 induces a change in the flexibility of the conjugate
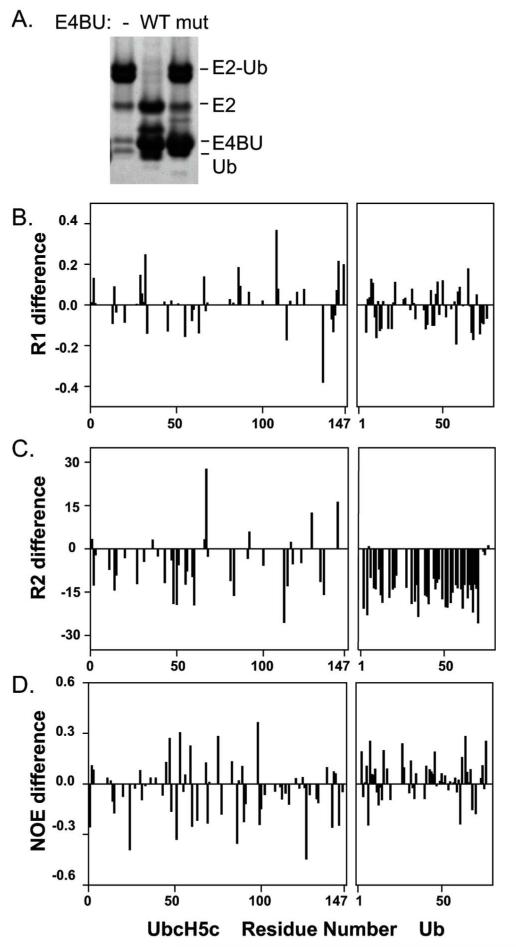
To overcome the inability to obtain accurate R2 values for the ternary complex, we turned to a mutant of E4BU, R1143A, which we have previously shown is much less effective at activating E2~Ub to discharge its Ub even though it has the same binding affinity for UbcH5c as WT E4BU (9). Notably, UbcH5c-O-Ub in a ternary complex with the mutant E4BU at a ratio of 3:1 is nearly as stable as the free UbcH5c-O-Ub conjugate (Figure 5A). Thus, use of E4BU R1143A allowed a full set of relaxation experiments to be performed on a single sample. To validate that the mutant is a viable model for the WT protein, we previously showed that titrations of UbcH5c with the R1143A E4BU yield the same pattern of chemical shift perturbations as the WT (9). The similarity extends to motions of WT and mutant E4BU in that the relaxation parameters measured at 600 MHz for the ternary complex with R1143A, other than the Ub R2 values, were remarkably consistent with the data obtained for the WT complex (Figure 5B-D, Table 1). The resulting τc values derived from these data are also consistent with the values found for the WT complex. This indicates that the motional properties of E2 and Ub in the ternary complex are not affected by the mutation in E4BU, and that use of the R1143A
Figure 5.
The allosteric mutant E4BU R1143A stabilizes Ub hydrolysis, but does not affect tumbling of the domain. (A) The ternary complex with E4BU R1143A is nearly as stable as the conjugate alone. (B-D) 15N NMR relaxation parameters are on average unchanged for each domain of the UbcH5c-O-Ub conjugate when bound to WT or R1143A E4BU. Shown are the differences between parameters measured for the WT and mutant ternary complexes at 600 MHz. The 15N R2 values for Ub could not be accurately obtained for the WT complex.
With accurate R2 values in hand, the relaxation data were fit to the extended model-free formalism as above (Figure 6, Table 2). To confirm the validity of the results obtained, the program relax was used to back-calculate 15N R1, R2 and NOE values for the model-free values. Overall, the simulated data agree well with experimental data (e.g. 15N R1 at 600 MHz is predicted to be 1.06 s−1 and was measured as 1.04 s−1 for UbcH5c residues in the conjugate, Table S1). The greatest difference was low values of the calculated NOE values for the E2 in the conjugate relative to the average observed value (0.65 vs 0.81). We have not found a specific explanation for this discrepancy, but attribute it to the limitations of applying the extended model-free formalism to describe the motions in this complex system.
Figure 6.
Ub is less flexible in the presence of E4BU. The determined extended model-free values for S2 are plotted for the conjugate E2-Ub with τc at 17.7 ns and the mutant ternary complex with τc at 21.8 ns.
The order parameters obtained for UbcH5c residues in the R1143A ternary complex, reflect a well-folded domain with no substantial inter-domain motion contributing to relaxation. The average S2s value of ~0.9 was similar to that seen for the conjugate and WT ternary complexes. Looking more deeply into the results, we found that the S2f values were somewhat higher than expected (0.88-0.95), but if these were fixed to the average expected for globular proteins (~0.85) (31), the average S2s value rose to ~1. As noted above, such high values of S2s and absence of inter-domain motion are expected for UbcH5c because the best fit for the global τc of the conjugate coincides with the apparent τc of this domain.
In contrast, there are clear differences in the inter-domain motion of Ub between the free conjugate and the ternary complex. The average S2 of 0.65 for the free conjugate indicates significant inter-domain flexibility. The value increases to 0.79 upon addition of E4BU R1143A, which corresponds to a reduction in inter-domain flexibility. The values of S2f are reasonably high (0.83-0.95), resulting in average S2s values of 0.78 for Ub in free conjugate and 0.85 in the ternary complex. If S2f values are fixed to the 0.85 value expected for a globular protein, the S2s values become 0.76 and 0.93, respectively. The high S2s value for the ternary complex implies little inter-domain motion, but we note this is inconsistent with the differences in UbcH5c and Ub relaxation parameters and correlation times (Table 1). Given the number of significant approximations implicit in applying the extended model-free formalism to this system, higher than expected values for S2s are not particularly surprising. The key point is that regardless of corrections needed to improve the fitting, a significant increase is observed in the S2s value of Ub upon binding of E4BU to UbcH5c-O-Ub. This increase in S2s corresponds to a reduction (but not complete loss) in the degree of inter-domain motion of Ub in the conjugate upon formation of the ternary complex.
Discussion
NMR relaxation analysis of free UbcH5c-O-Ub and in complex with E4BU demonstrates a reduction in Ub inter-domain flexibility upon binding of the conjugate to E4BU. The raw relaxation data alone for the ternary complex reveal a significant difference in the apparent correlation times of UbcH5c and Ub, directly demonstrating that the conjugate retains a significant degree of flexibility. Hence, the observation of Ub in a closed conformation in x-ray crystal structures provides only a partial description of the dynamic E3/E2~Ub complex. Our NMR relaxation analysis provides direct proof that E3 binding reduces the inter-domain motions of the E2~Ub conjugate, but does not result in a single static species. These findings provide strong support for the dynamic model of E2~Ub activation that is based on a combination of mutagenesis and NMR studies in which binding to the E3 ligase results in a shift in the conformational distribution of the flexible conjugate towards higher occupancy of closed states (Figure 7) (9). Furthermore, we show here that the E4BU mutant R1143A forms a complex with UbcH5c-O~Ub that is indistinguishable in its dynamical properties from the ternary complex formed with WT-E4BU, yet it is unable to activate E2~Ub. These findings reveal that the shift in population towards closed states upon binding to the E3 is necessary but not sufficient to activate the E2~Ub conjugate.
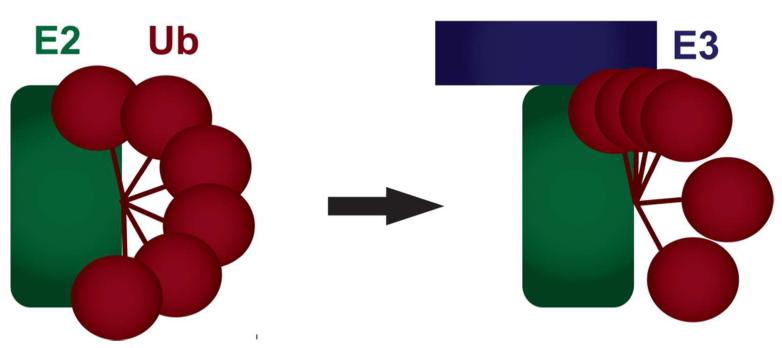
Figure 7.
Model of the change in population of inter-domain orientations of Ub in the E3/E2~Ub complex. Our data is consistent with a model of Ub flexibility allowing a large range of relative orientations in E2~Ub (left). Upon binding to a RING/U-box domain Ub is restricted somewhat such that it is not rigid but distribution of conformations is more populated in the ‘closed’ state (right).
Beyond the challenges of investigating this unstable and dynamic complex, a number of technical hurdles were encountered in applying relaxation-based analysis to this system. Describing inter-domain motion using NMR relaxation data requires an accurate global correlation time (τc). For flexibly tethered domains, this is complicated by the fact that the correlation time of the particle may be time-dependent and is often on a similar timescale to the inter-domain motion. One solution to this problem has been to focus simply on the apparent correlation time of each domain. However, a study with varied linker lengths found that even with an unstructured linker of 24 residues, two identical domains have different diffusion tensors (13). For both UbcH5c-O-Ub and E4BU/UbcH5c-O-Ub, we found that the correlation time for Ub was consistently smaller than that of the E2 indicating the presence of substantial inter-domain flexibility even when bound to the E3. Binding to E4BU slows tumbling of both the E2 and Ub, as the whole particle is now larger, therefore we expect increases in τc for both domains. Analysis of rotational diffusion complemented with simulations can reliably establish if flexibility exists between domains, but it appears that for more complicated systems, the ormalisms have not been sufficiently developed to quantify changes in a system as complex as E4BU/UbcH5c-O-Ub.
The approach we used to address this challenge was to obtain a global correlation time for the tethered domains and extract inter-domain motions. When one domain is much larger than the other, the global τc will be approximately equal to the apparent τc of the larger domain. This assumption appears to be valid in cases where the larger domain is several times larger than the second (14). In our work, a rough grid search showed that this assumption can be valid even for cases where the two domains are more similar in size (UbcH5c is approximately twice the mass of Ub). The timescale of inter-domain motion may provide an explanation for this observation.
A critical assumption in the extended model-free analysis is that there are no coupled motions. As described earlier, inter-domain motion with a significant change in shape may result in time-dependant rotational diffusion, so the measured global τc is then a time-averaged value. This study and previous work show that Ub can exist in a wide range of positions relative to the E2 and therefore is likely to exhibit time-dependent rotational diffusion. In addition, all model-free analyses rely on the assumption that the timescales of internal (τe or τs) and global (τc) motions are very different (28). Unfortunately, inter-domain motion is often on a timescale similar to that of global diffusion (nsec), and there is no rigorous theoretical treatment available (32). In such cases, extended model-free analysis inherently includes incorrect assumptions about coupled motions, and therefore, as we have seen in our study, the parameters extracted from the analysis reflecting the motions in this flexibly tethered system are not quantitatively accurate. Importantly, despite the limitations, the conclusions drawn about changes in inter-domain motion from comparative analyses as we have performed here will still be accurate.
A possible alternative to extended model-free is to utilize a basic model-free approach that assumes inter-conversion between two states (ITS), in which flexibility is described as an exchange between ‘open’ and ‘closed’ states (27, 32). Fast exchange would result in a population-weighted diffusion tensor, while slow exchange results in the slower domain reflecting global motion and inter-conversion is related to the difference in diffusion tensors (27). Good candidates for this analysis have no large change in shape unlike our system. A related formalism has been developed to account for the changes in global diffusion incurred during large conformational changes, but straightforward analysis is still limited to a two-state system (27). Unfortunately, more complicated systems such as the E3/E2~Ub complex with a continuum of different inter-domain orientations are not well fit by a two-state model.
In this work we have explored multiple methods to characterize inter-domain flexibility in a ternary complex. Identifying a suitable fitting algorithm proved to be a major hurdle, and while no fully accurate model is available, we have used the model that provides the best fit to the data. Assuming other types of conformational exchange do not exist, it should be possible to calculate the best fit parameters for each domain as a unit. Currently, both the relax and model-free programs fit S2, S2f, and τs for each residue. While this approach allows outliers to be found, the average value derived from residue-specific fitting does not necessarily provide an optimal fit for the domain as whole. A better strategy would be to perform domain-based minimization to search for a best fit for all residues simultaneously.
Interpretation of any formalism for analysis of relaxation data obtained for a system as complex as ours is not straightforward. Alone, the available relaxation data for UbcH5c~Ub reveal that the isolated E2~Ub samples a larger range of conformations than when E3 is present. This description can be refined based on previously published chemical shift and PRE analyses (Pruneda et al. 2012). Those data clearly indicate that in the ternary complex, Ub occupies a significant amount of time in ‘closed’ orientations. Combining all of the information allows us to model the system as shown in Figure 7 with Ub remaining flexibly attached to the E2 but shifting in the ternary complex to a distribution with a larger population of ‘closed’ orientations. We note that single molecule studies have the potential to provide a more detailed description of the specific motion of Ub in this system, for example by distinguishing a continuum of inter-domain orientations from population of a finite number of discrete states and providing a means to measure the lifetimes of Ub in any observable discreet states.
Conclusion
Changes in inter-domain motion of the nature we investigated here are difficult to examine with currently available experimental methods. As analyses of such systems are becoming more common, the need for additional data analysis formalisms will grow. Progress towards this objective is urgently required to enable model development for systems such as E2~Ub and the many modular proteins with multiple flexibly tethered domains. Our findings have significant implications for studies directed to elucidating the general mechanism of activation of E2~Ub conjugates. In particular, our results suggest that the static views derived from crystal structures may not capture the fundamentally dynamic character of ubiquitin in ternary complexes. Clearly, further investigations are required to test the competing models and obtain a complete picture of how binding to E3 catalyzes the reactivity of the E2~Ub conjugate.
Supplementary Material
Acknowledgements
The authors wish to thank Erik Zuiderweg for the use of the freemodel program, Jonathan Pruneda and Peter Brzovic for the UbcH5c-O-Ub backbone assignments, and Arthur G. Palmer III and Chris A. Brosey for helpful discussions.
This work was supported by the National Institutes of Health R01 GM075156 (WJC), R01 GM088055 (REK) and P30 CA068485 (Vanderbilt Ingram Cancer Center). The 900 MHz NMR instrument was purchased and access to other instrumentation was supported by grants MRI 0922863 from the NSF and 1S-10RR025677 from the NIH, and Vanderbilt University matching funds.
Abbreviations
- (Ub)
ubiquitin
- (E1)
E1 ubiquitin activating enzyme
- (E2)
E2 ubiquitin conjugating enzyme
- (E3)
E3 ubiquitin ligase
Footnotes
Supporting Information. Supporting information is available for chemical shift perturbations of E4BU titration into UbcH5c-O-Ub and the 15N NMR relaxation parameters recorded for UbcH5c-O-Ub alone and in complex with E4BU or E4BU R1143A. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 4.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage specific ubiquitin chain elongation by a single subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin ubiquitin conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in Motion: Structural studies of the E2~Ub conjugate. Biochemistry. 2011:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2~Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box Ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer AG. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 11.Baber JL, Szabo A, Tjandra N. Analysis of slow interdomain motion of macromolecules using NMR relaxation data. J. Am. Chem. Soc. 2001;123:3953–3959. doi: 10.1021/ja0041876. [DOI] [PubMed] [Google Scholar]
- 12.Amata I, Gallo M, Pennestri M, Paci M, Ragnini-Wilson A, Cicero DO. N-lobe dynamics of myosin light chain dictates its mode of interaction with myosin V IQ1. Biochemistry. 2008;47:12332–12345. doi: 10.1021/bi801178t. [DOI] [PubMed] [Google Scholar]
- 13.Walsh JD, Meier K, Ishima R, Gronenborn AM. NMR studies on domain diffusion and alignment in modular GB1 repeats. Biophys. J. 2010;99:2636–2646. doi: 10.1016/j.bpj.2010.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad A, Bhattacharya A, Mcdonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ERP. Heat shock protein 70 kDa chaperone / DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl. Acad. Sci. USA. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 16.d’Auvergne EJ, Gooley PR. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR. 2008;40:107–119. doi: 10.1007/s10858-007-9214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.d’Auvergne EJ, Gooley PR. Optimisation of NMR dynamic models II. A new methodology for the dual optimisation of the model-free parameters and the Brownian rotational diffusion tensor. J. Biomol. NMR. 2008;40:121–133. doi: 10.1007/s10858-007-9213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee LK, Rance M, Chazin WJ, Palmer AG. Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation. J. Biomol. NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- 19.García de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 20.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 21.Serniwka SA, Shaw GS. The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry. 2009;48:12169–12179. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- 22.Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Structural Biology. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Nordquist KA, Dimitrova YN, Brzovic PS, Ridenour WB, Munro KA, Soss SE, Caprioli RM, Klevit RE, Chazin WJ. Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry. 2010;49:347–355. doi: 10.1021/bi901620v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benirschke RC, Thompson JR, Nominé Y, Wasielewski E, Juranić N, Macura S, Hatakeyama S, Nakayama KI, Botuyan MV, Mer G. Molecular Basis for the Association of Human E4B U Box Ubiquitin Ligase with E2-Conjugating Enzymes UbcH5c and Ubc4. Structure. 2010;18:955–965. doi: 10.1016/j.str.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5 / Ubiquitin Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination. Mol. Cell. 2006;24:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Wong V, Case DA, Szabo A. Influence of the coupling of interdomain and overall motions on NMR relaxation. Proc. Natl. Acad. Sci. USA. 2009;106:11016–11021. doi: 10.1073/pnas.0809994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 29.Lipari G, Szabo A. Magnetic Resonance Relaxation in Macromolecules. 2. Analysis of Experimental Results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 30.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen 15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 31.Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium free calmodulin studied by 15N NMR relaxation measurements. FEBS J. 1995;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- 32.Ryabov YE, Fushman D. A model of interdomain mobility in a multidomain protein. Journal of the American Chemical Society. 2007;129:3315–3327. doi: 10.1021/ja067667r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.