Abstract
ErbB3 is a significant oncogenic target that is involved in the development of numerous malignancies. In the present in silico study, we evaluated the structural and functional impact of single nucleotide polymorphisms (SNPs) on the ErbB3 gene. The nonsynonymous SNPs (nsSNPs) are known to be deleterious or disease-causing variations because they alter protein sequence, structure, and function. Out of a total 531 SNPs in ErbB3, we investigated 77 coding nsSNPs and observed that 20 of them could be expected to alter the protein's function based on the predictions of both sequence homology–based (SIFT) and structural homology–based (Polyphen) algorithms. Thereafter, we computed the stability of mutants in units of free energy using I-Mutant 3.0, MuStab, and iPTree-STAB programs and identified seven crucial point mutations (V89M, V105G, C290Y, I418N, R669C, I744T, and A1131T) in epidermal growth factor receptor 3 that are manifested as nsSNPs. Furthermore, FASTSNP determined 14 synonymous SNPs that may have a profound impact on splicing regulation. The computational study identified seven novel hotspots predicted to maintain the native structural conformation and functional activity of ErbB3 and may account for cancer if mutated.
Key words: bioinformatics, cancer, ErbB3, nonsynonymous SNPs, single nucleotide polymorphism
Introduction
Epidermal growth factor receptor (EGFR) belongs to the receptor kinase I family. It is a transmembrane glycoprotein involved in many cell functions, including proliferation, differentiation, and adhesion.1,2 It has four isoforms or members, ErbB1, ErbB2, ErbB3, and ErbB4. In recent years, EGFR and its members have become well known as potential oncogenic drug targets. All four members share four common structural domains: ectodomain, juxtamembrane, kinase, and carboxy terminal domain.3 Activation of the ErbB receptor family occurs when specific ligands bind to the extracellular region, leading to dimerization. Consequently, autophosphorylation of tyrosine residues in the catalytic kinase domain occurs, forming a docking pocket for other adapter proteins and triggers for numerous different signaling cascades.3,4 However, ErbB3 is devoid of a catalytic kinase domain, which makes it unique from other members. Therefore, for activation, ErbB3 forms heterodimers with the other active ErbB receptors.5 It is well known that amplification, overexpression, mutation, or polymorphisms of ErbB3 can cause various cancers, including breast cancer and colon cancer.6 Hence, it is assumed that any alteration in the well-defined structural conformation may affect the functional activity of the gene.
Most recurrent genomic variations are manifested as single nucleotide polymorphisms (SNPs), and there is a strong correlation between certain polymorphisms and disease.7 Nonsynonymous SNPs (nsSNPs) are present in the coding region, which alters the amino acid composition and consequently has a profound impact on protein structure and function.8 Computational investigations of nsSNPs of ErbB1 and ErbB2 have previously been done,9,10 and in the present work, we identified critical deleterious nsSNPs and other functionally significant coding SNPs of the ErbB3 gene. We selected 77 nsSNPs of ErbB3 to determine their effect on the protein structure. Both SIFT (Sorting Intolerant from Tolerant) and PolyPhen v2 (Polymorphism Phenotyping) programs detected 20 destructive nsSNPs in ErbB3 protein.11,12 It is very important to evaluate point mutations that may disrupt structural conformation. Thus, we checked the protein stability upon substitution in terms of free energy by using three different web servers I-Mutant 3.0, MuStab, and iPTree-STAB.13–15 Consequently, we identified seven novel mutations of ErbB3 that may affect structural stability and alter expression of the protein. We also investigated 14 functionally important noncoding SNPs using the Function Analysis and Selection Tool for Single Nucleotide Polymorphisms (FASTSNP).16 The main advantage of this computational study is that it could lessen efforts needed for phenotyping–genotyping association studies. Moreover, the genomic analysis of the ErbB3 gene could explain diseases associated with ErbB3.
Materials and Methodology
Collection of the ErbB3 SNP dataset
The ErbB3 gene polymorphism data were mined from the dbSNP database (http://www.ncbi.nlm.nih.gov/snp).17 There were a total of 531 SNPs of human ErbB3, which included 79 nsSNPs (i.e., approximately 15%). Here, we considered 77 coding nsSNPs because they were associated with the same longest isoform protein (i.e., NP_001973.2) of ErbB3.
Assessment of the functional consequences of deleterious nsSNPs using a sequence homology–based method (SIFT)
The functional impacts of the 77 nsSNPs of the ErbB3 gene were detected using SIFT (http://sift.jcvi.org).11 The SIFT program predicts deleterious or nontolerated SNPs on the premise that some amino acids tend to be conserved in a protein family and any substitution at these positions would affect protein function and thus have a phenotypic effect. SIFT calculates the normalized probability in terms of SIFT score or tolerance index (TI) score for each mutation. The substitutions with normalized probabilities ≤0.05 are predicted to be nontolerated or deleterious amino acids substitutions, whereas those >0.05 are considered to be tolerated.
Investigation of the functional impact of coding nsSNPs using structure homology–based method (PolyPhen)
To analyze the possible impact of an amino acid substitution on the structure and function of an ErbB3 protein we used PolyPhen v2 (http://genetics.bwh.harvard.edu/pph2).12 The protein sequence with mutational position and two amino acid variants were submitted to the server. PolyPhen generates multiple sequence alignment of homologous protein structures, calculates the position-specific independent counts (PSIC) scores for each of the two variants, and then calculates the PSIC score difference between both the allelic variants. The higher the PSIC score difference, the higher the functional impact a particular amino acid substitution is likely to have or the more likely it is to be damaging. The PolyPhen server discriminates nsSNPs into three main categories, benign, possibly damaging, or probably damaging, and provides the corresponding specificity and sensitivity values. The probably damaging nsSNPs are those that are predicted with high confidence and are expected to affect protein structure or function. Therefore, we selected the nsSNPs that were determined to be probably damaging and possessed PSIC scores >0.951. Thereafter, we examined nsSNPs predicted to be deleterious or to cause disease both by the SIFT and PolyPhen programs.
Calculation of stability of predicted mutations by free energy
Mutations usually change the structural stability of a protein and thus affect its functional activity. In order to check the stability of a predicted 20 deleterious mutants in terms of energy we used three different web servers; namely, I-Mutant 3.0, iPTree-STAB, and MuStab.13–15 The I-Mutant 3.0 suite (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) is based on a support vector machine (SVM) algorithm that calculates protein stability related to a single mutation in units of free energy (i.e., ΔΔG values) and also predicts the deleterious SNPs from the human protein sequence.13 iPTree-STAB (http://210.60.98.19/IPTREEr/iptree.htm) is based on a decision tree along with a boosting algorithm that determines the stability changes (ΔΔG values) and thus predicts whether the substitutions are stabilizing or destabilizing.14 We also used MuStab (http://bioinfo.ggc.org/mustab), which is also based on an SVM, to detect the protein stability changes upon amino acid substitutions.15 The nsSNPs that were defined as unstable by any two of the programs and also possessed ΔΔG values of less than −1.0 kcal/mol were considered for the study.
Functional significance of SNPs in regulatory regions
The online tool FASTSNP (http://fastsnp.ibms.sinica.edu.tw/pages/input_SNPListAnalysis.jsp) was used to determine the functional impact of the synonymous SNPs, 3′ untranslated region (UTR) SNPs, 5′UTR SNPs, and intronic SNPs on the regulation of the ErbB3 gene.16 FastSNP follows the decision tree principle that predicts whether a noncoding SNP alters the transcription factor binding site of a gene or not. FastSNP generates the score on the basis of the risk level with a ranking from 0 to 5, which signifies the level of no risk to very high risk, respectively. The SNPs ranging from low risk (rank 2) to upper risk (rank 5) were considered to be functionally significant.
Results and Discussion
The SNP dataset of the ErbB3 gene
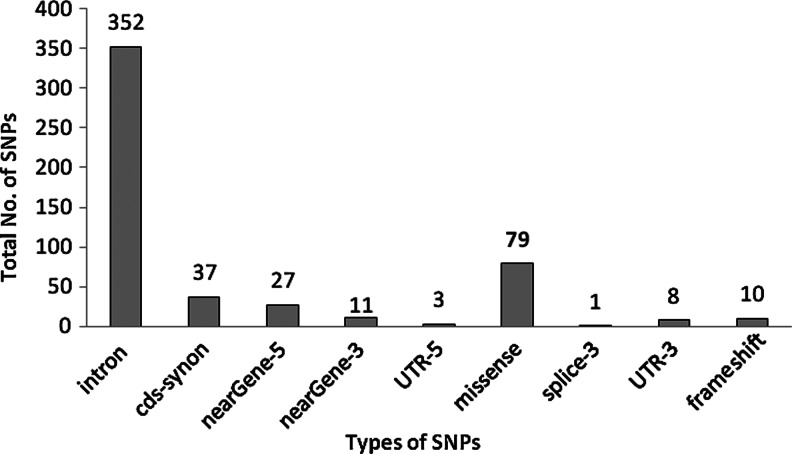
The polymorphism dataset of the ErbB3 gene was downloaded from dbSNP, which contained 531 SNPs. Out of 531 SNPs, records have been deleted for three (rs267603577, rs267603578, and rs267603579), so 528 SNPs remained. Of these 528 SNPs, 37 and 79 were synonymous and nonsynonymous (missense) SNPs, respectively. The remaining 412 SNPs were distributed in different regions, including three SNPs in the 5′UTR, eight SNPs in the 3′UTR, a single SNP in splice-3, 27 SNPs in near-gene 5′, 11 SNPs in near-gene 3′, 10 SNPs in frameshift, and 352 SNPs (66%) in the intronic region as shown in Figure 1. However, out of 79 missense SNPs we considered only 77 coding nsSNPs for our analysis because they belonged to the same longest isoform protein of the ErbB3 gene (i.e., NP_001973.2).
FIG. 1.
The division of ErbB3 SNPs in different regions. cds-synon, coding sequence synonymous.
Analysis of deleterious nsSNPs predicted by the SIFT program
SIFT is a sequence homology–based tool that determines whether a particular amino acid substitution has a tolerable impact or not based on its conservation level in the protein family. The residue change and mutational position of 77 missense nsSNPs along with their protein sequences were entered in the SIFT server to compute their TI scores, and the results are compiled in Table 1. According to the Ng and Henikoff11 classification, the TI score is inversely proportional to the functional impact of residue substitution. Among 77 nsSNP, 38 had a TI score of ≤0.05 and were predicted to be damaging or deleterious. Out of these 38 nsSNPs, 21 had a TI score of 0.0, five had a TI score of 0.1, three had a TI score of 0.02, four had a score of 0.03, two had a score of 0.04, and the remaining three nsSNPs had a score of 0.05. The amino acid change from Arg to Trp was found to occur the most frequently, which implies that there is an aberrant change from positively charged polar arginine residue to hydrophobic nonpolar residue tryptophan.
Table 1.
All 77 Coding Nonsynonymous Single Nucleotide Polymorphisms That Were Evaluated by Both SIFT and PolyPhen Algorithms
| |
|
|
SIFT |
PolyPhen |
||||
|---|---|---|---|---|---|---|---|---|
| S. no. | SNP ID | Mutation | Prediction | TI score | Prediction | Score | Sensitivity | Specificity |
| 1 | rs34379766 | S20Y | Damaging | 0.05 | Benign | 0.158 | 0.92 | 0.87 |
| 2 | rs56017157 | P30L | Tolerated | 0.08 | Benign | 0.007 | 0.96 | 0.75 |
| 3 | rs142735651 | T68M | Tolerated | 0.44 | Benign | 0.013 | 0.96 | 0.78 |
| 4 | rs143770796 | D73N | Tolerated | 0.13 | Probably damaging | 0.986 | 0.74 | 0.96 |
| D73Y | Damaging | 0 | Probably damaging | 1 | 0 | 1 | ||
| 5 | rs77228285 | V89M | Damaging | 0 | Probably damaging | 0.996 | 0.55 | 0.98 |
| 6 | rs200856864 | T96A | Tolerated | 0.55 | Benign | 0.002 | 0.99 | 0.3 |
| 7 | rs201479792 | N101S | Damaging | 0.01 | Benign | 0.281 | 0.91 | 0.88 |
| 8 | rs146486757 | R103C | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 9 | rs984896 | V105G | Damaging | 0 | Probably damaging | 0.995 | 0.68 | 0.97 |
| 10 | rs147905731 | D153N | Tolerated | 0.06 | Benign | 0.001 | 0.99 | 0.15 |
| 11 | rs141700623 | H157Y | Tolerated | 0.84 | Benign | 0.092 | 0.93 | 0.85 |
| 12 | rs188795493 | I161V | Tolerated | 0.23 | Probably damaging | 0.988 | 0.73 | 0.96 |
| 13 | rs200978269 | R170Q | Tolerated | 0.15 | Benign | 0 | 1 | 0 |
| 14 | rs150454821 | G198V | Damaging | 0.01 | Probably damaging | 0.998 | 0.27 | 0.99 |
| 15 | rs146860437 | E200K | Tolerated | 0.1 | Benign | 0 | 1 | 0 |
| 16 | rs56107455 | T204I | Tolerated | 0.44 | Benign | 0 | 1 | 0 |
| 17 | rs201079200 | D229N | Tolerated | 0.66 | Benign | 0.002 | 0.99 | 0.3 |
| 18 | rs140656187 | A232S | Damaging | 0 | Possibly damaging | 0.951 | 0.79 | 0.95 |
| 19 | rs149635848 | V285I | Tolerated | 0.53 | Benign | 0 | 1 | 0 |
| 20 | rs143406438 | C290Y | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 21 | rs137870123 | K314R | Tolerated | 0.11 | Benign | 0.039 | 0.94 | 0.83 |
| 22 | rs200211366 | N369S | Tolerated | 0.5 | Benign | 0.001 | 0.99 | 0.15 |
| 23 | rs12320176 | N385S | Tolerated | 0.32 | Benign | 0.001 | 0.99 | 0.15 |
| 24 | rs139868331 | R391W | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 25 | rs74763375 | N414H | Damaging | 0.01 | Probably damaging | 1 | 0 | 1 |
| 26 | rs201880960 | I418V | Damaging | 0 | Possibly damaging | 0.87 | 0.83 | 0.93 |
| 27 | rs141230043 | I418N | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 28 | rs144549266 | R453H | Tolerated | 0.16 | Probably damaging | 1 | 0 | 1 |
| 29 | rs200007116 | I456V | Tolerated | 0.07 | Possibly damaging | 0.743 | 0.85 | 0.92 |
| 30 | rs149951770 | R490H | Tolerated | 0.08 | Possibly damaging | 0.867 | 0.83 | 0.93 |
| 31 | rs182692782 | V494L | Tolerated | 0.32 | Benign | 0 | 1 | 0 |
| 32 | rs146593760 | K498I | Tolerated | 0.18 | Benign | 0.027 | 0.95 | 0.81 |
| 33 | rs145108143 | G513D | Damaging | 0.02 | Probably damaging | 0.999 | 0.14 | 0.99 |
| 34 | rs200670489 | T541S | Damaging | 0 | not run by the server | |||
| 35 | rs201942735 | S551F | Damaging | 0.03 | Benign | 0.013 | 0.96 | 0.78 |
| 36 | rs147888915 | C553R | Damaging | 0.01 | not run by the server | |||
| 37 | rs202048840 | G561S | Tolerated | 0.17 | Probably damaging | 0.989 | 0.72 | 0.97 |
| 38 | rs141636701 | A577T | Tolerated | 0.1 | Benign | 0.001 | 0.99 | 0.15 |
| 39 | rs200350558 | R580Q | Tolerated | 0.37 | Benign | 0.28 | 0.91 | 0.88 |
| 40 | rs200574817 | H614D | Damaging | 0.05 | Probably damaging | 0.995 | 0.68 | 0.97 |
| 41 | rs143726790 | E615K | Tolerated | 0.59 | Benign | 0.125 | 0.93 | 0.86 |
| 42 | rs151083303 | P624R | Tolerated | 0.07 | Probably damaging | 1 | 0 | 1 |
| 43 | rs141054346 | V635M | Tolerated | 0.15 | Benign | 0.063 | 0.94 | 0.84 |
| 44 | rs139022684 | G661S | Tolerated | 0.37 | Benign | 0 | 1 | 0 |
| 45 | rs200724560 | R669C | Damaging | 0 | Probably damaging | 0.999 | 0.14 | 0.99 |
| 46 | rs56387488 | R683W | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 47 | rs138548737 | S686R | Damaging | 0.05 | Probably damaging | 0.999 | 0.14 | 0.99 |
| 48 | rs181659329 | P692H | Damaging | 0 | Possibly damaging | 0.911 | 0.81 | 0.94 |
| 49 | rs35961836 | S717L | Damaging | 0.02 | Possibly damaging | 0.717 | 0.86 | 0.92 |
| 50 | rs189789018 | V723L | Damaging | 0.03 | Probably damaging | 0.999 | 0.14 | 0.99 |
| 51 | rs55787439 | I744T | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 52 | rs3891921 | D758H | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 53 | rs202221237 | G780E | Tolerated | 0.08 | Probably damaging | 1 | 0 | 1 |
| 54 | rs144510847 | L795V | Damaging | 0 | Benign | 0.123 | 0.93 | 0.86 |
| 55 | rs148448153 | H802Y | Damaging | 0 | Benign | 0.139 | 0.92 | 0.86 |
| 56 | rs182154425 | G804V | Damaging | 0 | Possibly damaging | 0.943 | 0.8 | 0.95 |
| 57 | rs80185484 | A805P | Tolerated | 0.08 | Benign | 0.091 | 0.93 | 0.85 |
| 58 | rs147206496 | P845A | Damaging | 0.03 | Benign | 0 | 1 | 0 |
| 59 | rs143021252 | S896N | Damaging | 0 | Probably damaging | 1 | 0 | 1 |
| 60 | rs144558290 | A913T | Tolerated | 0.1 | Benign | 0.007 | 0.96 | 0.75 |
| 61 | rs193920754 | Q934H | Damaging | 0.03 | Benign | 0.02 | 0.95 | 0.8 |
| 62 | rs60586767 | A962T | Tolerated | 0.08 | Probably damaging | 1 | 0 | 1 |
| 63 | rs56259600 | K998R | Tolerated | 0.44 | Benign | 0.056 | 0.94 | 0.84 |
| 64 | rs139267530 | E1019D | Tolerated | 0.51 | Benign | 0 | 1 | 0 |
| 65 | rs150001629 | T1024N | Tolerated | 0.07 | Benign | 0 | 1 | 0 |
| 66 | rs200017094 | R1040W | Damaging | 0 | Benign | 0.002 | 0.99 | 0.3 |
| 67 | rs149181380 | R1040Q | Damaging | 0.04 | Possibly damaging | 0.913 | 0.81 | 0.94 |
| 68 | rs151311358 | S1049G | Damaging | 0.04 | Benign | 0.088 | 0.93 | 0.85 |
| 69 | rs17118292 | M1055I | Tolerated | 0.59 | Benign | 0.005 | 0.97 | 0.74 |
| 70 | rs201958747 | R1118Q | Tolerated | 0.27 | Probably damaging | 0.986 | 0.74 | 0.96 |
| 71 | rs773123 | S1119C | Tolerated | 0.07 | Probably damaging | 1 | 0 | 1 |
| 72 | rs201486425 | P1126L | Tolerated | 0.16 | Benign | 0.104 | 0.93 | 0.86 |
| 73 | rs150312718 | A1131T | Damaging | 0.02 | Probably damaging | 0.996 | 0.55 | 0.98 |
| 74 | rs180986542 | R1173W | Damaging | 0.01 | Benign | 0 | 1 | 0 |
| 75 | rs55709407 | T1254K | Tolerated | 0.52 | Possibly damaging | 0.828 | 0.84 | 0.93 |
| 76 | rs201199014 | H1330Y | Damaging | 0 | Probably damaging | 0.997 | 0.41 | 0.98 |
| 77 | rs202205409 | P1335S | Tolerated | 0.59 | Benign | 0.001 | 0.99 | 0.15 |
TI, tolerance index.
Investigation of coding nsSNPs computed by the PolyPhen server
The PolyPhen program predicts the plausible consequences of an amino acid substitution on the structure and function of a human protein. The 77 point mutations marked as nsSNPs were submitted to the PolyPhen program, and the results are compiled in Table 1. The nsSNPs possessing a PSIC score difference of >0.951 were considered to be deleterious because they were all predicted to be probably damaging with high confidence. Out of 77 nsSNPs, 29 were identified as altering the native protein conformation. There was a significant association between the results obtained from both the SIFT and PolyPhen programs for 18 nsSNPs, suggesting that these nsSNPs may disrupt the protein at both sequence and structural levels. Out of the 29 nsSNPs, nine had a TI score of 0 and a PSIC score difference of 1; namely, rs143770796, rs146486757, rs143406438, rs139868331, rs141230043, rs56387488, rs55787439, rs3891921, and rs143021252. These nine nsSNPs were identified as the most damaging polymorphisms affecting protein activity as shown in Table 1. Thereafter, we selected 20 significant nsSNPs because they were predicted to be deleterious by both SIFT and PolyPhen programs. Out of these 20 nsSNPs, rs150454821 and rs74763375 were found to be the most destructive because they had low TI scores (0.01) and high PSIC scores (1 or approximately 0.99). Hence, the identification of these 20 damaging nsSNPs mutations are very important because they might cause disease.
Prediction of stability change on mutation of 18 nsSNPs
The main aim of the study was to identify the crucial coding nsSNPs that would be expected to disrupt the native structure of the protein and thus affect its function. We investigated the protein stability of 20 nsSNPs upon mutation in terms of free energy using I-Mutant 3.0, MuStab, and iPTree-STAB as shown in Table 2. There were a total of three mutants, V89M (rs77228285), V105G (rs984896), and I744T (rs55787439), that were predicted to be the most unstable as determined by all three programs. The mutation from valine to glycine at position 105 was found to be the most damaging because it exhibited the lowest free energy: −2.96 and −1.77 kcal/mol as determined by MuStab and iPTree-STAB, respectively. Four other mutations, C290Y (rs143406438), I418N (rs141230043), R669C (rs200724560), and A1131T (rs150912718), were predicted to be unstable by two severs. Of these seven mutants, V89M, V105G, C290Y, and I418N are present in the extracellular region where the specific ligand attaches, while R669C, I744T, and A1131T lie within the intracellular region, which contains the kinase domain.
Table 2.
The Free Energy or Stabilities of 20 Nonsynonymous Single Nucleotide Polymorphisms as Computed by I-Mutant 3.0, MuSTAB, and iPTree-STAB
| |
|
|
I-Mutant 3.0 |
MuSTAB |
iPTree-STAB |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. no. | SNP ID | Mutation | PHD | RI | DDG (kcal/mol) | SVM3 prediction | RI | Protein stability | PC (%) | Prediction | DDG (kcal/mol) |
| 1 | rs143770796 | D73Y | Disease | 5 | −0.02 | Large increase | 0 | Increased | 22.86 | Negative (destabilizing) | 0.62 |
| 2 | rs77228285a | V89M | Disease | 4 | −1.55 | Large decrease | 4 | Decreased | 86.07 | Negative (destabilizing) | −1.3492 |
| 3 | rs146486757 | R103C | Disease | 6 | −1.24 | Large decrease | 4 | Increased | 25.18 | Negative (destabilizing) | 1.945 |
| 4 | rs984896a | V105G | Disease | 7 | −2.96 | Large decrease | 9 | Decreased | 90.71 | Negative (destabilizing) | −1.7783 |
| 5 | rs150454821 | G198V | Disease | 5 | −0.37 | Neutral | 0 | Decreased | 82.32 | Negative (destabilizing) | −1.6632 |
| 6 | rs143406438a | C290Y | Disease | 6 | −0.18 | Large decrease | 2 | Decreased | 81.79 | Negative (destabilizing) | −1.66 |
| 7 | rs139868331 | R391W | Disease | 4 | −0.55 | Large decrease | 3 | Decreased | 79.64 | Negative (destabilizing) | 1.945 |
| 8 | rs74763375 | N414H | Disease | 4 | −0.97 | Large decrease | 4 | Decreased | 81.07 | Negative (destabilizing) | 0.9377 |
| 9 | rs141230043a | I418N | Disease | 6 | −2.29 | Large decrease | 7 | Decreased | 91.79 | Negative (destabilizing) | −0.4685 |
| 10 | rs145108143 | G513D | Disease | 5 | −0.35 | Neutral | 2 | Decreased | 82.32 | Negative (destabilizing) | −0.065 |
| 11 | rs200574817 | H614D | Neutral | 1 | −0.26 | Large decrease | 1 | Decreased | 80.54 | Negative (destabilizing) | −0.0846 |
| 12 | rs200724560a | R669C | Disease | 4 | −1.04 | Neutral | 1 | Decreased | 81.07 | Negative (destabilizing) | −1.72 |
| 13 | rs56387488 | R683W | Disease | 6 | −0.48 | Large decrease | 0 | Increased | 23.57 | Negative (destabilizing) | −0.0033 |
| 14 | rs138548737 | S686R | Disease | 3 | −0.04 | Neutral | 2 | Decreased | 83.75 | Negative (destabilizing) | −0.1221 |
| 15 | rs189789018 | V723L | Disease | 4 | −1.14 | Large increase | 3 | Decreased | 81.25 | Negative (destabilizing) | 0.6923 |
| 16 | rs55787439a | I744T | Disease | 4 | −2.03 | Large decrease | 7 | Decreased | 88.75 | Negative (destabilizing) | −1.324 |
| 17 | rs3891921 | D758H | Disease | 4 | −0.51 | Large decrease | 1 | Decreased | 81.61 | Negative (destabilizing) | −1.0233 |
| 18 | rs143021252 | S896N | Neutral | 2 | −0.26 | Neutral | 2 | Increased | 25.18 | Negative (destabilizing) | −1.1536 |
| 19 | rs150312718a | A1131T | Neutral | 5 | −0.74 | Large decrease | 0 | Decreased | 79.64 | Negative (destabilizing) | −4.2533 |
| 20 | rs201199014 | H1330Y | Disease | 6 | −0.09 | Neutral | 3 | Decreased | 81.25 | Negative (destabilizing) | −1.1536 |
The most crucial deleterious nsSNPs.
PHD, predictor of effect on human health; RI, reliability index; DDG, differences in the free energy; SVM, support vector machine; PC, prediction confidence.
Identification of functional SNPs in noncoding segments
We used FASTSNP to predict functionally significant SNPs. According to the FASTSNP results, 14 out of the 449 SNPs in the ErbB3 gene would be damaging (risks of 3–4 and 2–3 rank), with functional consequences for splicing regulation as shown in Table 3.
Table 3.
Record of All Functionally Significant Single Nucleotide Polymorphisms as Identified by FASTSNP
| S. no. | SNP ID | Noncoding region | Level of risk | Possible functional effects |
|---|---|---|---|---|
| 1 | rs67617070 | Frameshift | Low-medium (2–3) | Splicing regulation |
| 2 | rs67420827 | Frameshift | Low-medium (2–3) | Splicing regulation |
| 3 | rs66493360 | Frameshift | Low-medium (2–3) | Splicing regulation |
| 4 | rs56073151 | cds-synon | Low-medium (2–3) | Sense/synonymous; splicing regulation |
| 5 | rs55880327 | cds-synon | Low-medium (2–3) | Sense/synonymous; splicing regulation |
| 6 | rs55699040 | Intron | Low-medium (2–3) | Missense (conservative) |
| 7 | rs11171743 | Intron | Low-medium (2–3) | Missense (conservative) |
| 8 | rs2271189 | Intron | Low-medium (2–3) | Sense/synonymous; splicing regulation |
| 9 | rs2229046 | cds-synon | Low-medium (2–3) | Sense/synonymous; splicing regulation |
| 10 | rs66581925 | Intron | Medium-high (3–4) | Splicing site |
| 11 | rs2271194 | Intron | Medium-high (3–4) | Splicing site |
| 12 | rs2271188 | Intron | Medium-high (3–4) | Missense (nonconservative); splicing regulation |
| 13 | rs812826 | Intron | Medium-high (3–4) | Splicing site |
| 14 | rs773123 | Intron | Medium-high (3–4) | Missense (nonconservative); splicing regulation |
Conclusion
In the current work, the influence of functional SNPs in the ErbB3 oncogene was investigated through various computational methods. From a total of 531 SNPs in the ErbB3 gene, 79 SNPs were found to be nonsynonymous, 37 were synonymous, and 352 (66%) occurred in intronic regions. Out of 77 coding nsSNPs (which belonged to the same protein), 29 and 38 were found to be deleterious by PolyPhen and SIFT programs, respectively. An in silico evaluation using two different algorithms (SIFT and Polyphen) revealed that 20 nsSNPs were crucial for the structure or function of the EGFR3 protein. Further, we evaluated the protein stability based upon mutations caused by these 20 deleterious nsSNPs by using three distinct servers (I-Mutant 3.0, MuStab, and iPTree-STAB). Consequently, we determined that seven crucial mutations (V89M, V105G, I744T, C290Y, I418N, R669C, and A1131T) may disrupt the protein conformation. Of these seven, the mutants V89M, V105G, and I744T were identified as being the most unstable in terms of free energy. Moreover, there were 14 synonymous SNPs that were predicted to be functionally significant by the FASTSNP server. Our results suggest that these novel mutants have a potential functional impact and can thus be used for pharmacogenomic and pharmacokinetic studies. These proposed mutants could also be used as drug targets in screening studies because they might play an important role in causing malignancy.
Disclosure Statement
No competing financial interests exist.
References
- 1.Stein RA. Staros VJ. Insights into the evolution of the ErbB receptor family and their ligands from sequence analysis. BMC Evol Biol. 2006;6:79. doi: 10.1186/1471-2148-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han W. Lo HW. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318:124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorissen RN. Walker F. Poulit N, et al. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 4.Sukhramani PS. Sukhramani PS. Suthar MP. EGFR kinase: potential target for cancer therapy. J Pharmacol Toxicol. 2010;1:1–22. [Google Scholar]
- 5.Jathal MK. Chen L. Mudryj M, et al. Targeting ErbB3: the new RTK(id) on the prostate cancer block. Immunol Endocr Metab Agents Med Chem. 2011;11:131–149. doi: 10.2174/187152211795495643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sithanandam G. Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15:413–448. doi: 10.1038/cgt.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi Y. Mimori K. Mori M. Significance of genome-wide association study in cancer. Nihon Geka Gakkai Zasshi. 2012;113:210–214. [PubMed] [Google Scholar]
- 8.Wohlrab H. The human mitochondrial transport/carrier protein family nonsynonymous single nucleotide polymorphisms (nsSNPs) and mutations that lead to human diseases. Biochim Biophys Acta. 2006;1757:1263–1270. doi: 10.1016/j.bbabio.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 9.Choura M. Frikha F. Kharrat N, et al. Investigating the function of three non-synonymous SNPs in EGFR gene: structural modelling and association with breast cancer. Protein J. 2010;29:50–54. doi: 10.1007/s10930-009-9221-0. [DOI] [PubMed] [Google Scholar]
- 10.Rajasekaran R. George PDC. Sudandiradoss C, et al. Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. C R Biol. 2008;331:409–417. doi: 10.1016/j.crvi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Ng PC. Henikoff SS. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramensky V. Bork P. Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capriotti E. Fariselli P. Rossi I, et al. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics. 2008;9(Suppl. 2):S6. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng S. Srivastava AK. Wang L. Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics. 2010;11(Suppl. 2):S5. doi: 10.1186/1471-2164-11-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang LT. Gromiha MM. Ho SY. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics. 2007;23:1292–1293. doi: 10.1093/bioinformatics/btm100. [DOI] [PubMed] [Google Scholar]
- 16.Yuan HY. Chiou JJ. Tseng WH, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry ST. Ward MH. Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]