Abstract
Results from recent HIV-1 vaccine studies have indicated that high serum antibody (Ab) titers may not be necessary for Ab-mediated protection, and that Abs localized to mucosal sites might be critical for preventing infection. Enzyme-linked immunosorbent assay (ELISA) has been used for decades as the gold standard for Ab measurement, though recently, highly sensitive microsphere-based assays have become available, with potential utility for improved detection of Abs. In this study, we assessed the Bio-Plex® Suspension Array System for the detection of simian immunodeficiency virus (SIV)-specific Abs in rhesus macaques (RMs) chronically infected with SIV, whose serum or mucosal SIV-specific Ab titers were negative by ELISA. We developed a SIVmac239-specific 4-plex bead array for the simultaneous detection of Abs binding to Env, Gag, Pol, and Nef. The 4-plex assay was used to quantify SIV-specific serum IgG and rectal swab IgA titers from control (SIV-naive) and SIVmac239-infected RMs. The Bio-Plex assay specifically detected anti-SIV Abs in specimens from SIV-infected animals for all four analytes when compared to SIV-naive control samples (p≤0.04). Furthermore, in 70% of Env and 79% of Gag ELISA-negative serum samples, specific Ab was detected using the Bio-Plex assay. Similarly, 71% of Env and 48% of Gag ELISA-negative rectal swab samples were identified as positive using the Bio-Plex assay. Importantly, assay specificity (i.e., probability of true positives) was comparable to ELISA (94%–100%). The results reported here indicate that microsphere-based methods provide a substantial improvement over ELISA for the detection of Ab responses, aid in detecting specific Abs when analyzing samples containing low levels of Abs, such as during the early stages of a vaccine trial, and may be valuable in attempts to link protective efficacy of vaccines with induced Ab responses.
Key words: HIV, immunology, microbiology, SIV
Introduction
Results from the RV144 HIV-1 vaccine trial recently demonstrated that the elicitation of specific antibodies (Abs) may be linked to the protective efficacy observed. The precise location, activity, and levels of such Abs remain to be determined; however, the overall titers of the protective antibodies may have been quite low.1,2 Enzyme-linked immunosorbent assay (ELISA) titers were approximately 10% of those measured in the predecessor AIDSVAX trial, and roughly 1% of titers typically measured in HIV-1–infected subjects.3,4 Furthermore, nonneutralizing Abs associated with protection were found to be V2-directed and belonged to the IgG3 subset, suggesting Ab-dependent cellular cytotoxicity via Fc-receptor activities.5–9 Although mucosal samples were unavailable for testing, these findings have led to the speculation that the active Abs induced in RV144 might have been localized primarily at mucosal sites of potential infection, which is a phenomenon shown to correlate with protection in animal models.10,11 These data have highlighted the necessity for careful measurement of Ab specificity, type, and level in both serum and mucosal samples, which can present technical challenges, particularly if Ab titers are low.
Over the past decade, soluble microsphere (bead)-based arrays have become an increasingly popular alternative for quantifying Abs.12–21 Such assays are advantageous as replacements for traditional ELISAs, because they can easily be multiplexed, reducing the sample volume required to evaluate multiple Abs, provide soluble-phase binding of antigen–antibody pairs, and offer a vastly increased dynamic range compared to the ELISA optical density scale. One of the most popular microsphere-based assays relies on Luminex® xMAP® technology (Luminex Corp., Austin, TX). In this system, different microsphere populations are internally labeled with varying concentrations of fluorescent dyes, giving each microsphere type in the array a unique fluorescence signature that can be identified with an appropriate instrument, such as the Bio-Plex® 200 System plate reader (Bio-Rad, Hercules, CA). As a result, when proteins of interest are covalently linked to unique bead populations, up to 100 different analytes can theoretically be measured in a single Bio-Plex plate well. Such multiplexed assays are particularly useful when simultaneous measurement of numerous analytes in a given sample is desired, as is often the case with cytokines, for which numerous assays have been developed.15,16,19,20,22–29 In addition, multiplexed bead-based immunoassays have been established and commercial kits are available for simultaneous measurement of specific Abs against various disease, atopic, autoimmune, and cancer-related antigens.17,21,28,30–34 These assays have been employed to measure analytes in cultured cells or in various bodily fluids such as serum, semen, and cervical mucus from humans or numerous experimental animal models.
Microsphere-based assays have demonstrated good correlation with ELISA data for most, but not all analytes. This correlation appears largely dependent on analyte concentration in the sample, with microsphere-based assays often detecting analytes present in low concentrations that ELISA does not, which is interpreted as a false positive when ELISA is used as the gold-standard.23–25,31,35,36 However, this common observation might indicate that microsphere-based assays are superior for detecting low levels of analytes compared with ELISA. To determine if microsphere-based assays such as the Bio-Plex system provide increased sensitivity for detection and quantification of Abs in serum and mucosal samples, we used the Bio-Plex Suspension Array System to develop a 4-plex bead array to detect Abs specific for SIVmac239 Env, Gag, Pol, and Nef. This assay was used to reassess endpoint titers for serum IgG and rectal swab IgA from specimens collected from rhesus macaques (RMs) infected with SIVmac239, which exhibited negative ELISA titers despite detectable viral loads.
Methods
Samples
Serum and rectal swabs were obtained from Indian RMs as part of ongoing vaccine studies. Of the RMs from which rectal swabs were obtained, 24 had been vaccinated by the intratonsillar or intramuscular route using a 3× SIVmac239 Env/Gag/Pol DNA prime+SIVmac239-recombinant adenovirus boost regimen. All RMs, excluding the SIV-naive control animals, were challenged with low-dose SIVmac239 by the intrarectal route and confirmed to be infected by a positive SIVmac239 viral load. All samples assayed herein were obtained between 7 and 123 days postinfection. Sera were obtained in 2004–2005 or in 2012 and stored at −80°C. Rectal swabs were obtained in 2011–2012 and stored at −80°C. Viral loads were determined within 20 days of sampling and were found to be between 103 and 108 genome copy equivalents per milliliter.
Rectal swabs
Rectal swabs were collected as previously described using Weck-Cel® Eye Spears (Beaver-Visitec, Waltham, MA).37 Sample collection minimized bleeding and subsequent elution was performed according to the published protocol.37 Prior to use, samples were tested for the presence of blood using Hemoccult® Test Cards (Beckman Coulter, Brea, CA) according to the manufacturer's protocol using 20 μL of eluate. If blood was detected, samples were not used for this study.
Bead coupling
One milliliter of Bio-Plex COOH (carboxylated) beads were conjugated to SIV proteins according to the manufacturer's protocol using the Bio-Plex amine coupling kit (Bio-Rad). Beads were counted just prior to conjugation using a Vi-Cell Viability Analyzer (Beckman-Coulter) to ensure a consistent bead-to-protein ratio since bead loss occurs during the conjugation process.38 Protein was added at a ratio of 25 μg of protein/5×106 beads for Envelope gp130 (produced in-house), Gag p55 (Protein Sciences Corp., Meriden, CT), and Pol (Immune Technology, New York, NY), or 75 μg/5×106 beads for Nef (Immune Technology). Final volume was adjusted to 5 mL in phosphate-buffered saline (PBS), and tubes were agitated for 2 h. Beads were then washed with 5 mL of PBS, resuspended in 2.5 mL of Stabilguard blocking buffer (SurModics, Eden Prairie, MN), and agitated for 30 min.39 Beads were washed, resuspended in 400 μL of storage buffer, counted, aliquoted, and stored at −80°C.
ELISA
Polystyrene, flat-bottom, high-binding, half-area plates (Corning, Kennebunk, ME) were coated overnight at 4°C by adding 17.5 ng of Env gp130, 125 ng of Gag p55 (IgG ELISA), 150 ng of Env gp130 or Gag p55 (IgA ELISA) per well. Plates were washed in PBS/0.02% Tween-20 and blocked for 1 h at 37°C with 120 μL of PBS/3% bovine serum albumin (BSA; IgG ELISA) or PBS/3% milk (IgA ELISA). Plates were washed, and serum or swab eluate was added at 1:100 or 1:10 diluted in PBS/1% BSA (IgG ELISA) or PBS/1% milk (IgA ELISA), respectively. Serum was diluted threefold across the plate, while swab eluate was diluted twofold. Plates were incubated and washed as above before the addition of biotin-conjugated goat antimonkey IgG for serum (0.125 μg/mL in PBS/1% BSA; Rockland Immunochemicals, Gilbertsville, PA) or goat antimonkey IgA for swabs (0.66 μg/mL for Env ELISA, 4 μg/mL for Gag ELISA, in PBS/1% milk; Rockland Immunochemicals). Plates were incubated and washed as before prior to the addition of streptavidin–horse radish peroxidase (1 μg/mL; Biolegend, San Diego, CA). After incubation and washing, TMB Substrate Reagent Set (3,3′,5,5′-tetramethyl benzidine; Biolegend) was added according to the manufacturer's protocol. The reaction was stopped with 2 N H2SO4 and plates were read at 450 nm using a VERSAmax ELISA reader (Molecular Devices, Sunnyvale, CA).
Bio-Plex assay
Analysis of serum and swab eluate with Bio-Plex assays were performed using conditions suggested by the manufacturer (Bio-Rad). One hundred fifty microliters of assay buffer (PBS, 0.5% casein, 0.1% BSA, 0.02% Tween-20, 0.05% sodium azide, pH 7.4) was added to each well of a Multiscreen HTS, HV clear plate (Millipore, Billerica, MA) before the plates were incubated for 10 min with shaking. All subsequent sample or assay dilutions were carried out in assay buffer. Samples were thawed and centrifuged at 20,000 g for 1 min to pellet any debris. Plates were drained by vacuum pressure before 1:50 diluted serum samples or 1:10 diluted swab samples were added. Serum samples were titrated threefold across the plate, while swab samples were titrated twofold across the plate. An equal volume of conjugated bead set for all four SIV proteins (50 beads/μL; 1250 beads/well) was added to diluted samples before the plate was incubated at room temperature for 1 h with agitation. Plates were washed twice by vacuum with 150 μL assay buffer before the addition of goat antimonkey IgG (0.07 μg/mL) or IgA (1 μg/mL) biotin-conjugated secondary antibody (Rockland Immunochemicals) and were incubated at room temperature for 1 h with agitation. Plates were washed twice by vacuum with 150 μL assay buffer before incubation with 50 μL of Phycolink Streptavidin-R-Phycoerythrin (1 μg/mL, Thermo Scientific, Waltham, MA) at room temperature for 1 h with agitation. Plates were finally washed twice in casein-free assay buffer before resuspension in 80 μL of casein-free assay buffer with agitation at room temperature for 5 min. Plates were read using a Bio-Plex 200 System plate reader (Bio-Rad).
Data analysis
Background was subtracted before data points were fit to four-parameter curves using Prism version 5.03 (GraphPad Software, Inc.). Endpoint titers were interpolated from the curves using the median RLU (Bio-Plex) or optical density (ELISA) of the SIV-naive samples at the lowest sample dilution (1:100 for serum, 1:10 or 1:20 for rectal swabs) as a cutoff value. Statistical analyses were carried out with GraphPad Prism software, using a two-tailed unpaired t-test. Titers were considered positive for a given assay if they were greater than the mean of the control sample endpoint titers+3 standard deviations (positive cutoff value).40 Specificity of the ELISA and Bio-Plex assays were calculated as (number of true negatives [i.e., SIV-naive samples])/(number of true negatives+number of false positives).
Results
Determination of serum and swab anti-SIV IgG and IgA endpoint titers by ELISA
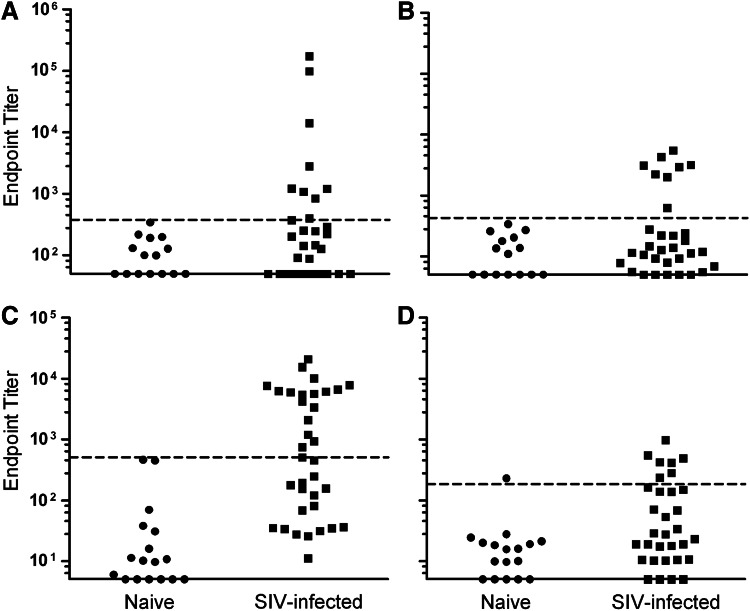
Thirty-two serum samples from SIVmac239-infected RMs with detectable viral load and 15 serum samples obtained from SIV-naive control RMs were tested for Env- and Gag-specific IgG titers by ELISA. It was found that the naive control sera exhibited a mean Env-specific ELISA endpoint titer of 117, thus samples exhibiting endpoint titers greater than 379 were considered positive for Env-specific IgG (Fig. 1A, dotted line). Among the sera obtained from SIV-infected RMs, 9 of 32 samples were found to be positive for anti-Env IgG by ELISA, with endpoint titers ranging from 402 to 172,809, while the remaining 23 titers fell below the assigned cutoff value, exhibiting titers of 50–376 (Fig. 1A). Furthermore, naive control sera exhibited a mean Gag-specific ELISA endpoint titer of 131, resulting in a positive cutoff of 423. Eight SIV-infected samples exhibited positive Gag-specific titers ranging from 625 to 5444, while the remaining negative sample titers ranged from 50 to 274 (Fig. 1B).
FIG. 1.
ELISA endpoint titers. (A) Env-specific and (B) Gag-specific IgG titers in serum obtained from 32 SIV-naive and SIV-infected rhesus macaques. (C) Env-specific and (D) Gag-specific IgA titers in eluates from rectal swabs obtained from 35 SIV-naive and SIV-infected rhesus macaques. Dotted lines indicate positive cutoff values. ELISA, enzyme-linked immunosorbent assay; SIV, simian immunodeficiency virus.
Eluates from rectal swabs obtained from 35 infected RMs and 17 SIV-naive RMs were similarly analyzed by IgA ELISA. Naive control samples exhibited a mean Env-specific endpoint titer of 67 resulting in a cutoff value being assigned at 505. Accordingly, 18 of 35 SIV-infected samples were scored positive for Env-specific IgA, with titers ranging from 506 to 20,600 (Fig. 1C). Negative samples exhibited titers of 11–449. Additionally, naive control samples exhibited a mean Gag-specific endpoint titer of 26, which corresponded to a positive cutoff value at 184. Only 7 of 32 samples were found to be positive, with titers ranging from 233 to 969, while negative titers ranged from 5 to 163 (Fig. 1D).
Determination of serum and swab anti-SIV IgG and IgA endpoint titers by Bio-Plex assay
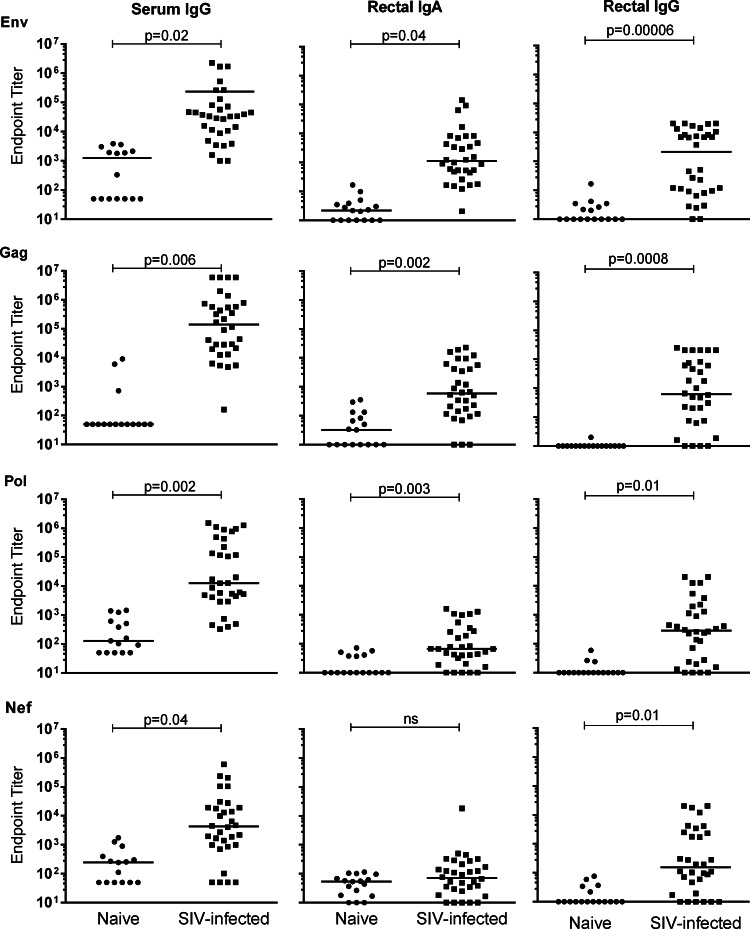
In order to compare the Bio-Plex data with those obtained by ELISA, cutoffs for endpoint titer determination and positive endpoint cutoffs were calculated by the same methods for both assays. As well, both assays employed the same antigens and biotinylated secondary Abs. Serum samples were tested for SIV-specific IgG titers, while rectal swab eluates were tested for SIV-specific IgG and IgA. Overall, the novel Bio-Plex assay was found to specifically detect the analytes tested as judged by the significantly higher antigen-specific Ab titers in serum samples obtained from SIV-infected RMs compared to the naive control samples for all four antigens in the Bio-Plex assay (Fig. 2A–D). Similarly, rectal swab eluates obtained from SIV-infected RMs exhibited significantly higher antigen-specific IgG titers compared to naive control samples for all four analytes and significantly higher Env, Gag, and Pol-specific IgA titers; however, Nef-specific IgA titers were not significantly different between the two groups (Fig. 2E–L).
FIG. 2.
Bio-Plex detection of anti-SIV specific antibodies in rhesus macaque samples. Lines indicate mean endpoint values.
Bio-Plex assay sensitivity improves detection of positive samples compared to ELISA
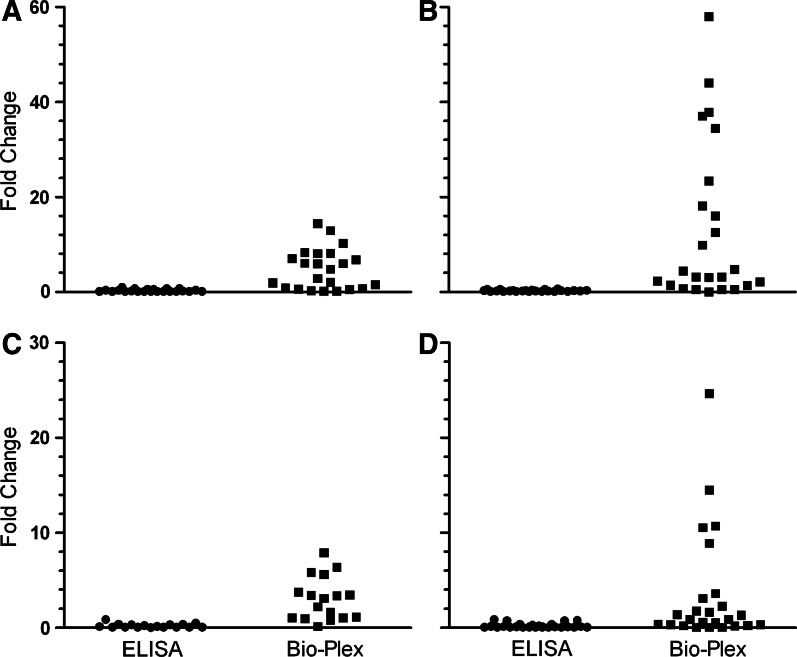
Many of the samples from infected animals that had exhibited negative Env- or Gag-specific endpoint titers by ELISA were found to be positive using the Bio-Plex assay. Overall, the Bio-Plex assay found 25 and 27 of 32 infected serum samples to be positive for Env- and Gag-specific Abs, respectively. Of the 23 serum samples negative for Env-specific IgG by ELISA, 16 samples (70%) were found to be positive by the Bio-Plex assay, exhibiting titers 1.6- to 14-fold higher than the Bio-Plex endpoint cutoff value (5544; Fig. 3A). Similarly, 18 of the serum samples (79%) found negative for Gag-specific titers by ELISA exhibited detectable titers by Bio-Plex, which were 1.4- to 58-fold higher than the cutoff value (9181; Fig. 3B). Env- and Gag-specific rectal swab IgA titers measured by Bio-Plex were also compared to ELISA data. Overall, the Bio-Plex assay found 30 and 18 of 35 infected rectal swab samples to be positive for Env- and Gag-specific IgA, respectively. Of the 17 samples found to be negative in the Env-specific IgA ELISA, 12 (71%) of these samples were found to be positive by Bio-Plex, exhibiting titers 1.05- to 7.9-fold higher than the cutoff value (157; Fig. 3C). Of the 25 samples found negative by ELISA for Gag-specific IgA, 12 samples (48%) were found to be positive by Bio-Plex, exhibiting titers 1.3- to 25-fold higher than the cutoff value (386; Fig. 3D).
FIG. 3.
Samples found to be negative for SIV-specific antibody (Ab) titers by ELISA exhibit detectable titers using the Bio-Plex assay. Fold change in endpoint titer compared with positive cutoff value for each ELISA-negative sample is shown. (A) Env-specific and (B) Gag-specific serum IgG assays. (C) Env-specific and (D) Gag-specific rectal swab eluate IgA assays.
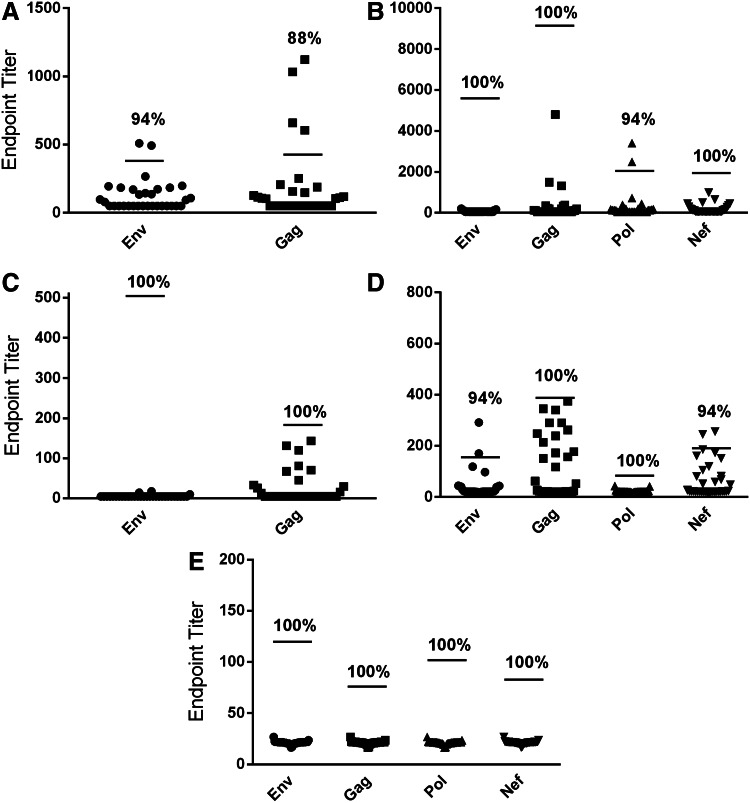
Importantly, the specificity of the Bio-Plex assay (i.e., the proportion of viral load-negative samples predicted to test negative for SIV-specific Abs) was assessed to ensure that this assay was truly superior to ELISA for SIV-specific Ab detection and was not increasing the false-positive rate. Thirty-two additional SIV-naive serum and rectal swab samples were tested, and positive endpoint cutoff values were applied as already described. It was found that the serum ELISA exhibited 94% and 88% specificity for the Env and Gag IgG assays, respectively, while the serum Bio-Plex assay exhibited 100% specificity for the Env, Gag, and Nef IgG assays, and 94% specificity for the Pol IgG assay (Fig. 4A, B). Furthermore, it was found that the rectal swab ELISA exhibited 100% specificity for both the Env and Gag IgA assays, while the Bio-Plex rectal swab assay exhibited 100% specificity for the Env, Gag, Pol, and Nef IgG and Gag and Pol IgA assays, while the Env and Nef IgA assays exhibited 94% specificity (Fig. 4C–E).
FIG. 4.
The Bio-Plex assay exhibits comparable specificity to ELISA. Thirty-two additional SIV-naive rhesus macaque serum samples and rectal swabs were analyzed using the same endpoint titer and positive cutoff values as determined for the initial sample set. (A) Specificity of serum IgG ELISA and (B) Bio-Plex assay. (C) Specificity of rectal swab eluate IgA ELISA and (D) Bio-Plex assay. (E) Specificity of rectal swab eluate IgG Bio-Plex assay. Lines indicate positive cutoff values. Percent specificities are shown.
Discussion
Numerous fluorescent bead–based immunoassays have been developed over the past decade, and this technology is quickly becoming commonplace. These assays are beneficial because they provide a simple platform with which ELISA-type assays can be multiplexed, allowing for rapid, simultaneous detection of numerous analytes in bodily fluids. Furthermore, these assays offer a greatly increased dynamic range and lower detection limit compared with ELISA and require minimal sample volume. Numerous studies have been performed comparing data obtained using a microsphere-based assay with that obtained by ELISA. In general, these assays have shown fair correlation with each other and with traditional ELISA, though this has not been found to be uniformly true, especially with analytes at low concentrations.23–25,35 Previous studies that have determined the sensitivity and specificity of Luminex-based assays have typically used ELISA data as the gold standard, often finding comparable or improved sensitivity but reduced specificity (i.e., an increased number of false positives compared with ELISA).24,35,40 Importantly, it has been suggested that this apparent increase in the false-positive rate might be attributable to the ability of microsphere-based assays to detect a lower level of analyte as compared with ELISA.31,40 Using specimens from RMs shown to be positive or negative for infection by analysis of virus loads, we determined whether the increased rate of positives found by Bio-Plex correlated with infection or was truly a false positive. In our survey of RMs shown to be infected but exhibiting negative ELISA titers, the Bio-Plex assay detected positive endpoint titers in 48%–79% of these animals. This result is in agreement with previous studies of microsphere assays for detection of human leukocyte antigen–specific Ab (HLA-Ab) in organ transplant recipients.41–43 These studies have found that such assays are far more sensitive than the conventional HLA-Ab assay for the detection of HLA-reactive Abs, whose presence was associated with worsened transplant outcomes. As with the present study, the utility of the microsphere-based assay in these studies was determined not by comparison to a typical gold standard Ab assay, but to more relevant disease markers.
Our results suggest that in the case of SIV and potentially HIV, ELISA cannot be used as a gold standard to link the presence of specific Abs with infection. Importantly, in our comparison of these assays, endpoint and positive cutoff values were calculated by the same method, and the same antigen–Ab pairs and reagents were used in both assay platforms where possible, in order to achieve a fair comparison of data.23–25,40 Numerous factors explain the superior sensitivity of the Bio-Plex assay. Because the SIV antigens were covalently bound to the microspheres, higher assay avidity and lower background signal were achieved as compared with ELISA, in which antigen is noncovalently coated to plate wells. Covalent binding also minimizes antigen loss during washing steps, and due to the significantly reduced surface area, washes are likely more efficient than in an ELISA, further reducing the background signal. Additionally, the ∼3-log surface area reduction compared to an ELISA most likely contributes significantly to a reduction in nonspecific binding.27,44–46
In conclusion, these data demonstrate that microsphere-based platforms are advantageous beyond their multiplexing capacity, offering vastly increased sensitivity for specific Ab detection in bodily fluids compared with ELISA. Multiplexed, microsphere-based assays that differentiate low levels from negative levels of Abs may be critical to the interpretation of vaccine trial data, especially when samples are difficult to obtain. The results reported here also suggest the potential to use these methods for the prompt diagnosis of disease and seroconversion to various pathogens.43,47,48
Acknowledgments
The authors are grateful for the samples provided by the laboratory of Louis J. Picker, Oregon Health and Science University, Portland, OR. The authors also acknowledge Alexei Carpov for production of SIVmac239 Env (International AIDS Vaccine Initiative). This work was supported by the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery and USAID.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rerks-Ngarm S. Pitisuttithum P. Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Montefiori DC. Karnasuta C. Huang Y, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore JP. HIV-1 Env antibodies: are we in a bind or going blind? Nat Med. 2012;18:346–347. doi: 10.1038/nm.2689. author reply 347–348. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert PB. Peterson ML. Follmann D, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 5.Karasavvas N. Billings E. Rao M, et al. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses. 2012;28:1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao HX. Bonsignori M. Alam SM, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolland M. Edlefsen PT. Larsen BB, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alter G. Building the evidence base for nonneutralizing antibody-based vaccines. 7th Annual Collaboration for AIDS Vaccine Discovery Meeting; Seattle, WA. 2012. [Google Scholar]
- 9.Bonsignori M. Pollara J. Moody MA, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86:11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hur EM. Patel SN. Shimizu S, et al. Inhibitory effect of HIV-specific neutralizing IgA on mucosal transmission of HIV in humanized mice. Blood. 2012;120:4571–4582. doi: 10.1182/blood-2012-04-422303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letvin NL. Rao SS. Montefiori DC, et al. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci Transl Med. 2011;3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Q. Guo H. Tang M, et al. Simultaneous detection of antibodies to five simian viruses in nonhuman primates using recombinant viral protein based multiplex microbead immunoassays. J Virol Methods. 2011;178:143–152. doi: 10.1016/j.jviromet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan IH. Mendoza S. Yee J, et al. Simultaneous detection of antibodies to six nonhuman-primate viruses by multiplex microbead immunoassay. Clin Vaccine Immunol. 2006;13:45–52. doi: 10.1128/CVI.13.1.45-52.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D. Chiu H. Gupta V, et al. Validation of a multiplex immunoassay for serum angiogenic factors as biomarkers for aggressive prostate cancer. Clin Chim Acta. 2012;413:1506–1511. doi: 10.1016/j.cca.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toedter G. Hayden K. Wagner C, et al. Simultaneous detection of eight analytes in human serum by two commercially available platforms for multiplex cytokine analysis. Clin Vaccine Immunol. 2008;15:42–48. doi: 10.1128/CVI.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heijmans-Antonissen C. Wesseldijk F. Munnikes RJ, et al. Multiplex bead array assay for detection of 25 soluble cytokines in blister fluid of patients with complex regional pain syndrome type 1. Mediators Inflamm. 2006;2006:28398. doi: 10.1155/MI/2006/28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaras GD. Yates NL. Liu P, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pochechueva T. Chinarev A. Spengler M, et al. Multiplex suspension array for human anti-carbohydrate antibody profiling. Analyst. 2011;136:560–569. doi: 10.1039/c0an00758g. [DOI] [PubMed] [Google Scholar]
- 19.Gopichandran N. Ekbote UV. Walker JJ, et al. Multiplex determination of murine seminal fluid cytokine profiles. Reproduction. 2006;131:613–621. doi: 10.1530/rep.1.00959. [DOI] [PubMed] [Google Scholar]
- 20.Orsi NM. Ekbote UV. Walker JJ, et al. Uterine and serum cytokine arrays in the mouse during estrus. Anim Reprod Sci. 2007;100:301–310. doi: 10.1016/j.anireprosci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Opalka D. Lachman CE. MacMullen SA, et al. Simultaneous quantitation of antibodies to neutralizing epitopes on virus-like particles for human papillomavirus types 6, 11, 16, and 18 by a multiplexed luminex assay. Clin Diagn Lab Immunol. 2003;10:108–115. doi: 10.1128/CDLI.10.1.108-115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dabitao D. Margolick JB. Lopez J, et al. Multiplex measurement of proinflammatory cytokines in human serum: comparison of the Meso Scale Discovery electrochemiluminescence assay and the Cytometric Bead Array. J Immunol Methods. 2011;372:71–77. doi: 10.1016/j.jim.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dossus L. Becker S. Achaintre D, et al. Validity of multiplex-based assays for cytokine measurements in serum and plasma from “non-diseased” subjects: comparison with ELISA. J Immunol Methods. 2009;350:125–132. doi: 10.1016/j.jim.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 24.dupont NC. Wang K. Wadhwa PD, et al. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66:175–191. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elshal MF. McCoy JP. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Q. Zhu J. Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010;56:314–318. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellar KL. Douglass JP. Multiplexed microsphere-based flow cytometric immunoassays for human cytokines. J Immunol Methods. 2003;279:277–285. doi: 10.1016/s0022-1759(03)00248-5. [DOI] [PubMed] [Google Scholar]
- 28.Liu J. Keele BF. Li H, et al. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol. 2010;84:10406–10412. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young SH. Antonini JM. Roberts JR, et al. Performance evaluation of cytometric bead assays for the measurement of lung cytokines in two rodent models. J Immunol Methods. 2008;331:59–68. doi: 10.1016/j.jim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Bigbee WL. Gopalakrishnan V. Weissfeld JL, et al. A multiplexed serum biomarker immunoassay panel discriminates clinical lung cancer patients from high-risk individuals found to be cancer-free by CT screening. J Thorac Oncol. 2012;7:698–708. doi: 10.1097/JTO.0b013e31824ab6b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biagini RE. Sammons DL. Smith JP, et al. Comparison of a multiplexed fluorescent covalent microsphere immunoassay and an enzyme-linked immunosorbent assay for measurement of human immunoglobulin G antibodies to anthrax toxins. Clin Diagn Lab Immunol. 2004;11:50–55. doi: 10.1128/CDLI.11.1.50-55.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson AJ. Cheshier RC. Cosentino G, et al. Validation of a microsphere-based immunoassay for detection of anti–West Nile virus and anti–St. Louis encephalitis virus immunoglobulin M antibodies. Clin Vaccine Immunol. 2007;14:1084–1093. doi: 10.1128/CVI.00115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickering JW. Martins TB. Greer RW, et al. A multiplexed fluorescent microsphere immunoassay for antibodies to pneumococcal capsular polysaccharides. Am J Clin Pathol. 2002;117:589–596. doi: 10.1309/lmch-c4q2-vfl9-3t1a. [DOI] [PubMed] [Google Scholar]
- 34.Shoma S. Verkaik NJ. de Vogel CP, et al. Development of a multiplexed bead-based immunoassay for the simultaneous detection of antibodies to 17 pneumococcal proteins. Eur J Clin Microbiol Infect Dis. 2011;30:521–526. doi: 10.1007/s10096-010-1113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nifli AP. Notas G. Mamoulaki M, et al. Comparison of a multiplex, bead-based fluorescent assay and immunofluorescence methods for the detection of ANA and ANCA autoantibodies in human serum. J Immunol Methods. 2006;311:189–197. doi: 10.1016/j.jim.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Stenger RM. Smits M. Kuipers B, et al. Fast, antigen-saving multiplex immunoassay to determine levels and avidity of mouse serum antibodies to pertussis, diphtheria, and tetanus antigens. Clin Vaccine Immunol. 2011;18:595–603. doi: 10.1128/CVI.00061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozlowski PA. Lynch RM. Patterson RR, et al. Modified wick method using Weck-Cel sponges for collection of human rectal secretions and analysis of mucosal HIV antibody. J Acquir Immune Defic Syndr. 2000;24:297–309. doi: 10.1097/00126334-200008010-00001. [DOI] [PubMed] [Google Scholar]
- 38.Dasso J. Lee J. Bach H, et al. A comparison of ELISA and flow microsphere-based assays for quantification of immunoglobulins. J Immunol Methods. 2002;263:23–33. doi: 10.1016/s0022-1759(02)00028-5. [DOI] [PubMed] [Google Scholar]
- 39.Pickering JW. Larson MT. Martins TB, et al. Elimination of false-positive results in a luminex assay for pneumococcal antibodies. Clin Vaccine Immunol. 2010;17:185–189. doi: 10.1128/CVI.00329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang S. Smith J. Onley D, et al. A comparability study of the emerging protein array platforms with established ELISA procedures. J Immunol Methods. 2005;302:1–12. doi: 10.1016/j.jim.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Gibney EM. Cagle LR. Freed B, et al. Detection of donor-specific antibodies using HLA-coated microspheres: another tool for kidney transplant risk stratification. Nephrol Dial Transplant. 2006;21:2625–2629. doi: 10.1093/ndt/gfl202. [DOI] [PubMed] [Google Scholar]
- 42.Smith JD. Hamour IM. Banner NR, et al. C4d fixing, luminex binding antibodies—a new tool for prediction of graft failure after heart transplantation. Am J Transplant. 2007;7:2809–2815. doi: 10.1111/j.1600-6143.2007.01991.x. [DOI] [PubMed] [Google Scholar]
- 43.Tait BD. Hudson F. Cantwell L, et al. Review article: Luminex technology for HLA antibody detection in organ transplantation. Nephrology (Carlton). 2009;14:247–254. doi: 10.1111/j.1440-1797.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 44.Kellar KL. Iannone MA. Multiplexed microsphere-based flow cytometric assays. Exp Hematol. 2002;30:1227–1237. doi: 10.1016/s0301-472x(02)00922-0. [DOI] [PubMed] [Google Scholar]
- 45.Kellar KL. Mahmutovic AJ. Bandyopadhyay K. Multiplexed microsphere-based flow cytometric immunoassays. Curr Protoc Cytom. 2006;Chapter 13(Unit13):11. doi: 10.1002/0471142956.cy1301s35. [DOI] [PubMed] [Google Scholar]
- 46.Carson RT. Vignali DA. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J Immunol Methods. 1999;227:41–52. doi: 10.1016/s0022-1759(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 47.Bartolo I. Camacho R. Barroso H, et al. Rapid clinical progression to AIDS and death in a persistently seronegative HIV-1 infected heterosexual young man. AIDS. 2009;23:2359–2362. doi: 10.1097/QAD.0b013e328332d5e1. [DOI] [PubMed] [Google Scholar]
- 48.Roques PA. Gras G. Parnet-Mathieu F, et al. Clearance of HIV infection in 12 perinatally infected children: clinical, virological and immunological data. AIDS. 1995;9:F19–26. [PubMed] [Google Scholar]