Abstract
The purpose of this study was to explore how genetic deletion and pharmacological antagonism of the P2X7 receptor (P2rx7) alter mood-related behaviour, gene expression and stress reactivity in the brain. The forced swim test (FST), tail suspension test (TST) and amphetamine-induced hyperlocomotion (AH) tests were used in wild-type (P2rx7+/+) and P2rx7-deficient (P2rx7−/−) mice. Biogenic amine levels were analysed in the amygdala and striatum, adrenocorticotropic hormone (ACTH) and corticosterone levels were measured in the plasma and pituitary after restraint stress. Chimeric mice were generated by bone marrow transplantation. A whole genome microarray analysis with real-time polymerase chain reaction validation was performed on the amygdala. In the absence of P2rx7s decreased behavioural despair in the FST, reduced immobility in the TST and attenuated amphetamine-induced hyperactivity were detected. Basal norepinephrine levels were elevated in the amygdala, whereas stress-induced ACTH and corticosterone responses were alleviated in P2rx7−/− mice. Sub-acute treatment with the selective P2rx7 antagonist, Brilliant Blue G, reproduced the effect of genetic deletion in the TST and AH test in P2rx7+/+ but not P2rx7−/− mice. No change in behavioural phenotype was observed in chimeras lacking the P2rx7 in their haematopoietic compartment. Whole genome microarray analysis indicated a widespread up- and down-regulation of genes crucial for synaptic function and neuroplasticity by genetic deletion. Here, we present evidence that the absence of P2rx7s on non-haematopoietic cells leads to a mood-stabilizing phenotype in several behavioural models and suggest a therapeutic potential of P2rx7 antagonists for the treatment of mood disorders.
Key words: Antagonists, bipolar disorder, depression, knockout, microarray, P2X7 receptor
Introduction
P2X7 receptors (P2rx7) belong to the ionotropic P2X receptors that are sensitive to ATP and other purine and pyrimidine nucleotides. The homo-oligomeric P2rx7 (Surprenant et al. 1996) has distinct structural, functional and pharmacological features within the P2X receptor family (Jarvis & Khakh, 2009; Sperlagh et al. 2006): (1) its intracellular carboxyl-terminal domain is longer than those of other P2X receptor subunits; (2) it has several splice variants that display different functionality in P2rx7+/+ and knockout mice lines (Nicke et al. 2009); (3) its persistent activation elicits the opening of a membrane pore permeable to high molecular weight substances; (4) it needs high micromolar concentrations of ATP to be activated.
P2rx7s are widely distributed in different cells, including cells of haematopoietic origin, neurons, microglia and astrocytes. Accordingly, P2rx7s are involved in the regulation of different aspects of the host-defence reaction (Chen & Brosnan, 2006; di Virgilio et al. 2009). A major immunomodulatory function of P2rx7 activation is that it acts as a necessary co-stimulus for the post-translational processing and subsequent release of the pro-inflammatory cytokine interleukin (IL)-1β in peripheral immune cells in response to bacterial endotoxin (Ferrari et al. 2006; Solle et al. 2001).
The primary role of P2rx7s in the brain is the modulation of neurotransmitter release (Anderson & Nedergaard, 2006; Sperlagh et al. 2006, 2007a). The activation of P2rx7s elicits Ca2+ influx (Miras-Portugal et al. 2003), followed by increased glutamate and subsequent γ-aminobutyric acid (GABA) release (Alloisio et al. 2008; Papp et al. 2004; Patti et al. 2006; Sperlagh et al. 2002). By regulating the activation and proliferation of microglia and the subsequent pathological process leading to neuronal death (Monif et al. 2009; Skaper et al. 2006), P2rx7s might also act as a ‘danger signal’ and contribute to neurodegenerative and neuroplasticity events underlying a variety of central nervous system disorders from Alzheimer's disease to mood disorders (Burnstock, 2008; Skaper et al. 2010; Sperlagh et al. 2006).
Gene polymorphism studies have revealed that non-synonymous single nucleotide polymorphisms in the human P2X7 gene (P2RX7) affect receptor function (Roger et al. 2010; Stokes et al. 2010) and are associated with bipolar disorder (Barden et al. 2006; Hejjas et al. 2009; McQuillin et al. 2009; Nagy et al. 2008; Soronen et al. 2011) and major depressive disorder (Lucae et al. 2006; Soronen et al. 2011; but see Green et al. 2009; Grigoroiu-Serbanescu et al. 2010; Lavebratt et al. 2010; Viikki et al. 2011). These mutations might underlie the susceptibility to genetically related mood disorders (Harvey et al. 2007; Sluyter et al. 2010). Moreover, recent studies found an antidepressant phenotype in mice genetically deficient in P2rx7s (Basso et al. 2009; Boucher et al. 2011). Nevertheless, it is not known, how P2rx7 affects mania-like behaviour and whether behavioural alterations detected in the absence of P2rx7s can be reproduced by pharmacological blockade. In addition, the mechanism, whereby the activity of P2rx7 leads to alterations in mood also remains to be explored.
We report here that, in addition to the known antidepressant-like effect, the genetic deletion of P2rx7 (P2rx7−/−) results in a mood stabilizing-like phenotype and P2rx7 antagonists reproduce these behavioural changes. The lack of P2rx7s also leads to decreased stress response to restraint stress and region-specific alterations in brain monoamine levels, in particular in the amygdala, an intrinsic part of the limbic system. Experiments performed on bone marrow chimeric mice demonstrated that behavioural alterations detected in P2rx7−/− mice are not related to P2rx7s on haematopoietic cells. Therefore, a whole genome microarray analysis was performed in the amygdala, which revealed widespread alterations in the expression of genes responsible for synaptic signalling and neuroplasticity, in the absence of P2rx7s.
Method
Animals
All studies were conducted in accordance with the principles and procedures outlined in the NIH Guide for the Care and Use of Laboratory Animals and were approved by the local Animal Care Committee of the IEM HAS. This study used 2- to 3-month old (approx. 30 g), drug- and test-naive male wild-type (P2rx7+/+) and knockout mice (P2rx7−/−), which were housed in a light-controlled (12-h light/dark cycle; lights on 07:00 hours), humidity-controlled (60 ± 10%) and temperature-controlled (23 ± 2 °C) room with food and water available ad libitum. Homozygous P2rx7+/+ mice were bred on a background of C57BL/6J. The original breeding pairs of P2rx7−/− mice (C57BL/6J-based) were kindly supplied by Christopher Gabel (Pfizer Inc., USA). The animals contained the DNA construct [P2X7-F1 (5′-CGGCGTGCGTTTTGACATCCT-3′) and P2X7-R2 (5′-AGGGCCCTGCGGTTCTC-3′)], previously shown to produce the genetic deletion of P2rx7 (Solle et al. 2001). Offspring of this mouse line were cross-bred with P2rx7+/+ mice and the resulting heterozygotes were used as breeding stock for the F1 generation offspring employed in the behaviour studies. The genotypes of animals were confirmed by polymerase chain reaction (PCR) analysis, as described previously (Solle et al. 2001). Mice carrying the CD45.1 allele on the C57BL/6J genetic background were obtained from the Jackson Laboratory (USA).
Behaviour experiments
Mice establish strong dominance hierarchies in both their natural habitats and the laboratory (Capanna et al. 1984; Ferrari et al. 1998; Poshivalov 1980). Therefore, to avoid confounds from social status, subjects were kept in individual cages (15 × 45 × 20 cm) for 1 wk before experimentation. In contrast to rats, isolation does not increase but slightly decreases depression-like behaviour in mice, at least in the Swiss strain of this species (Hilakivi et al. 1989).
All experiments and treatments were carried out during the light phase (07:00–19:00 hours). P2rx7+/+ and P2rx7−/− mice were submitted to testing in alternation. Except for the elevated plus maze (EPM) test, which immediately followed the open field (OF) test, the animals used in this study were subjected to behaviour tests only once. Treatments with the P2rx7 antagonist, Brilliant Blue G (BBG; Sigma-Aldrich, Hungary) or the selective serotonin reuptake inhibitor, citalopram (Sigma-Aldrich), were applied i.p. 30 min before testing. The doses were chosen based on literature data (Diaz-Hernandez et al. 2009) and preliminary tests.
In the forced swim test (FST), the OF test and the EPM test, behaviour was video-recorded and analysed later with a computer-based event recorder by an experimenter blind to the genotype.
Forced swim test
Behavioural despair was analysed as described previously (Porsolt et al. 1977). Each mouse was placed in a transparent glass cylinder filled with water (22 ± 0.5 °C) and submitted to 15 min of forced swim, divided into three equal trial periods (5 min each) and followed by a single 5-min test period the next day. Fresh water was used for each mouse and four animals were tested simultaneously. After the test period, the mice were removed from the cylinder, dried with paper towels and a clean towel was left in the home cage for 1 h to avoid cooling. The time of immobility (floating) was expressed in seconds.
Automated tail suspension test (TST)
This test was performed using an automated tail suspension device (BIO-TST2, Bioseb, France), following the method described in Cryan et al. (2005), which was connected to a computer that recorded the activity of the animals during test sessions. Three mice were individually suspended by the tail onto the hooks of the device using adhesive tape (distance from tip of tail was 1–2 cm). The automatic measurements were started within 5–10 s after placing the last animal into the chamber and each measurement lasted 6 min. During the test, the animals showed several escaping behaviours with temporary periods of immobility. The threshold was set at level 6. The time of immobility was expressed in seconds. In accordance with the observations of Mayorga & Lucki (2001), a proportion of the animals (8.9% on average, 0–16%, depending on the experiment) displayed tail-climbing behaviour. The data of these animals were post-hoc excluded from the calculations.
OF test and EPM test
For technical details, see Supplement 1 (available online).
Amphetamine-induced hyperlocomotion (AH) in the OF test
Experiments were performed in the light phase under dimmed lights (∼3 lx). At least 3 d before the tests, animals were transferred to the experimental room.
Each animal was placed in the centre of a non-transparent Plexiglas arena (dimensions: 40 × 40 × 40 cm) for a habituation period of 30 min and then removed for 2 min into their home cages for i.p. saline or d-amphetamine-sulfate (equivalent to 2.5 mg/kg free base, A5880; Sigma-Aldrich, Hungary) treatment. Immediately after amphetamine (or saline) injection, each mouse was placed back into the box and the locomotor activity of the animals was recorded for 90 min using a video camera positioned above the arena. To measure locomotor activity, video files were analysed offline by converting them into single frames (25 frames/s) and a custom-written motion tracking algorithm was applied within the image processing software ImageJ. The total distance (m) was provided for the 90 min of the experiment. Hyperactivity induced by amphetamine was expressed in percentage of locomotor activity measured in saline-treated mice under an identical period.
IL-1β experiments
All animals were given an i.p. injection of sterile saline (0.9% NaCl) or bacterial lipopolysaccharide (LPS, 250 μg/kg i.p.) with an injection volume of 0.1 ml/mouse). Animals were killed by decapitation 6 h after LPS injection. The amygdalae were collected, frozen on dry ice and stored at −70 °C until further investigation. The IL-1β assays were performed as described previously (Csölle & Sperlágh, 2010).
High performance liquid chromatography (HPLC) analysis of endogenous biogenic amine levels
After various treatments (saline/2.5 mg/kg amphetamine i.p.; 30 min restraint; saline/50 mg/kg.d BBG i.p. for 7 d), animals were killed by decapitation and native amygdalae and striata were frozen in liquid nitrogen. The weighted frozen tissue was homogenized in ice-cold 0.1 m perchloric acid containing theophylline (10 μm; internal standard) and 0.5 mm sodium metabisulphite. The suspension was centrifuged at 300 g for 10 min at 4 °C. The perchloric anion was precipitated by addition of 1 m KOH, removed by centrifugation and the protein content of the pellet was determined according to the method of Lowry et al. (1951) . The supernatant was kept at −20 °C until analysis. Biogenic amines were measured by a liquid–liquid, two-dimensional reversed phase and ion pair-reversed phase chromatographic separation, as described earlier (Baranyi et al. 2006) using a Gilson liquid chromatographic system (Gilson Medical Electronics Inc., USA) equipped with an Applied Biosystems 785/A UV and BAS CC-4 amperometric detector in a cascade line. Data were expressed as pmol per mg protein.
[3H]Dopamine release experiments
[3H]Dopamine release experiments on acute striatal slices were performed according to the method described in Milusheva et al. (2010) . For the detailed protocol of release experiments, see Supplement 2.
Stress studies
Restraint stress consisted of placing mice for 30 min in ventilated polyethylene tubes (inner diameter: 2.5 cm; length: 10 cm), closed with plastic tape (Lolait et al. 2007), and was immediately (within 15 s) followed by decapitation. Controls were decapitated at the same time under basal conditions. Trunk blood was collected in ice-cold sodium-EDTA (20%) during the light phase between 09:00 and 13:00 hours and centrifuged at 3000 g for 20 min at −4 °C. The plasma was stored at −20 °C until the hormone assays were performed. The pituitaries were dissected into four pieces and incubated at 37 °C in 1 ml Dulbecco's minimal essential medium (DMEM, pH 7.4; Sigma) containing 2.5% bovine serum albumin in a 95% O2/5% CO2 atmosphere (one gland/tube) for 60 min. After a further pre-incubation with fresh DMEM for 60 min, 15 min samples were collected three times with corticotropin-releasing hormone (CRH)-containing (5 × 10−8 m; Sigma) medium in the second fraction.
Hormone assays
Adrenocorticotropic hormone (ACTH) and corticosterone in the trunk blood and in acutely isolated pituitaries were measured by radioimmunoassay in 50-μl unextracted samples as described previously (Zelena et al. 2008).
Bone marrow chimeras
Bone marrow chimeras were generated by transplantation of P2rx7+/+ or P2rx7−/− bone marrow cells (carrying the CD45.2 allele) into lethally irradiated recipients carrying the CD45.1 allele as described previously (Jakus et al. 2009). The repopulation of the peripheral leukocyte compartment by donor-derived cells was tested 8 wk after transplantation by flow cytometric analysis of blood samples stained for the granulocyte-specific Gr1 and the donor-specific CD45.2 markers (Jakus et al. 2009). Percent repopulation was defined as the percentage of CD45.2-positive cells among Gr1-positive cells with typical forward and side-scatter characteristics.
Preparation of microglia
Microglia was prepared following the method described by Racz et al. (2008), with slight modifications. Briefly, single-cell suspensions from mouse brain were generated using a commercially available enzymatic kit (Miltenyi Neural Tissue Dissociation kit ‘P’; Miltenyi Biotec, Germany) and filtered through an 83-μm sieve. Percoll (GE Healthcare, UK) gradients of 75/25% were performed for fractionation of cells at 800 g for 25 min and 800 g for 10 min and brain mononuclear cells were collected from the interface.
Immunohistochemistry
Light and eletronmicrosopic immunostaining for P2rx7s and the microglia marker CD11b was performed using standard protocols described earlier (Csölle et al. 2008). For the detailed protocol of immunohistochemistry, see Supplement 3.
Microarray experiments and real-time PCR validation
Mice were randomly assigned to experimental groups and injected i.p. with sterile saline (0.9% NaCl) or LPS (250 μg/kg) and killed by decapitation 6 h after injection. Microarray profiling on amygdala samples was performed in the Agilent Microarray Core Facility (Semmelweis University, Budapest; http://www.dgci.sote.hu/microarray). The complete microarray expression data are deposited at NCBI's Gene Expression Omnibus (Edgar et al. 2002) through GEO series accession number GSE21218 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=rzwxlikqskwayfk&acc=GSE21218). The microarray data were further analysed using the bioinformatics analysis tool GeneCodis (Carmona-Saez et al. 2007; Nogales-Cadenas et al. 2009).
Real-time PCR validation
TaqMan low-density arrays (TLDA microfluidic card; Applied Biosystems, USA) were performed on 92 selected genes of interest and four housekeeping genes (18S rRNA, Gapdh, Hprt, B2m) (Vandesompele et al. 2002).
See Supplement 4 for the technical details of the microarray and real-time PCR experiments.
Statistics
All data shown are the means±s.e.m. of n determinations. OF and EPM data were evaluated by analysis of variance (ANOVA). TST and AH data were analysed by one-way ANOVA followed by Dunnett's test. Hormone levels, monoamine contents, IL-1β, microarray and PCR data were analysed by two-way ANOVA. The FST and in vitro ACTH secretion was analysed by repeated measures ANOVA. The Student's t test and Fischer's LSD test were used for pairwise comparisons. Unless otherwise stated, statistical analyses were performed using the Statistica program (Statsoft, USA). The level of significance was set at p < 0.05.
Results
Genetic deletion of P2rx7 leads to mood-stabilizing behavioural phenotype
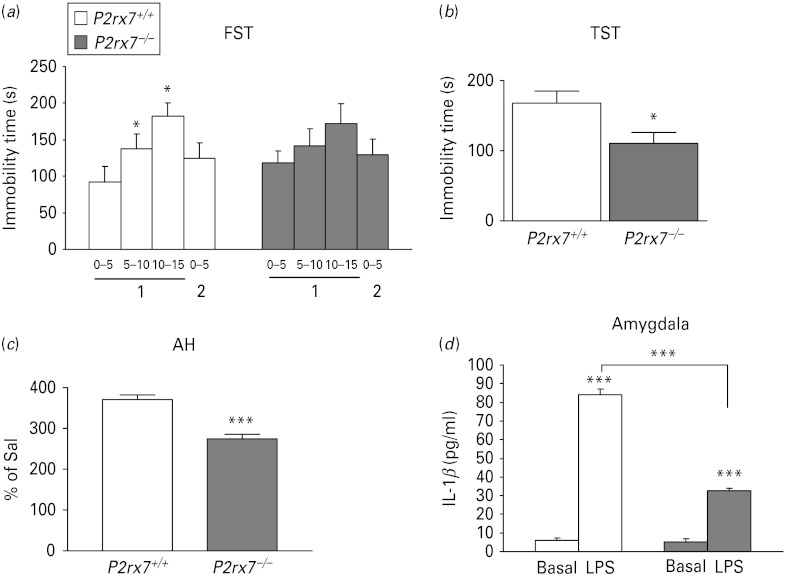
To model depressive-like behaviour, occurring during major depressive disorder and the depressive pole of bipolar disorder, FST and TST were used. In the FST, we have used the widely used behavioural despair paradigm, originally described by Porsolt et al. (1977), which involves subjects being forced to swim twice on two consecutive days. P2rx7+/+ mice displayed a time-dependent increase in the time of immobility during consecutive 5-min periods of the first trial, indicating that they developed behavioural despair, a well-known depressive-like behaviour (Fig. 1 a). P2rx7−/− mice were less prone to develop depression-like behaviour than P2rx7+/+ mice (Fig. 1 a). During the first trial, the duration of immobility was similar in the two genotypes (92.43 ± 21.12 s, for wild-type vs. 118.38 ± 16.46 s for P2rx7−/−; genotype: F1,18 = 0.05, p < 0.8). In P2rx7+/+mice, the time spent in immobility increased over time during the first trial (F3,36 = 3.61, p < 0.02). In P2rx7−/− mice, parallel changes were seen, but these were not significant (F3,36 = 0.30, p > 0.8). When the two genotypes were considered together, the time × genotype interaction was significant (F3,54 = 2.92, p < 0.04) and significant time-related changes were seen in P2rx7+/+ mice only.
Fig. 1.
Genetic deletion of P2rx7s in mice leads to an antidepressant-like phenotype in the forced swim test (FST) and tail suspension test (TST) and mood stabilizing-like phenotype in amphetamine-induced hyperlocomotion test (AH), but does not affect basal interleukin (IL)-1β levels in the amygdala. (a) P2rx7−/− mice failed to develop the depression-like behaviour typical to the FST. The time of immobility is expressed in seconds. * Indicates significant changes from immobility values observed during min 0–5 of the first day, n = 20/group; * p < 0.05. (b) Genetic disruption decreased basal immobility in the TST (n = 9–11, * p < 0.05 vs. P2rx7+/+, Student's t test). The immobility time is expressed in seconds. The total test period was 360 s. (c) Amphetamine-induced hyperactivity is significantly attenuated in P2rx7−/− mice. Mice were placed into the open field arena for a 30-min habituation period and then injected with i.p. saline (Sal) or d-amphetamine-sulfate (2.5 mg/kg). Locomotor activity was assessed for 90 min immediately after the injection and expressed as the percentage of Sal-treated mice, n = 9–10, *** p < 0.001 vs. P2rx7+/+, Student's t test. Drug- and test-naive male homozygous mice (P2rx7+/+ and P2rx7−/−, aged 2–3 months) weighing approximately 30 g were used in the experiments. (d) IL-1β protein level in the amygdala of P2rx7+/+ and P2rx7−/− mice after Sal and lipopolysaccharide (LPS) treatment. The IL-1β protein level was similar in the amygdalae of Sal-treated P2rx7+/+ and P2rx7−/− mice. Injection of LPS (E. coli; 250 μg/kg i.p.) significantly increased the level of IL-1β in the amygdalae of P2rx7+/+ mice 6 h after treatment. IL-1β protein level was less elevated in the amygdalae of P2rx7−/− mice in response to systemic endotoxin. The levels of IL-1β were quantified in the supernatants by ELISA. Data are given as the mean level of cytokines±s.e.m., expressed in pg/ml. * Indicates significant differences between Sal- and LPS-treated and between P2rx7+/+ and P2rx7−/− mice (n = 4 per group, *** p < 0.001, two-way analysis of variance).
The basal time of immobility in the TST was 167.90 ± 17.13 s (n = 9) in P2rx7+/+ mice (Fig. 1 b). In line with previous data (Basso et al. 2009), P2rx7−/− mice spent significantly less time with immobility (i.e. they exhibited antidepressant-like phenotype in this test as well; 110.24 ± 15.84 s, n = 11, p < 0.05, Fig. 1b). Neither basal locomotor activity nor anxiety-like behaviour, examined in the OF and EPM tests, was affected by the genetic disruption of the P2rx7 gene (Supplementary Fig. 1 a, b).
As a behavioural model of manic pole of bipolar disorder (Einat, 2007), the AH test was used in our study. The locomotor activity of saline-treated mice, expressed as the total covered distance during the test period, was similar in P2rx7+/+ and P2rx7−/− mice (120.17 ± 24.71 m, n = 9 and 156.53 ± 18.48 m, n = 10, respectively, p > 0.05). By contrast, amphetamine (2.5 mg/kg i.p.) induced hyperactivity was significantly attenuated in mice genetically deficient in P2rx7 (Fig. 1 c).
Neurochemical changes and stress-induced hormone response in the absence of P2rx7
The level of IL-1β 6 h after saline administration was 6.06 ± 1.18 pg/ml in the amygdalae of P2rx7+/+ mice (Fig. 1 d, n = 4). Systemic LPS administration (250 μg/kg i.p.) caused a significant elevation in IL-1β levels (83.94 ± 3.01 pg/ml, n = 8, F1,28 = 45.2, p < 0.001, Fig. 1d). The IL-1β level in the amygdalae of saline-treated P2rx7−/− mice was 5.29 ± 1.48 pg/ml (n = 4), not significantly different from P2rx7+/+ mice (p > 0.05, Fig. 1 d). The LPS-induced elevation of IL-1β level was significantly attenuated in the amygdalae of P2rx7−/− mice (32.53 ± 1.55 pg/ml, n = 8; genotype: F1,28 = 183.3, p < 0.001; interaction: F1,28 = 42.55, p < 0.001, Fig. 1d).
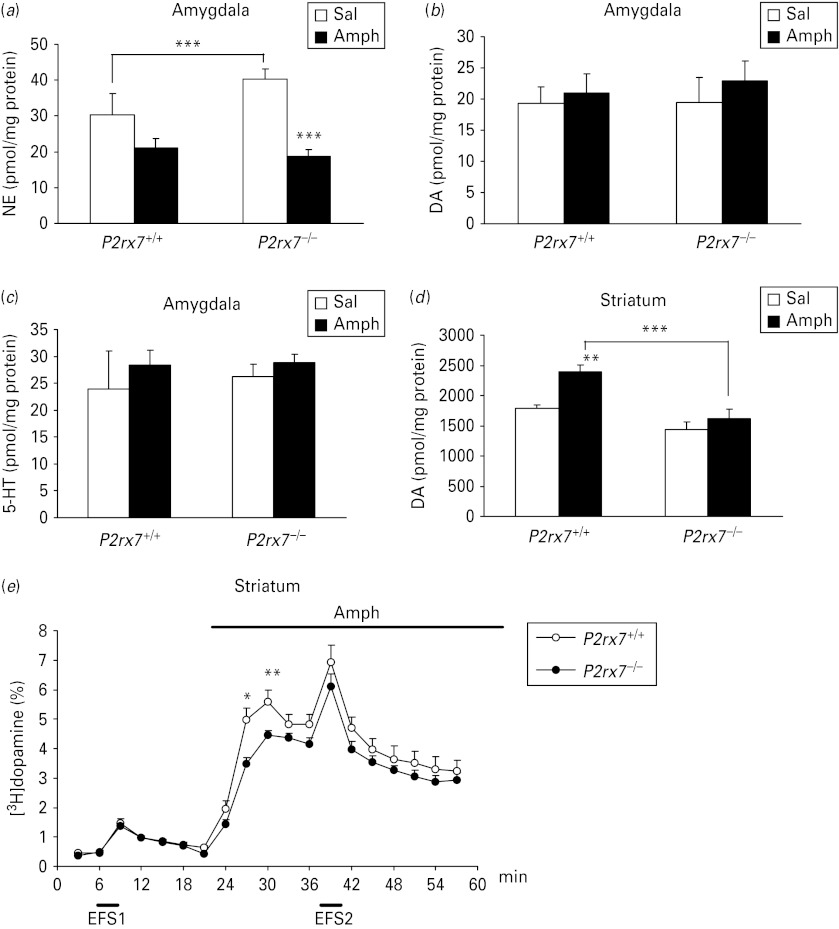
To explore neurochemical correlates of changes in behaviour, the level of norepinephrine, dopamine and serotonin (5-HT) was also analysed in the amygdala and striatum of saline and amphetamine (2.5 mg/kg i.p.) treated P2rx7+/+ and P2rx7−/− mice. The basal level of norepinephrine was elevated in the absence of P2rx7 in the amygdala (genotype: F1,27 = 3.30, p = 0.08; interaction: F1,27 = 6.52, p = 0.016; post-hoc comparison by Fischer's LSD test, p < 0.005), whereas no change in basal norepinephrine level was found in the striatum (29.37 ± 13.79, n = 7 and 18.21 ± 3.83 pmol/mg protein, n = 8 in P2rx7+/+ and P2rx7−/− mice, respectively; interaction: F1,27 = 0.05, p > 0.8). Likewise, basal dopamine and 5-HT levels were not affected by the genetic deletion either in the amygdala (Fig. 2 a–c, dopamine: F1,27 = 0.05, p > 0.8; 5-HT: F1,27 = 1.07, p > 0.3) or in the striatum of saline-treated mice (Fig. 2 d and data not shown). Consistent with the observed increase in locomotor activity, amphetamine treatment caused an elevation in the dopamine content of the striatum of P2rx7+/+ mice (Fig. 2 d, F1,27 = 10.08; p < 0.004), a reduction of norepinephrine level in the amygdala (F1,27 = 17.17, p < 0.001), without changing the 5-HT and dopamine levels in the amygdala (Fig. 2 a–c, 5-HT: F1,27 = 0.23; p > 0.6, dopamine: F1,27 = 0.43, p > 0.5) and with a tendency for lower norepinephrine levels in the striatum (26.4 ± 3.23, n = 7 and 12.2 ± 3.07 pmol/mg protein, n = 8 in P2rx7+/+ and P2rx7−/− mice, respectively; genotype: F1,27 = 3.36, p = 0.07). In line with these findings, when acute striatal slices treated in vitro with amphetamine (30 μm), a profound increase in [3H]dopamine efflux was observed (Fig. 2e). Both the amphetamine-induced increase in dopamine content (Fig. 2d; interaction: F1,27 = 2.91, p = 0.09, post-hoc comparison by Fischer's LSD test: p < 0.001) and [3H]dopamine efflux (Fig. 2e) were significantly attenuated in P2rx7−/− mice, without the alteration of basal and electrical field stimulation-evoked [3H]dopamine release (Fig. 2 e).
Fig. 2.
Changes in the content of norepinephrine (NE) (a), dopamine (DA) (b), 5-HT (c) in the amygdala; dopamine content in the striatum (d) and release of [3H]dopamine (e) in acute striatal slices in the absence of P2rx7. (a–d) P2rx7+/+ and P2rx7−/− mice were treated with saline (Sal) or amphetamine (Amph, 2.5 mg/kg i.p.) and 30 min after the treatment were decapitated. NE, DA and 5-HT levels were analysed by high performance liquid chromatography in the amygdala and striatum and are expressed in pmol/mg protein. * Indicates significant differences between P2rx7+/+ and P2rx7−/− mice and between Sal- and Amph-treated groups, as indicated (n = 7–8/group, ** p < 0.01, *** p < 0.001, two-way analysis of variance followed by Fischer's LSD test). (e) Basal, electrical field stimulation (EFS)- and Amph-induced [3H]dopamine efflux from striatal slices of P2rx7+/+ and P2rx7−/− mice. Striatal slices were incubated with [3H]dopamine and superfused with Krebs’ solution. EFS (20 V, 2 Hz, 240 shocks) and Amph (30 μm) were applied as indicated by the horizontal bars. The efflux of [3H]dopamine is expressed as fractional release, which represents the tritium content in a sample as a percentage of the actual total tritium content. Amph-induced [3H]dopamine release was significantly decreased in striatal slices of P2rx7−/− mice, whereas basal and electrical stimulation-induced efflux remained unchanged. n = 8/group, * p < 0.05, ** p < 0.01, Student's t test.
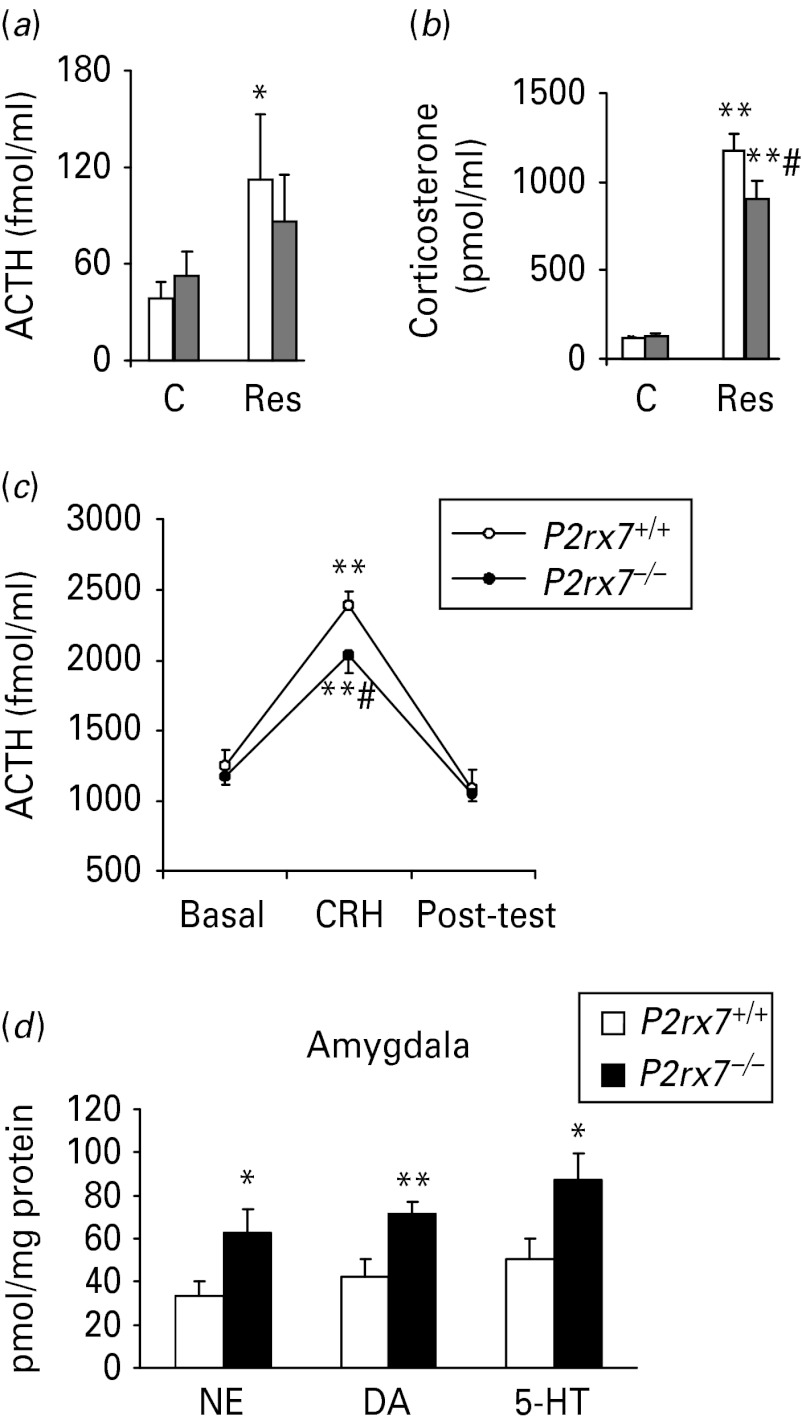
Body, thymus and adrenal gland weights remained unchanged by P2rx7 gene disruption (Supplementary Table S1). Similarly, resting ACTH and corticosterone levels were not affected by genotype (Fig. 3 a, b; ACTH: F1,34 = 0.1, p > 0.9; corticosterone: F1,33 = 1.06, p > 0.3). Restraint stress increased the level of both hormones (ACTH: F1,34 = 6.9, p = 0.013; corticosterone: F1,33 = 272.7, p < 0.001), but the elevation of ACTH levels reached the level of significance in P2rx7+/+animals only (p = 0.03; Fig. 3 a). In addition, the corticosterone increase was significantly lower in P2rx7−/− mice, than in P2rx7+/+ mice (p = 0.048; Fig. 3 b). In isolated in vitro pituitaries, CRH treatment significantly increased ACTH secretion (Fig. 3 c). However, this effect depended on genotype (F1,17 = 4.65, p = 0.045), with a significantly smaller response in P2rx7−/− mice (Fig. 3c). After restraint stress, norepinephrine, dopamine and 5-HT levels were all elevated in the amygdala of P2rx7−/− mice when compared to P2rx7+/+littermates (Fig. 3d).
Fig. 3.
P2rx7−/− mice [grey bars in (a) and (b)] respond with a decreased elevation of adrenocorticotropic hormone (ACTH) and corticosterone in the plasma (a, b) and pituitary (c) and increased norepinephrine (NE) level (d) in the amygdala to stress. Hormonal secretion was evaluated after 30 min of restraint. Plasma levels of ACTH (a) and corticosterone (b) were measured by radioimmunoassay of trunk blood (n = 9–10). The resting ACTH and corticosterone levels were not affected by genotype. Restraint significantly increased the level of both hormones, but the elevation of ACTH levels reached the level of significance in P2rx7+/+ animals (clear bars) only (p = 0.03). (c) The ACTH response to 5 × 10−8 m corticotropin-releasing hormone (CRH) was studied in vitro (n = 10 per group). Fifteen-min fractions were collected. CRH treatment significantly increased ACTH secretion. This effect was dependent on the genotype and there was a significantly smaller response in P2rx7−/− mice. C, control; Res, exposed to 30-min restraint. Post-hoc comparisons: * p < 0.05; ** p < 0.01 vs. control; #p < 0.05 vs. P2rx7+/+. (d) The level of NE, dopamine (DA) and 5-HT was analysed by high performance liquid chromatography in the amygdalae of P2rx7+/+ and P2rx7−/− mice after 30 min restraint. The results are expressed as pmol/mg protein. * Represents a significant difference between P2rx7+/+ and P2rx7−/− animals, n = 10–12/group, * p < 0.05, ** p < 0.01, Student's t test.
P2rx7 antagonist treatment reproduces the effect of genetic deletion
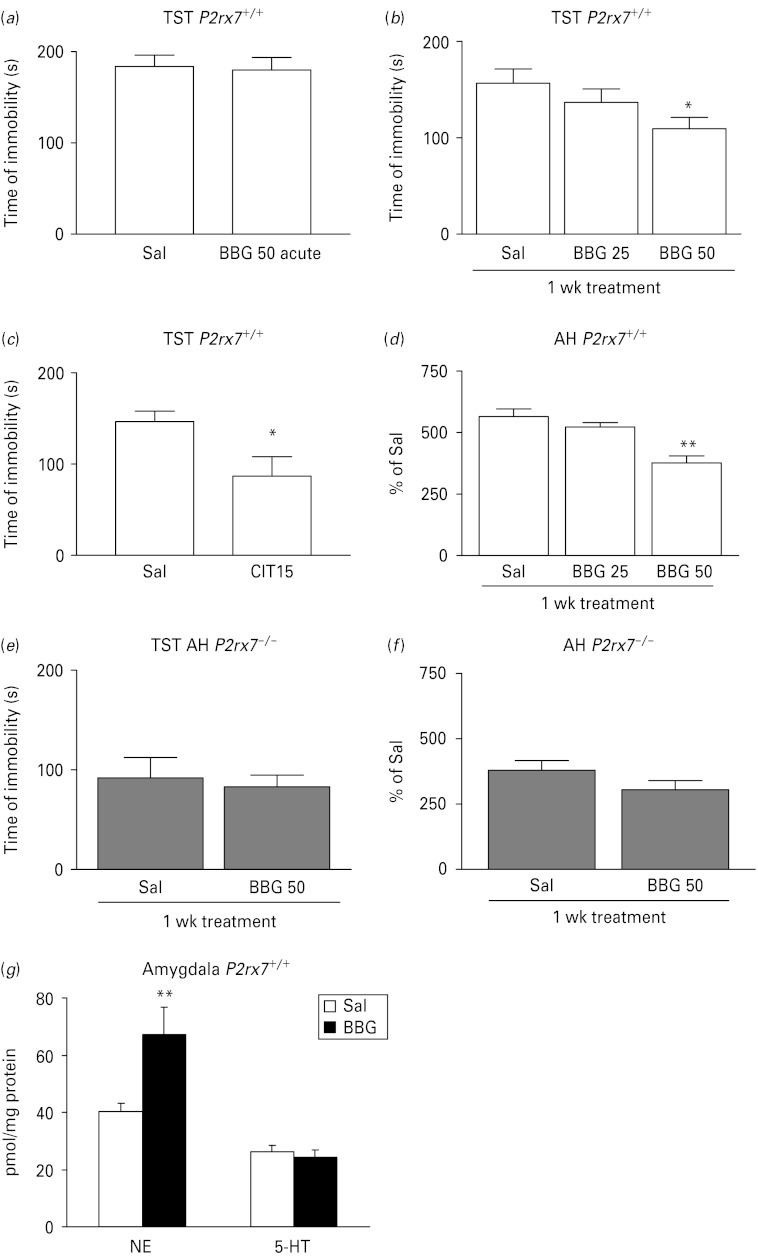
Next, we asked whether the behavioural phenotype detected in P2rx7−/− mice could be reproduced by the systemic administration of a P2rx7 antagonist. BBG, a selective P2rx7 antagonist, was ineffective upon acute application (50 mg/kg i.p.), but it dose-dependently (25–50 mg/kg i.p.) inhibited depressive-like behaviour after a 1-wk sub-acute treatment in the TST (Fig. 4 a, b). Citalopram (acute treatment, 15 mg/kg i.p.), which was used as a reference compound, elicited a 40.6% decrease in the immobility time in the TST in P2rx7+/+ mice (Fig. 4 c). Sub-acute (Fig. 4 d), but not acute (data not shown) BBG treatment (25–50 mg/kg i.p.) also reproduced the effect of genetic deletion in the AH test and elevated norepinephrine levels in the amygdala (Fig. 4 g). When sub-acute BBG treatment was applied in P2rx7−/− mice, no significant change either in immobility in the TST (Fig. 4 e) or in AH (Fig. 4 f) was detected.
Fig. 4.
The selective P2rx7 antagonist Brilliant Blue G (BBG) exhibits antidepressant-like activity in the tail suspension test (TST), decreases amphetamine-induced hyperactivity and elevates norepinephrine (NE) level in the amygdala in P2rx7+/+, but not P2rx7−/− mice. (a) The effect of acute BBG treatment on basal immobility in P2rx7+/+ mice (n = 13–15). (b) Effect of sub-acute, 1-wk treatment with BBG on basal immobility in P2rx7+/+ mice. Mice were treated with BBG for 7 d with the daily doses indicated on the abscissa or with saline (Sal) and then submitted to the TST (n = 13–15, * p < 0.05 vs. SAL, one-way analysis of variance (ANOVA), followed by Dunnett's test). (c) Citalopram (Cit; 15 mg/kg), the potent selective serotonin reuptake inhibitor antidepressant, decreased basal immobility in the TST in P2rx7+/+mice (n = 11/group, * p < 0.05 vs. Sal, Student's t test). (d) Effect of 1-wk daily treatment with BBG (25–50 mg/kg i.p.) or Sal on amphetamine induced hyperactivity in P2rx7+/+mice. (n = 10–12/group, ** p < 0.001 vs. Sal, one-way ANOVA followed by Dunnett's test). Citalopram and BBG were administered i.p. 30 min before testing at the doses (mg/kg) indicated on the abscissa. Sal-treated mice were injected with an equal volume of Sal. (e) Effect of sub-acute, 1-wk treatment with BBG (50 mg/kg i.p.) on basal immobility in P2rx7−/− mice. Mice were treated with BBG for 7 d or with Sal and then submitted to the TST (n = 8–10/group). (f) Effect of 1-wk daily treatment with BBG (50 mg/kg i.p.) or Sal on amphetamine-induced hyperactivity in P2rx7−/− mice (n = 10/group). Note that the basal immobility (e) and amphetamine-induced hyperactivity (f) is lower in Sal-treated P2rx7−/− mice than in Sal-treated P2rx7+/+ mice. (g) Sub-acute, 1-wk treatment with BBG (50 mg/kg i.p.) elevates NE, but not 5-HT levels in the amygdala in P2rx7+/+ mice, when compared to Sal-treated mice. Tissue content of NE and 5-HT were analysed by high performance liquid chromatography and are expressed in pmol/mg protein, n = 11–12, ** p < 0.01, Student's t test.
Alteration of behavioural phenotype of P2rx7−/− mice is not related to P2rx7s expressed on cells of haematopoietic origin
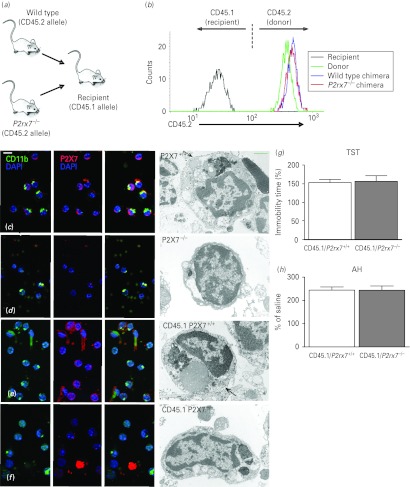
To test whether the alteration of behavioural phenotype detected in P2rx7−/− mice was due to the lack of P2rx7s on cells of haematopoietic origin, we generated bone marrow chimeras that lacked the P2rx7 only in their haematopoietic compartment (Fig. 5 a). Bone marrow cells harvested from P2rx7+/+ or P2rx7−/− mice (expressing the CD45.2 leukocyte marker) were injected i.v. into lethally irradiated congenic recipients carrying the CD45.1 marker. The efficiency of the reconstitution was tested 8 wk after transplantation by flow cytometric analysis of the expression of the donor-specific CD45.2 marker in peripheral blood leukocytes. As shown in Fig. 5b, CD45.2 was present on granulocytes of the donor strain but not on those of the recipient strain. Importantly, granulocytes isolated from bone marrow chimeras used for the behavioural studies, engrafted with either P2rx7+/+ or P2rx7−/− bone marrow cells, were practically exclusively (>99%) of donor (CD45.2-positive) origin. The recruitment of donor-derived cells was also assessed by immunocytochemical staining of P2rx7s in microglia cells isolated from the brains of intact mice and bone marrow chimeras. As shown by fluorescent and electron microscopic images, the cell membranes of microglia (i.e. cells positive for the microglia marker CD11b) of intact P2rx7+/+ mice and of bone marrow chimeras engrafted with P2rx7+/+ bone marrow cells (CD45.1/P2rx7+/+) showed intense P2rx7 immunoreactivity (Fig. 5 c, e). Because not all microglia are positive for CD11b and not only microglia express the P2rx7, CD11b-negative/P2rx7-positive and CD11b-positive/P2rx7-negative cells were also found in the preparation. In contrast, no P2rx7 immunoreactivity was observed on microglial cells isolated from intact P2rx7−/− mice (Fig. 5 d). Microglia cells of bone marrow chimeras engrafted with P2rx7−/− bone marrow cells (CD45.1/P2rx7−/−) also displayed scarce P2rx7 immunoreactivity, whereas other non-haematopoietic cells of the preparation were stained with the antibody against the P2rx7 (Fig. 5 f). Bone marrow chimeric mice were subjected to the TST and AH as described above. No difference was found in the time of immobility in either the TST (Fig. 5 g) or in the hyperlocomotion in the AH (Fig. 5 h), showing that the mood stabilizing-like phenotype found in P2rx7−/− mice was not transferred to P2rx7+/+ recipients with the engraftment of the P2rx7−/− bone marrow cells.
Fig. 5.
Mood-stabilizing phenotype is not detectable in chimeric mice transplanted with the bone marrow of P2rx7−/− mice. (a) General scheme of bone marrow transplantation. (b) Flow cytometric analysis of the expression of the donor-specific CD45.2 allele on peripheral blood granulocytes of an intact C57BL/6J mouse (donor), a CD45.1-expressing congenic mouse (recipient) and bone marrow chimeras engrafted with P2rx7+/+ and P2rx7−/− bone marrow cells (P2rx7+/+ and P2rx7−/− chimeras, respectively). (c–f) Immunocytochemical demonstration of the P2rx7 in mouse microglial cells. Co-localization of P2rx7s (labelled red) with microglial labelling CD11b (green) was found in microglia cells retrieved from P2rx7+/+ mouse brain tissue (c) and in chimeras transplanted with P2rx7+/+ bone marrow [(e) CD45.1/ P2rx7+/+]. Microglial cells of P2rx7−/− animals (d) did not show P2rx7 immunoreactivity. In chimeras transplanted with P2rx7−/− bone marrow [(f), CD45.1/P2rx7−/−], the P2rx7 (labelled red) was not found on microglial cells (labelled green) but was in some unidentified cells and cell debris in the cell suspension/fraction. Electron microscopy supported this finding. The cell membrane of morphologically characterized microglial cells – dark nuclei, either oval or bean shaped, electron dense cytoplasm, long cisternae of granular endoplasmic reticulum and large inclusions of phagocytosed material in the cell bodies that are commonly found in old animals – were covered by diaminobenzidine precipitates, demonstrating P2rx7 immunoreactivity (c and e rows). Cell processes of unidentified cells also showed P2rx7 immunoreactivity (e). Microglial cells of P2rx7 gene knockout animals (d) or microglial cells from CD45.1/P2rx7−/− animals (f) were free of precipitate. (g, h) Basal immobility time in the tail suspension test (g) and amphetamine-induced hyperactivity (h) were not different in CD45.1/P2rx7+/+ and CD45.1/P2rx7−/− mice. Mice were submitted to behaviour tests 8 wk after engraftment (n = 13–14/group).
Alteration of the gene expression profile in the amygdalae of P2rx7−/− mice
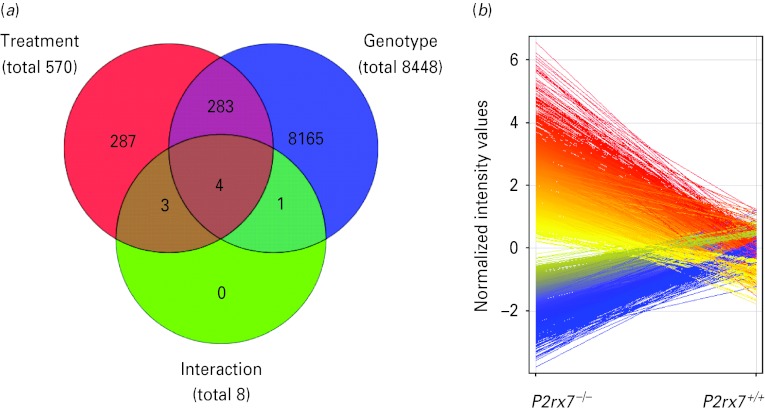
Because previous experiments and literature data indicated that monoamine transmitter levels and c-Fos expression (Boucher et al. 2011) are regulated by P2rx7 in the amygdala, we next examined how global gene expression is changed in the absence of P2rx7 in this region and LPS treatment was used as a reference stimulus. Four amygdala samples/group collected from P2rx7+/+ and P2rx7−/− mice 6 h after LPS (250 μg/kg i.p.) or saline treatment were subjected to whole genome microarray analysis. Out of the 41041 transcripts printed on the microarrays, the expression of a total of 8739 transcripts was significantly altered with the applied statistical filters (>2.0-fold change; p < 0.05; Benjamini–Hochberg's multiple correction; Fig. 6 a). These transcripts were sorted according to their main effect, namely, genotype and treatment, as illustrated in the Venn diagram in Fig. 6 a. Deficiency of P2rx7 had the most profound effect on the expression profile, changing the expression of 8448 transcripts (3133 increased, 5315 decreased, Fig. 6b) independently from the treatment, followed by the effect of LPS, which affected the expression of 570 transcripts, (415 increased, 155 decreased) independently from the genotype. Four transcripts, as illustrated by the red/blue/green intersection in the Venn diagram, were differentially altered by the treatment, depending on the genotype.
Fig. 6.
Summary of results of the whole genome microarray analysis performed on amygdala samples of P2rx7+/+ and P2rx7−/− mice subjected to saline or lipopolysaccharide (LPS) treatments (n = 4/group). (a) Venn diagram represents the main effects of the two-way analysis of variance with Benjamini–Hochberg's multiple correction comparison test for unequal replications and using the selection filters of >2.0-fold change: LPS treatment (red circle), genotype (blue circle) and their interaction (green circle). Numbers inside the compartments represent the number of transcripts that are significant for that effect. The intersections of the sets represent significantly changed genes for each of the effects involved in the intersection. Significance threshold was 0.05 and the p value computation was asymptotic. In the primary microarray analysis, the expression of a total of 8739 transcripts was found to be significantly altered with the applied statistical filters. These 8739 genes sum up from the genes significantly affected by treatment independently from genotype (287 + 283 = 570, illustrated by the numbers in those areas of the red circle, which is covered only by the red area, and covered by the red and blue areas), by genotype independently from treatment (8165 + 283 = 8448, illustrated by the numbers in those areas of the blue circle, which is covered only by the blue area, and covered by the red and blue areas) and by their interaction (4, as illustrated by the numbers within the intersection of the red/blue and green circles), i.e. 287 + 283 + 8165 + 4=8739. The intersection of the red and green circles represents three transcripts, which were not only significantly affected by LPS treatment but may also be altered by genotype, whereas the intersection of the blue and green circles represents one transcript, which is not only significantly affected by knockout genotype but may also be altered by LPS treatment. (b) Deficiency of the P2rx7 had the most profound effect on the expression profile, changing the expression of 8448 transcripts. Among them, 3133 were increased (red) and 5315 were decreased (blue), as illustrated by the deviation of the lines according to the normalized intensity values.
To further understand the functional impact of the biological processes affected by P2rx7 deficiency, microarray data were further subjected to gene ontology analysis. Because the total number of modified genes affected only by P2rx7 deletion (8165) was relatively large, 588 genes showing the top rank of up- and down-regulation were selected for gene ontology analysis by GeneCodis (Supplementary Table S2 a). This allowed us to investigate the association of the 588 genes using the defined algorithm to determine biological annotations or combinations of annotations that were over-represented with regard to a reference list of Mus musculus deposited in the NCBI GenBank database. The GeneCodis analysis generated a relationship of 373 annotation groups characterized with the Gene Ontology (GO) Biological Process, including transport, ion transport, signal transduction, synaptic transmission, G-protein coupled receptor protein signalling pathway, ATP synthesis coupled proton transport, transcription, regulation of transcription and GABA signalling pathway (Supplementary Table S2 b, c). The genes involved in each annotation group are listed in Supplementary Table S2 with reference to their involved GO categories. The annotation groups were further sorted using a permutation test for p value correction.
Using the data generated by the GO analyses and searching biological plausibility for depression (e.g. synaptic transmission and neuroplasticity), we chose 60 genes for further validation out of the 8165 transcripts that were changed significantly by P2rx7 deficiency, regardless of the effect of LPS treatment, and showed a fold change (FC) of ⩾2.0 (Table 1). Among the 287 transcripts that had a significant LPS treatment effect, regardless of the effect of genotype, 17 transcripts were selected that had biological plausibility in inflammatory and immune responses (Table 2).
Table 1.
Genes that were up- and down-regulated by the deficiency of P2rx7 in the mouse amygdala
| Symbol | Gene ID | Gene name (accession number) | Agilent (FC) | Real-time PCR (RQ) | TaqMan ID |
|---|---|---|---|---|---|
| (a) | |||||
| Adra2a | 11 551 | Adrenergic receptor α2a (NM_007417) | 2.72 | 1.34 ± 0.38 p = 0.0171 | Mm00845383_s1 |
| Adrb1 | 11 554 | Adrenergic receptor β1 (NM_007419) | 4.77 | Mm00431701_s1 | |
| Ak5 | 229 949 | Adenylate kinase 5 (NM_001081277) | 5.71 | Mm00461978_m1 | |
| Aplp2 | 11 804 | Amyloid β (A4) precursor-like protein 2 (NM_009691) | 5.55 | Mm00507819_m1 | |
| Bdnf | 12 064 | Brain derived neurotrophic factor isoform 1 (NM_007540) | 3.89 | Mm01334047_m1 | |
| Casp8 | 12 370 | Caspase 8 (NM_009812) | 9.57 | Mm00802247_m1 | |
| Chkb | 12 651 | Choline kinase β (NM_007692) | 4.160 | Mm00432498_m1 | |
| Cnr1 | 12 801 | Cannabinoid receptor 1 (NM_007726) | 2.081 | 1.31 ± 0.28 p = 0.0087 | Mm01212171_s1 |
| Ddc | 13 195 | Dopa decarboxylase (NM_016672) | 2.051 | 1.58 ± 0.84 p = 0.0249 | Mm00516688_m1 |
| Drd2 | 13 489 | Dopamine receptor 2 (NM_010077) | 2.37 | 2.44 ± 0.47 p = 0.0120 | Mm00438545_m1 |
| Gabarapl1 | 57 436 | GABAA receptor-associated protein-like 1 (NM_020590) | 4.47 | Mm00457880_m1 | |
| Gabra1 | 14 394 | GABAA receptor subunit α1 (NM_010250) | 4.130 | Mm00439046_m1 | |
| Gabra5 | 110 886 | GABAA receptor subunit α5 (NM_176942) | 7.79 | Mm00621092_m1 | |
| Gabrb2 | 14 401 | GABAA receptor subunit β2 (NM_008070) | 2.141 | 2.51 ± 1.12 p = 0.0036 | Mm00549788_s1 |
| Gabrb3 | 14 402 | GABAA receptor subunit β3 (NM_008071) | 2.041 | 1.34 ± 0.55 p < 0.0005 | Mm00433473_m1 |
| Gabrg1 | 14 405 | GABAA receptor subunit γ1 (NM_010252) | 2.55 | 3.38 ± 1.19 p = 0.0005 | Mm00439047_m1 |
| Gabrg2 | 14 406 | GABAA receptor subunit γ2 (NM_008073) | 2.43 | 2.04 ± 0.89 p = 0.0012 | Mm00433489_m1 |
| Gad1 | 14 415 | Glutamic acid decarboxylase 1 (NM_008077) | 2.171 | 1.10 ± 0.06 p = 0.0009 | Mm00725661_s1 |
| Gfra1 | 14 585 | Glial cell line derived neurotrophic factor family receptor α1 (NM_010279) | 2.231 | 1.82 ± 0.54 p = 0.0308 | Mm00833897_m1 |
| Gja1 | 14 609 | Gap junction protein α1 (NM_010288) | 4.097 | Mm00621092_m1 | |
| Gjb6 | 14 623 | Gap junction protein β6 (NM_001010937) | 4.48 | 1.40 ± 0.28 p < 0.0016 | Mm00433661_s1 |
| Gjc1 | 14 615 | Gap junction protein γ1 (NM_008122) | 2.161 | 1.247 ± 0.11 p = 0.0050 | Mm01253027_m1 |
| Gjd2 | 14 617 | Gap junction protein δ2 (NM_010290) | 4.61 | Mm00439121_m1 | |
| Gje1 | 76 743 | Gap junction membrane channel protein ε1 (NM_080450) | 2.00 | 1.49 ± 0.18 p = 0.0022 | Mm00519120_s1 |
| Glra2 | 237 213 | Glycine receptor α2 subunit (NM_183427) | 2.34 | 1.30 ± 0.25 p = 0.0157 | Mm00439140_m1 |
| Glrb | 14 658 | Glycine receptor β subunit (NM_010298) | 5.089 | 1.16 ± 0.25 p = 0.0018 | Mm00439140_m1 |
| Gria2 | 14 800 | Glutamate receptor, ionotropic, AMPA2 (α2) | 2.59 | 1.13 ± 0.05 p < 0.0002 | Mm00442822_m1 |
| Gria4 | 14 802 | Glutamate receptor, ionotropic, AMPA4 (α4), transcript variant 1 (NM_019691) | 2.74 | 1.41 ± 0.66 p = 0.0014 | Mm00444754_m1 |
| Grm7 | 108 073 | Glutamate receptor, metabotropic 7 (NM_177328) | 35.81 | 1.07 ± 0.16 p = 0.0317 | Mm01189424_m1 |
| Gstm4 | 14 865 | Glutathione S-transferase μ4 (NM_026764) | 3.34 | Mm00728197_s1 | |
| Klhl12 | 240 756 | Kelch-like 12 (Drosophila) (NM_153128) | 6.98 | 1.19 ± 0.31 p = 0.0074 | Mm00462323_m1 |
| Narg1l | 66 897 | NMDA receptor regulated 1-like (NM_025832) | 6.98 | 1.77 ± 0.21 p = 0.0032 | Mm00462323_m1 |
| Ncam1 | 17 967 | Neural cell adhesion molecule 1 (NM_010875) | 3.310 | Mm00456815_m1 | |
| Npy5r | 18 168 | Neuropeptide Y receptor Y5 (NM_016708) | 6.68 | Mm02620267_s1 | |
| P2ry1 | 18 441 | Purinergic receptor P2Y, G-protein coupled 1 (NM_008772) | 3.89 | 1.47 ± 0.73 p = 0.0467 | Mm00435471_m1 |
| Pcdh10 | 18 526 | Protocadherin 10 (NM_001098171) | 14.93 | Mm00477987_s1 | |
| Pcmt1 | 18 537 | Protein-l-isoaspartate (d-aspartate) O-methyltransferase 1 (NM_008786) | 2.251 | 1.23 ± 0.28 p = 0.0073 | Mm00476600_m1 |
| Pde1a | 18 573 | Phosphodiesterase 1A calmodulin- dependent (NM_016744) | 2.191 | 1.24 ± 0.74 p = 0.0123 | Mm00450244_m1 |
| Plp1 | 18 823 | Proteolipid protein (myelin) 1 (NM_011123) | 9.031 | 1.51 ± 0.18 p = 0.0033 | Mm00456892_m1 |
| Ryr3 | 20 192 | Ryanodine receptor 3 (BC116740) | 2.80 | 1.79 ± 0.22 p < 0.0002 | Mm01335482_m1 |
| (b) | |||||
| Adcy8 | 11 514 | Adenylate cyclase (NM_009623) | 17.56 | Mm00507722_m1 | |
| Adrbk1 | 110 355 | Adrenergic receptor kinase β1 (NM_130863) | 5.55 | Mm00804778_m1 | |
| Adrbk2 | 320 129 | Adrenergic receptor kinase β2 (NM_177080) | 4.38 | Mm00622037_m1 | |
| Aloxe3 | 23 801 | Arachidonate lipoxygenase 3 (NM_011786) | 5.121 | Mm004778628_m1 | |
| Anxa7 | 11 750 | Annexin A7 (NM_009674) | 3.94 | Mm00477549_m1 | |
| Avp | 11 998 | Arginin vasopressine (NM_009732) | 57.00 | Mm00437761_g1 | |
| Bai1 | 107 831 | Brain-specific angiogenesis inhibitor 1 (NM_174991) | 2.52 | Mm00558144_m1 | |
| Bdrkb1 | 12 061 | Bradykinin receptor β1 (NM_007539) | 27.79 | 1.60 ± 0.43 p = 0.0498 | Mm00432059_s1 |
| Crhr2 | 12 922 | Corticotropin releasing hormone receptor 2 (NM_007539) | 5.110 | 5.89 ± 0.43 p = 0.0040 | Mm00438303_m1 |
| Drd4 | 13 491 | Dopamine receptor 4 (NM_007878) | 4.580 | Mm004382893_m1 | |
| Gabrd | 14 403 | GABAA receptor subunit delta (NM_008072) | 2.72 | Mm00433476_m1 | |
| Gabrr1 | 14 408 | GABAC receptor, subunit rho (NM_008075) | 4.123 | Mm00433499_m1 | |
| Gcat | 26 912 | Glycine C-acetyltransferase (2-amino-3 ketobutyrate-coenzyme A ligase) (NM_013847) | 25.45 | Mm00496962_m1 | |
| Glul | 14 645 | glutamate-ammonia ligase (glutamine synthetase) (NM_008131) | 4.56 | Mm00725701_s1 | |
| Grik1 | 14 805 | Glutamate receptor, ionotropic, kainate 2 (NM_010348) | 12.130 | Mm00446882_m1 | |
| Grik5 | 14 809 | Glutamate receptor, ionotropic, kainate 5 (γ2) (NM_008168) | 9.95 | Mm00435701_m1 | |
| Grin2b | 14 812 | Glutamate receptor, ionotropic, NMDA2B (ε2) (NM_008171) | 2.77 | 1.10 ± 0.13 p = 0.0062 | Mm00433820_m1 |
| Grin2d | 14 814 | Glutamate receptor, ionotropic, NMDA2D (ε4) (NM_008171) | 7.168 | Mm00433822_m1 | |
| Grm1 | 14 816 | Glutamate receptor metabotropic 1 (NM_016976) | 33.64 | Mm00810231_s1 | |
| Grm5 | 108 071 | Glutamate receptor metabotropic 5 (NM_001081414) | 6.110 | Mm00690332_m1 | |
| Nrxn2 | 18 190 | Neurexin II (NM_020253) | 7.131 | 1.28 ± 0.18 p < 0.0328 | Mm01236851_m1 |
| P2rx2 | 231 602 | Purinergic receptor P2X, ligand-gated ion channel, 2 (NM_153400) | 11.180 | Mm00462952_m1 | |
GABA, γ-aminobutyric acid; NMDA, N-Methyl-d-aspartate.
The table shows the comparison of gene expression data from microarray and real-time polymerase chain reaction (PCR) studies. The detected changes in the expression level are expressed in fold change (FC) in the microarray study and relative quantity (RQ) in the PCR study. Among the representative list of 60 genes of interest, 39 were down-regulated (a) and 21 were up regulated (b) exclusively by P2rx7 deletion in the mouse amygdala using the selection filters of >2.0-FC in the microarray study. Significant differences were calculated by a two-way analysis of variance (ANOVA) followed by Benjamini–Hochberg's multiple correction, with p < 0.05. Significant down- or up-regulatory effects of P2rx7 deficiency were confirmed for 29 of the genes by TaqMan-based quantitative real-time PCR (TLDA). Samples of RNA from the microarray experiment (n = 4) and from an independent experiment (n = 4) were measured for each group on TLDA and the results were evaluated by RealTime StatMiner software. (a) Twenty-five genes among the total of 39 were changed significantly (two-way ANOVA, p < 0.05) and showed similarity in size and direction by both methods. (b) Four out of 21 genes were changed significantly (two-way ANOVA, p < 0.05) and showed similarity in size and direction by both methods. The TaqMan ID gives the product number of Applied Biosystems Gene Expression Assay used for the validation of the corresponding gene.
Table 2.
Genes that were up-regulated by lipopolysaccharide (LPS) treatment in the mouse amygdala
| Symbol | Gene ID | Gene name (accession number) | Agilent (FC) | Real-time PCR (RQ) | TaqMan ID |
|---|---|---|---|---|---|
| Ifit1 | 15 957 | Interferon-induced protein with tetratricopeptide repeats 1 (NM_008331) | >1000 | 25.77 p < 0.0001 |
Mm00515153_m1 |
| Casp4 | 12 363 | Caspase 4, apoptosis-related cysteine peptidase (NM_007609) | >1000 | 19.24 p = 0.0007 |
Mm00432307_m1 |
| Ccl5 | 20 340 | Chemokine (C—C motif) ligand 5 (NM_013653) | >1000 | 745.12 p = 0.0002 |
Mm01302428_m1 |
| Ccl9 | 20 308 | Chemokine (C—C motif) ligand 9 (NM_011338) | >1000 | 20.22 p = 0.0001 |
Mm00441260_m1 |
| Cxcl10 | 15 945 | Chemokine (C—X—C motif) ligand 10 (NM_021274) | >1000 | 518.39 p = 0.0002 |
Mm99999072_m1 |
| Cxcl16 | 66 102 | Chemokine (C—X—C motif) ligand 16 (NM_023158) | 55.71 | 7.94 p = 0.0007 |
Mm00469712_m1 |
| Gbp2 | 14 469 | Guanylate nucleotide binding protein 2 (NM_010260) | >1000 | 68.13 p = 0.0001 |
Mm00494575_m1 |
| Gpr65 | 14 744 | G-protein coupled receptor 65 (NM_008152) | >1000 | 10.44 p = 0.0021 |
Mm02619732_s1 |
| Igtp | 16 145 | Interferon γ induced GTPase (NM_018738) | >1000 | 10.43 p = 0.0002 |
Mm00497611_m1 |
| Iigp1 | 60 440 | Interferon inducible GTPase 1 (NM_021792) | >1000 | 46.86 p = 0.0001 |
Mm00649928_s1 |
| Il1rn | 16 181 | Interleukin 1 receptor antagonist (NM_001039701) | >1000 | >1000 p = 0.0001 |
Mm01337566_m1 |
| Il4ra | 16 190 | Interleukin 4 receptor, α (NM_001008700) | 23.58 | 6.81 p < 0.0001 |
Mm00439634_m1 |
| Irgm | 15 944 | Immunity-related GTPase family (NM_008326) | >1000 | 21.93 p = 0.0024 |
Mm00492596_m1 |
| Isg20 | 57 444 | Interferon- stimulated protein (NM_020583) | >1000 | 12.29 = 0.0001 |
Mm00469585_m1 |
| Mpa2l | 100 702 | Macrophage activation 2 like (NM_194336) | >1000 | 61.41 p = 0.0001 |
Mm00843395_m1 |
| Saa1 | 20 208 | Serum amyloid A 1 (NM_009117) | >1000 | 34.91 p = 0.0026 |
Mm00656927_g1 |
| Tlr2 | 24 088 | Toll like receptor 2 (NM_0119057) | >1000 | 36.57 p = 0.0004 |
Mm00442346_m1 |
The table shows the comparison of gene expression data from microarray and real-time polymerase chain reaction (PCR) studies. Seventeen genes were selected among the exclusively up-regulated genes by systemic LPS (250 μg/kg i.p.) in the mouse amygdala (P2rx7+/+ and P2rx7−/−) using the selection filters of >2.0-fold change (FC) in the microarray study. Significant differences were calculated by two-way analysis of variance (ANOVA) followed by Benjamini–Hochberg's multiple correction, with p < 0.05. These changes were validated by TaqMan-based quantitative real-time PCR (TLDA). The detected changes in the expression level are expressed in FC in the microarray study and relative quantity (RQ) in the PCR study. The change of all 17 genes was statistically significant (two-way ANOVA, p < 0.05) and showed similarity in direction by both methods. Samples of RNA from the microarray experiment (n = 4) and from an independent experiment (n = 4) were measured for each group on TLDA and the results were evaluated by RealTime StatMiner software. The TaqMan ID gives the product number of Applied Biosystems Gene Expression Assay used for the validation of the corresponding gene.
Microarray experiments were validated by assessing the expression of genes of interest using quantitative TLDA real-time PCR, applying both sample and treatment validation. Significant down- or up-regulatory effects of P2rx7 deficiency were confirmed for 29 genes (Table 1 a, b), which corresponded to 48% of the total genes tested by real-time PCR. Among these genes, 25 genes were down-regulated in P2rx7−/− mice, including genes encoding various subunits of the GABAA receptor (Gabrb2, Gabrb3, Gabrg2, Gabrg3), ionotropic glutamate receptors (AMPA2, AMPA4), the metabotropic glutamate receptor 7 (Grm7), the α2 adrenergic receptor (Adra2a), the CB1 cannabinoid receptor (Cnr1), the D2 dopamine receptor (Drd2), dopa decarboxylase (Ddc) and subunits of the glycine receptor (Glra2, Glrb; Table 1 a). In contrast, four validated genes of the 29, including β bradykinin receptor (Bdkrb1) and the NMDA2B ionotropic glutamate receptor (Grin2b), were up-regulated (Table 1 b). All genes selected for the validation of endotoxin treatment were significantly up-regulated in the real-time PCR assay (Table 2).
Discussion
According to the current view, major depressive disorder and bipolar disorder are caused by plastic alterations of distributed networks involving multiple neurotransmitters and pathways (Stone et al. 2008). There is still much controversy concerning the changes in the brain that underlie the therapeutic actions of antidepressants and mood stabilizers. Moreover, despite the emerging knowledge of the pathophysiology of mood disorders, a relatively high proportion of patients do not respond to existing medications (Martinowich et al. 2009; Sanacora et al. 2008), which urges the discovery and validation of new potential therapeutic targets. One such target could be the P2rx7s. Therefore, we tested this hypothesis by the evaluation of the genetic deletion and pharmacological antagonism of P2rx7s in several behavioural paradigms used for testing antidepressants and mood-stabilizing drugs. P2rx7−/− mice did not develop behavioural despair in the FST and reduced immobility was detected in the TST. In addition, we report here for the first time an attenuated response of P2rx7−/− mice in the AH test, a widely used model of the manic pole of bipolar disorder. Because there was no difference between the two genotypes in the OF and EPM tests, the observed changes in behaviour could not be accounted for by changes in spontaneous locomotor activity or anxiety. These observations confirm and extend previous data, which showed decreased immobility in the TST (Basso et al. 2009) and decreased floating time in the FST on the second day (Boucher et al. 2011) after genetic deletion. However, partly differing from the study of Basso et al., in our experiments there was no significant difference in the basal immobility in the FST between the two genotypes, which might be explained by different housing conditions and purely homozygous mouse strain of a different origin used by the above study.
Changes in behaviour were accompanied by corresponding alterations in brain monoamine levels in the amygdala and the striatum of P2rx7−/− mice. In line with the decreased behavioural despair in the FST and decreased immobility in the TST, an increase in basal norepinephrine level was found in the amygdala, which could be reproduced by P2rx7 antagonist treatment. As a neurochemical correlate of decreased hyperlocomotion, amphetamine-induced elevation of dopamine content and release in the striatum were also alleviated in P2rx7−/− mice.
P2rx7−/− mice responded with a decreased elevation of plasma ACTH and corticosterone in response to restraint stress, which suggests that P2rx7−/− mice react with an attenuated response to external stress, the opposite of what is found in depressive patients. Functional P2rx7s are expressed on pituitary cells (Chung et al. 2000), which, together with our in vitro results, suggests a direct ATP effect on ACTH secretion. Nevertheless, an indirect action through other neuronal pathways converging on the hypothalamic–pituitary–adrenal axis cannot be excluded either. Interestingly, norepinephrine, dopamine and 5-HT levels were higher in the amygdala of P2rx7−/− mice after restraint, which indicates an interaction between stress and the brain monoaminergic system, which is regulated by P2rx7.
Importantly, we could reproduce the effect of genetic deletion by the systemic application of a specific antagonist of P2rx7s. BBG is a non-toxic and fairly selective antagonist of P2rx7s, which is able to penetrate the blood–brain barrier (Peng et al. 2009). Although it was active only using a sub-acute application, BBG dose-dependently decreased immobility in the TST and attenuated hyperactivity elicited by amphetamine in P2rx7+/+ mice. Consistent with the involvement of P2rx7 in these effects, an identical BBG treatment was ineffective in both tests in P2rx7−/− mice.
Recent studies have shown that certain behavioural patterns, such as pathological grooming, can be transferred from one animal to another by immune cell transplantation (Chen et al. 2010). Circulating cytokines, including IL-1β, are important mediators of depressive-like behaviour (Dantzer et al. 2008) and, therefore, peripheral IL-1β could be a mediator of P2rx7 activation on mood-related changes. In bone marrow chimera mice transplanted with the P2rx7−/− bone marrow, the changes in behaviour in the TST and AH could not be reproduced despite the >99% reconstitution of donor cells among recipient leukocytes. These data suggest that the lack of P2rx7 expressed on peripheral immune cells is not responsible for the behavioural phenotype found in P2rx7−/− mice. The majority of brain microglial cells derived from P2rx7−/−/CD45.1 mice were negative for the P2rx7, which implies that the observed changes of mood-related behaviour were also independent from microglial P2rx7s that might regulate IL-1β production (Sperlagh & Illes, 2007b). Moreover, the basal IL-1β level was not subject to regulation by P2rx7, either in the amygdala (present study) or in the serum and hippocampus (Csölle & Sperlágh, 2010), which is against the role of IL-1β as a mediator of behavioural changes shown here.
Consequently, P2rx7s expressed on other cell types, most likely on neurons or astrocytes, are responsible for P2rx7-related changes in mood. Our findings also indicated P2rx7-dependent changes in monoamine levels in the amygdala, which is an intrinsic part of the neuronal circuitry regulating emotions and the site of action of antidepressants (Castro et al. 2010). Moreover, a recent study (Boucher et al. 2011) showed that c-Fos expression induced by a repeated FST in the amygdala was attenuated by the genetic deletion of P2rx7. Therefore, to gain insight into the impact of P2rx7s on gene expression in this region, a whole genome microarray analysis was performed. Here we should note that the amygdala is a heterogeneous brain nucleus, comprising anatomically and functionally distinct subnuclei. Among them, the basolateral amygdala has primarily been associated with emotional processing, whereas our analyses were performed on the whole amygdala. Our data show that the expression of a surprisingly high number of genes (>8000) was changed significantly by genetic deletion of P2rx7, which is in line with the results of Boucher et al. (2011), who found a relatively selective decrease of c-Fos activation in the basolateral amygdala of P2rx7−/− mice after repeated FST. Twenty-nine out of the 60 genes selected for validation were significantly down- or up-regulated in the real-time PCR assay. The majority of these genes have biological plausibility in synaptic signalling and neuroplasticity and could be linked to the pathogenesis of depression. Interestingly, a down-regulation of the dopa decarboxylase enzyme was found in the absence of P2rx7, which indicates that elevated norepinephrine levels in the amygdala are not due to increased synthesis but decreased metabolism or utilization. In addition, D2 receptors were also down-regulated in the amygdala of P2rx7−/− mice. Consistent with the presumed dysfunction of glutamatergic transmission and consequent neuroplasticity changes in depression (Sanacora, 2010; Sanacora et al. 2008), a down-regulation of different α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) and metabotropic glutamate receptor subunits (Gria 2, Gria 4, Grm 7) and the glial cell-derived neurotrophic factor was detected in P2rx7−/− mice. In contrast, an up-regulation of the NR2B subunit of N-methyl-d-aspartate receptors (Grin2b) and CRHR2 receptors (crhr2) were found in P2rx7−/− mice. These changes in gene expression profile might also underlie the behavioural alterations and decreased stress reactivity found in the absence of P2rx7. Genes related to GABAergic transmission were also affected by genetic deletion, causing a subtype-specific up- and down-regulation of various GABAA receptor subunits and the down-regulation of the GABA synthesizing enzyme, glutamic acid decarboxylase. These changes are consistent with previous results showing that the activation of P2rx7s in the brain leads to an increased glutamate and GABA release from nerve terminals (Papp et al. 2004; Patti et al. 2006; Sperlagh et al. 2002) and astrocytes (Duan et al. 2003). Therefore, it is tempting to speculate that the increased activation due to gain-of-function mutations of the P2RX7 gene (Stokes et al. 2010) may lead to glutamatergic over-activation. This over-activation would lead to depressive symptoms, whereas the lack of these effects in the absence of the P2rx7 results in compensatory changes in the expression of glutamate and GABA receptors. This assumption is strengthened by observations from animal and human studies, indicating that excessive glutamate transmission might be involved in the pathophysiology of depressive disorders. These include abnormalities in the level of glutamate (Hashimoto et al. 2007), genetic polymorphisms (Mundo et al. 2003), the differential expression of glutamate receptor subunits in bipolar and major depressive disorders (Meador-Woodruff et al. 2001; Toro & Deakin, 2005), the antidepressant properties of glutamate receptor antagonists and modulators (Sanacora et al. 2008) and the regulation of glutamate receptor subunits by clinically used antidepressants (Nowak et al. 1998; Skolnick et al. 1996).
As a control stimulus, gene expression changes were also examined after a relatively modest LPS stimulus. This treatment elicited the significant change of 570 transcripts with >2FC, which is comparable to the gene expression changes found by others in the brain after systemic endotoxin treatment (Marques et al. 2009; Marsh et al. 2009; Mastronardi et al. 2007; Perreau et al. 2007). Therefore, genes encoding chemokines and other signalling proteins of immune response were primarily up-regulated by the endotoxin.
In conclusion, our data show that the genetic deletion and pharmacological antagonism of P2rx7s leads to selective alteration in mood-related behaviour and related changes in brain monoamine levels, stress reactivity and gene expression. Although the precise translation of P2rx7 function to behavioural changes needs further investigation, our data may open up new avenues in the treatment of mood disorders.
Supplementary Material
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Acknowledgements
This study was supported by grants from the Hungarian Research and Development Fund (Grants 046785 to J. H., 048783 to D. Z. and NN79957 to B. S.), the Hungarian Medical Research Council (ETT 05-102 to B.S.), the Pfizer Hungary Partnership programme (to B.S.), the Wellcome Trust (087782 to A. M.) and the Richter Gedeon Plc. (KK/1135/2007 to B. S.). The authors are grateful to István Likó for participation in Taqman array experiments. The manuscript text was edited by the Elsevier Language Editing Service. The authors are entirely responsible for the scientific content of the paper.
Note
Supplementary material accompanies this paper on the Journal's website (http://journals.cambridge.org/pnp).
Statement of Interest
None.
References
- Alloisio S, Cervetto C, Passalacqua M, Barbieri R, et al. (2008). Functional evidence for presynaptic P2X7 receptors in adult rat cerebrocortical nerve terminals. FEBS Letters 582, 3948–3953 [DOI] [PubMed] [Google Scholar]
- Anderson CM, Nedergaard M (2006). Emerging challenges of assigning P2X7 receptor function and immunoreactivity in neurons. Trends in Neurosciences 29, 257–262 [DOI] [PubMed] [Google Scholar]
- Baranyi M, Milusheva E, Vizi ES, Sperlagh B (2006). Chromatographic analysis of dopamine metabolism in a Parkinsonian model. Journal of Chromatography A 1120, 13–20 [DOI] [PubMed] [Google Scholar]
- Barden N, Harvey M, Gagne B, Shink E, et al. (2006). Analysis of single nucleotide polymorphisms in genes in the chromosome 12Q24.31 region points to P2RX7 as a susceptibility gene to bipolar affective disorder. American Journal of Medical Genetics Part B 141, 374–382 [DOI] [PubMed] [Google Scholar]
- Basso AM, Bratcher NA, Harris RR, Jarvis MF, et al. (2009). Behavioral profile of P2X7 receptor knockout mice in animal models of depression and anxiety: relevance for neuropsychiatric disorders. Behavioral Brain Research 198, 83–90 [DOI] [PubMed] [Google Scholar]
- Boucher AA, Arnold JC, Hunt GE, Spiro A, et al. (2011). Resilience and reduced c-Fos expression in P2X7 receptor knockout mice exposed to repeated forced swim test. Neuroscience 189, 170–177 [DOI] [PubMed] [Google Scholar]
- Burnstock G (2008). Purinergic signalling and disorders of the central nervous system. Nature Reviews Drug Discovery 7, 575–590 [DOI] [PubMed] [Google Scholar]
- Capanna E, Corti M, Mainardi D, Parmigiani S, et al. (1984). Karyotype and intermale aggression in wild house mice: ecology and speciation. Behavior Genetics 14, 195–208 [DOI] [PubMed] [Google Scholar]
- Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, et al. (2007). GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biology 8, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro JE, Varea E, Marquez C, Cordero MI, et al. (2010). Role of the amygdala in antidepressant effects on hippocampal cell proliferation and survival and on depression-like behavior in the rat. PLoS ONE 5, e8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Brosnan CF (2006). Regulation of immune response by P2X7 receptor. Critical Reviews in Immunology 26, 499–513 [DOI] [PubMed] [Google Scholar]
- Chen SK, Tvrdik P, Peden E, Cho S, et al. (2010). Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Park KS, Cha SK, Kong ID, et al. (2000). ATP-induced [Ca2+]i changes and depolarization in GH3 cells. British Journal of Pharmacology 130, 1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Mombereau C, Vassout A (2005). The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neuroscience and Biobehavioral Reviews 29, 571–625 [DOI] [PubMed] [Google Scholar]
- Csölle C, Heinrich A, Kittel A, Sperlágh B (2008). P2Y receptor mediated inhibitory modulation of noradrenaline release in response to electrical field stimulation and ischemic conditions in superfused rat hippocampus slices. Journal of Neurochemistry 106, 347–360 [DOI] [PubMed] [Google Scholar]
- Csölle C, Sperlágh B (2010). Peripheral origin of IL-1beta production in the rodent hippocampus under in vivo systemic bacterial lipopolysaccharide (LPS) challenge and its regulation by P2X(7) receptors. Journal of Neuroimmunology 219, 38–46 [DOI] [PubMed] [Google Scholar]
- Dantzer R, O'Connor JC, Freund GG, Johnson RW, et al. (2008). From inflammation to sickness and depression: when the immune system subjugates the brain. Nature Reviews Neuroscience 9, 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Virgilio F, Ceruti S, Bramanti P, Abbracchio MP (2009). Purinergic signalling in inflammation of the central nervous system. Trends in Neurosciences 32, 79–87 [DOI] [PubMed] [Google Scholar]
- Diaz-Hernandez M, Diez-Zaera M, Sanchez-Nogueiro J, Gomez-Villafuertes R, et al. (2009). Altered P2X7-receptor level and function in mouse models of Huntington's disease and therapeutic efficacy of antagonist administration. FASEB Journal 23, 1893–1906 [DOI] [PubMed] [Google Scholar]
- Duan S, Anderson CM, Keung EC, Chen Y, et al. (2003). P2X7 receptor-mediated release of excitatory amino acids from astrocytes. Journal of Neuroscience 23, 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einat H (2007). Establishment of a battery of simple models for facets of bipolar disorder: a practical approach to achieve increased validity, better screening and possible insights into endophenotypes of disease. Behavioral Genetics 37, 244–255 [DOI] [PubMed] [Google Scholar]
- Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, et al. (2006). The P2X7 receptor: a key player in IL-1 processing and release. Journal of Immunology (Baltimore, MD) 176, 3877–3883 [DOI] [PubMed] [Google Scholar]
- Ferrari PF, Palanza P, Parmigiani S, Rodgers RJ (1998). Interindividual variability in Swiss male mice: relationship between social factors, aggression, and anxiety. Physiology and Behavior 63, 821–827 [DOI] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Raybould R, Elvidge G, et al. (2009). P2RX7: a bipolar and unipolar disorder candidate susceptibility gene? American Journal of Medical Genetics 150B, 1063–1069 [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Herms S, Mühleisen TW, Georgi A, et al. (2010). Variation in P2RX7 candidate gene (rs2230912) is not associated with bipolar I disorder and unipolar major depression in four European samples. American Journal of Medical Genetics 150B, 1017–1021 [DOI] [PubMed] [Google Scholar]
- Harvey M, Belleau P, Barden N (2007). Gene interactions in depression: pathways out of darkness. Trends in Genetics 23, 547–556 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Sawa A, Iyo M (2007). Increased levels of glutamate in brains from patients with mood disorders. Biological Psychiatry 62, 1310–1316 [DOI] [PubMed] [Google Scholar]
- Hejjas K, Szekely A, Domotor E, Halmai Z, et al. (2009). Association between depression and the Gln460Arg polymorphism of P2RX7 gene: a dimensional approach. American Journal of Medical Genetics 150B, 295–299 [DOI] [PubMed] [Google Scholar]
- Hilakivi LA, Ota M, Lister RG (1989). Effect of isolation on brain monoamines and the behavior of mice in tests of exploration, locomotion, anxiety and behavioral ‘despair’. Pharmacology, Biochemistry and Behavior 33, 371–374 [DOI] [PubMed] [Google Scholar]
- Hogg S (1996). A review of the validity and variability of the elevated plus-maze as an animal model of anxiety. Pharmacology, Biochemistry and Behavior 54, 21–30 [DOI] [PubMed] [Google Scholar]
- Jakus Z, Simon E, Frommhold D, Sperandio M, et al. (2009). Critical role of phospholipase Cgamma2 in integrin and Fc receptor-mediated neutrophil functions and the effector phase of autoimmune arthritis. Journal of Experimental Medicine 206, 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MF, Khakh BS (2009). ATP-gated P2X cation-channels. Neuropharmacology 56, 208–215 [DOI] [PubMed] [Google Scholar]
- Lavebratt C, Aberg E, Sjöholm LK, Forsell Y (2010). Variations in FKBP5 and BDNF genes are suggestively associated with depression in a Swedish population-based cohort. Journal of Affective Disorders 125, 249–255 [DOI] [PubMed] [Google Scholar]
- Lolait SJ, Stewart LQ, Jessop DS, Young WS III, et al. (2007). The hypothalamic-pituitary-adrenal axis response to stress in mice lacking functional vasopressin V1b receptors. Endocrinology 148, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951). Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry 193, 265–275 [PubMed] [Google Scholar]
- Lucae S, Salyakina D, Barden N, Harvey M, et al. (2006). P2RX7, a gene coding for a purinergic ligand-gated ion channel, is associated with major depressive disorder. Human Molecular Genetics 15, 2438–2445 [DOI] [PubMed] [Google Scholar]
- McQuillin A, Bass NJ, Choudhury K, Puri V, et al. (2009). Case-control studies show that a non-conservative amino-acid change from a glutamine to arginine in the P2RX7 purinergic receptor protein is associated with both bipolar- and unipolar-affective disorders. Molecular Psychiatry 14, 614–620 [DOI] [PubMed] [Google Scholar]
- Marques F, Sousa JC, Coppola G, Falcao AM, et al. (2009). Kinetic profile of the transcriptome changes induced in the choroid plexus by peripheral inflammation. Journal of Cerebral Blood Flow and Metabolism 29, 921–932 [DOI] [PubMed] [Google Scholar]
- Marsh B, Stevens SL, Packard AE, Gopalan B, et al. (2009). Systemic lipopolysaccharide protects the brain from ischemic injury by reprogramming the response of the brain to stroke: a critical role for IRF3. Journal of Neuroscience 29, 9839–9849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Schloesser RJ, Manji HK (2009). Bipolar disorder: from genes to behavior pathways. Journal of Clinical Investigation 119, 726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi C, Whelan F, Yildiz OA, Hannestad J, et al. (2007). Caspase 1 deficiency reduces inflammation-induced brain transcription. Proceedings of the National Academy of Sciences USA 104, 7205–7210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorga AJ, Lucki I (2001). Limitations on the use of the C57BL/6 mouse in the tail suspension test. Psychopharmacology (Berlin) 155, 110–112 [DOI] [PubMed] [Google Scholar]
- Meador-Woodruff JH, Hogg AJ, Smith RE (2001). Striatal ionotropic glutamate receptor expression in schizophrenia, bipolar disorder, and major depressive disorder. Brain Research Bulletin 55, 631–640 [DOI] [PubMed] [Google Scholar]
- Milusheva E, Baranyi M, Kormos E, Hracskó Z, et al. (2010). The effect of antiparkinsonian drugs on oxidative stress induced pathological [3H]dopamine efflux after in vitro rotenone exposure in rat striatal slices. Neuropharmacology 58, 816–825 [DOI] [PubMed] [Google Scholar]
- Miras-Portugal MT, Diaz-Hernandez M, Giraldez L, Hervas C, et al. (2003). P2X7 receptors in rat brain: presence in synaptic terminals and granule cells. Neurochemical Research 28, 1597–1605 [DOI] [PubMed] [Google Scholar]
- Monif M, Reid CA, Powell KL, Smart ML, et al. (2009). The P2X7 receptor drives microglial activation and proliferation: a trophic role for P2X7R pore. Journal of Neuroscience 29, 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundo E, Tharmalingham S, Neves-Pereira M, Dalton EJ, et al. (2003). Evidence that the N-methyl-D-aspartate subunit 1 receptor gene (GRIN1) confers susceptibility to bipolar disorder. Molecular Psychiatry 8, 241–245 [DOI] [PubMed] [Google Scholar]
- Nagy G, Ronai Z, Somogyi A, Sasvari-Szekely M, et al. (2008). P2RX7 Gln460Arg polymorphism is associated with depression among diabetic patients. Progress in Neuro-Psychopharmacology and Biological Psychiatry 32, 1884–1888 [DOI] [PubMed] [Google Scholar]
- Nicke A, Kuan YH, Masin M, Rettinger J, et al. (2009). A functional P2X7 splice variant with an alternative transmembrane domain 1 escapes gene inactivation in P2X7 knock-out mice. Journal of Biological Chemistry 284, 25813–25822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, et al. (2009). GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Research 37, W317–W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak G, Legutko B, Skolnick P, Popik P (1998). Adaptation of cortical NMDA receptors by chronic treatment with specific serotonin reuptake inhibitors. European Journal of Pharmacology 342, 367–370 [DOI] [PubMed] [Google Scholar]
- Papp L, Vizi ES, Sperlagh B (2004). Lack of ATP-evoked GABA and glutamate release in the hippocampus of P2X7 receptor−/− mice. Neuroreport 15, 2387–2391 [DOI] [PubMed] [Google Scholar]
- Patti L, Raiteri L, Grilli M, Parodi M, et al. (2006). P2X(7) receptors exert a permissive role on the activation of release-enhancing presynaptic alpha7 nicotinic receptors co-existing on rat neocortex glutamatergic terminals. Neuropharmacology 50, 705–713 [DOI] [PubMed] [Google Scholar]
- Peng W, Cotrina ML, Han X, Yu H, et al. (2009). Systemic administration of an antagonist of the ATP-sensitive receptor P2X7 improves recovery after spinal cord injury. Proceedings of the National Academy of Sciences USA 106, 12489–12493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau VM, Bondy SC, Cotman CW, Sharman KG, et al. (2007). Melatonin treatment in old mice enables a more youthful response to LPS in the brain. Journal of Neuroimmunology 182, 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsolt RD, Bertin A, Jalfre M (1977). Behavioral despair in mice: a primary screening test for antidepressants. Archives Internationales de Pharmacodynamie et de Therapie 229, 327–336 [PubMed] [Google Scholar]
- Poshivalov VP (1980). The integrity of the social hierarchy in mice following administration of psychotropic drugs. British Journal of Pharmacology 70, 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz I, Nadal X, Alferink J, Banos JE, et al. (2008). Crucial role of CB(2) cannabinoid receptor in the regulation of central immune responses during neuropathic pain. Journal of Neuroscience 28, 12125–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger S, Mei ZZ, Baldwin JM, Dong L, et al. (2010). Single nucleotide polymorphisms that were identified in affective mood disorders affect ATP-activated P2X7 receptor functions. Journal of Psychiatric Research 44, 347–355 [DOI] [PubMed] [Google Scholar]
- Sanacora G (2010). Cortical inhibition, gamma-aminobutyric acid, and major depression: there is plenty of smoke but is there fire? Biological Psychiatry 67, 397–398 [DOI] [PubMed] [Google Scholar]
- Sanacora G, Zarate CA, Krystal JH, Manji HK (2008). Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nature Reviews Drug Discovery 7, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaper SD, Debetto P, Giusti P (2010). The P2X7 purinergic receptor: from physiology to neurological disorders. FASEB Journal 24, 337–345 [DOI] [PubMed] [Google Scholar]
- Skaper SD, Facci L, Culbert AA, Evans NA, et al. (2006). P2X(7) receptors on microglial cells mediate injury to cortical neurons in vitro. Glia 54, 234–242 [DOI] [PubMed] [Google Scholar]
- Skolnick P, Layer RT, Popik P, Nowak G, et al. (1996). Adaptation of N-methyl-D-aspartate (NMDA) receptors following antidepressant treatment: implications for the pharmacotherapy of depression. Pharmacopsychiatry 29, 23–26 [DOI] [PubMed] [Google Scholar]
- Sluyter R, Stokes L, Fuller SJ, Skarratt KK, et al. (2010). Functional significance of P2RX7 polymorphisms associated with affective mood disorders. Journal of Psychiatric Research 44, 1116–1117 [DOI] [PubMed] [Google Scholar]
- Solle M, Labasi J, Perregaux DG, Stam E, et al. (2001). Altered cytokine production in mice lacking P2X(7) receptors. Journal of Biological Chemistry 276, 125–132 [DOI] [PubMed] [Google Scholar]
- Soronen P, Mantere O, Melartin T, Suominen K, et al. (2011). P2RX7 gene is associated consistently with mood disorders and predicts clinical outcome in three clinical cohorts. American Journal of Medical Genetics Part B 156, 435–447 [DOI] [PubMed] [Google Scholar]
- Sperlagh B, Heinrich A, Csolle C (2007a). P2 receptor-mediated modulation of neurotransmitter release-an update. Purinergic Signalling 3, 269–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlagh B, Illes P (2007b). Purinergic modulation of microglial cell activation. Purinergic Signalling 3, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlagh B, Kofalvi A, Deuchars J, Atkinson L, et al. (2002). Involvement of P2X7 receptors in the regulation of neurotransmitter release in the rat hippocampus. Journal of Neurochemistry 81, 1196–1211 [DOI] [PubMed] [Google Scholar]
- Sperlagh B, Vizi ES, Wirkner K, Illes P (2006). P2X7 receptors in the nervous system. Progress in Neurobiology 78, 327–346 [DOI] [PubMed] [Google Scholar]
- Stokes L, Fuller SJ, Sluyter R, Skarratt KK, et al. (2010). Two haplotypes of the P2X7 receptor containing the Ala-348 to Thr polymorphism exhibit a gain-of-function effect and enhanced interleukin-1β secretion. FASEB Journal 24, 2916–2927 [DOI] [PubMed] [Google Scholar]
- Stone EA, Lin Y, Quartermain D (2008). A final common pathway for depression? Progress toward a general conceptual framework. Neuroscience and Biobehavioral Reviews 32, 508–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surprenant A, Rassendren F, Kawashima E, North RA, et al. (1996). The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7). Science 272, 735–738 [DOI] [PubMed] [Google Scholar]
- Toro C, Deakin JF (2005). NMDA receptor subunit NRI and postsynaptic protein PSD-95 in hippocampus and orbitofrontal cortex in schizophrenia and mood disorder. Schizophrenia Research 80, 323–330 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, de Preter K, Pattyn F, Poppe B, et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viikki M, Kampman O, Anttila S, Illi A, et al. (2011). P2RX7 polymorphisms Gln460Arg and His155Tyr are not associated with major depressive disorder or remission after SSRI or ECT. Neuroscience Letters 493, 127–130 [DOI] [PubMed] [Google Scholar]
- Zelena D, Domokos A, Barna I, Mergl Z, et al. (2008). Control of the hypothalamo-pituitary-adrenal axis in the neonatal period: adrenocorticotropin and corticosterone stress responses dissociate in vasopressin-deficient Brattleboro rats. Endocrinology 149, 2576–2583 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.