Abstract
The role of miRNAs in neuroectoderm specification is largely unknown. We screened miRNA profiles that are differentially changed when human embryonic stem cells (hESCs) were differentiated to neuroectodermal precursors (NEP), but not to epidermal (EPI) cells and found that two miRNA families, miR-200 and miR-96, were uniquely downregulated in the NEP cells. We confirmed zinc-finger E-box-binding homeobox (ZEB) transcription factors as a target of the miR-200 family members and identified paired box 6 (PAX6) transcription factor as the new target of miR-96 family members via gain- and loss-of-function analyses. Given the essential roles of ZEBs and PAX6 in neural induction, we propose a model by which miR-200 and miR-96 families coordinate to regulate neural induction.
Keywords: MicroRNA, Neural induction, Human embryonic stem cell, ZEB, PAX6
INTRODUCTION
MicroRNAs (miRNAs) play crucial roles in post-transcriptional gene regulation. The number of miRNAs expressed in the nervous system is larger than that in any other systems and accumulating data suggest roles of miRNAs in neurogenesis and gliogenesis (Fineberg et al., 2009; Shi et al., 2010). miR-9 and miR-124 regulate the transition from neural stem cells to differentiated neurons by silencing the essential regulators REST and TLX (Conaco et al., 2006; Cheng et al., 2009; Zhao et al., 2009). miR17-3p controls the patterning of ventral spinal progenitor domains through repression of the key patterning gene Olig2 (Chen et al., 2011). The miR-132 cluster participates in the cAMP response element-binding (CREB) transcriptional pathway to regulate the dendritic growth, synaptogenesis and synaptic activity (Vo et al., 2005; Magill et al., 2010). The miRNA-mediated regulation of glial differentiation, including that of astrocytes, is less well known (Zheng et al., 2012), although miR-219 is found to control oligodendrocyte maturation and myelination (Dugas et al., 2010; Zhao et al., 2010). However, the effect of miRNAs in the earliest step of neural development, neural induction, remains exclusive. This is complicated by the relative stability of existing miRNAs and lack of suitable drivers expressed in prospective neural precursors when performing the selective disruption of miRNAs in mouse studies (Davis et al., 2008).
Neural induction represents the earliest step in the determination of the ectoderm fate. In vertebrates, bone morphogenetic proteins (BMPs) act as signals for epidermal acquisition, and inhibition of the BMP signaling pathway in the ectoderm confers neural induction (Muñoz-Sanjuán and Brivanlou, 2002). Zinc finger E-box-binding homeobox (ZEB) transcription factor family represses the expression of BMP and its downstream genes, facilitating the induction of the neural fate (Postigo et al., 2003; Nitta et al., 2004). In mammals, embryonic stem cells (ESCs) offer a dynamic model to dissect the mechanism of neural induction (Zhang, 2006), including the role of miRNAs. The miR-302 family and miR-371 family members were reported to be repressed during neural differentiation from human ESCs (Rosa et al., 2009; Kim et al., 2011). However, these miRNAs are highly expressed in and are required for maintaining undifferentiated hESCs (Laurent et al., 2008). It is hence expected that these miRNAs are downregulated upon hESC differentiation, but the cause-effect relationship between the repression of these miRNAs and neural induction remains to be established.
We took a different approach to compare the miRNA profile during hESC differentiation to neuroectodermal (NEP) cells with that during hESC differentiation to epidermal (EPI) cells and identified two miRNA families, miR-200 and miR-96, that were specifically downregulated in NEP but not in EPI. Gain- and loss-of-function analyses indicated that miR-200 regulates the level of its target ZEB, whereas miR-96 regulates PAX6 (paired box 6), two key transcription factors essential for human neuroectoderm specification.
MATERIALS AND METHODS
Culture and differentiation of human ESCs
Human ESCs, H9 and H1 lines (WiCell Institute, Madison, WI, USA, NIH Code 0062 and 0043, passages 18-35) were cultured on irradiated mouse embryonic fibroblasts (MEFs) as described in the standard protocol http://www.wicell.org. A ZEB2-inducible ESC line was established by inserting a ZEB2-inducible expression cassette in the AAVS1 genomic locus with the TALEN technology as described previously (Hockemeyer et al., 2011). Neural differentiation and meso-endodermal differentiation of hESCs was performed according to published protocols (Zhang et al., 2001; Pankratz et al., 2007; Du et al., 2009). To induce the epidermal fate, 0.5 μM PD0325901 was added to the medium from day 4 to 10. The procedure for dual Nodal/BMP inhibition in monolayer culture was modified from the published protocol (Li et al., 2011) using the neural medium containing 0.5 μM LDN193189, 2 μM SB431542 and 3 μM CHIR99021 (all small molecules are from Stemgent, Cambridge, MA, USA).
miRNA extraction, miRNA profiling and qPCR
For miRNA profiling and qPCR, total RNA containing miRNA was extracted using a miRNeasy kit (Qiagen, Hilden, Germany). Total RNA and miRNA were reverse transcribed using the miScript reverse transcription kit (Qiagen), according to the manufacturer's instructions. miRNA profiling was performed using the Human miScript Primer Assay set V10.2 kit (Qiagen). Primers used for miRNAs were ordered from Qiagen and primers used for RNA qPCR are described in a previous publication (Zhang et al., 2010) or in the supplementary material Table S2. All the qPCR analysis was repeated in triplicates, and relative difference in gene expression was normalized to GAPDH.
Vector construction
Lentiviral-expressing vectors were modified from the plasmid pLVCT-tTRKRAB (Szulc et al., 2006, Addgene plasmid #11643, Addgene, Cambridge, MA, USA) by replacing the GFP-IRES-tTRKRAB fragment with GFP-IRES-Bsr or mcherry-T2A-Pur fragment. miR-200, miR-96 and miR-182 DNA fragments were cloned from BAC genomic clones (Life Technologies, Grand Island, NY, USA) using the primer sets (supplementary material Table S2) and then inserted into lentiviral-expressing vector before the WPRE element. The detailed protocol will be provided upon request.
Lentivirus production and transduction of hESCs
Lentivirus was produced and transduced as described previously (Du and Zhang, 2010). For transduction of ESCs, hESCs were pre-treated with ROCK inhibitor before they were trypsinized to single cells and collected by brief centrifugation. Cell pellets were then incubated with lentivirus at 37°C for 1 hour. The virus and cell mixture was then transferred to the MEF feeder layer overnight before changing medium on the next day. Forty-eight hours after infection, blasticidin or puromycin (both from InvivoGen, San Diego, CA, USA) was added to the cells for selecting drug-resistant clones. The final concentration of blasticidin or puromycin was 2 μg/ml or 0.5 μg/ml, respectively.
Western blotting
Cells were lysed with the lysis buffer [10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% NP-40 with 1× protease inhibitor from Roche, Indianapolis, IN, USA]. Proteins were quantitated with BCA assay (Bio-Rad, Hercules, CA, USA). Either 50 μg or 100 μg of protein lysates were separated by SDS-PAGE, transferred to a nitrocellulose membrane and probed with primary antibodies and IRDye secondary antibodies (1:10,000, Li-Cor Bioscience, Lincoln, NE, USA). The following primary antibodies were used: Zeb2 (rIgG, 1:1000, Sigma, St Louis, MO, USA), Sox2 (mIgG, 1:2000, R&D, Minneapolis, MN, USA), actin (mIgG, 1:2500, Sigma), Pax6 (rIgG, 1:800, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and Pou5f1 (mIgG, 1:2000, Santa Cruz Biotechnology). The membranes were scanned with Odyssey Infrared Imaging System (Li-Cor Biosciences) and quantitatively analyzed with Odyssey V3.0.
Immunostaining and microscopy
Immunohistochemical staining was performed according to Zhang et al. (Zhang et al., 2001). The following primary antibodies were used: Pax6 (mIgG, 1:5000, DSHB, Iowa City, IA, USA), keratin 18 (mIgG, 1:500, EMD Millipore, Billerica, MA, USA), Pou5f1 (mIgG, 1:1000, Santa Cruz Biotechnology), Sox17 (mIgG, 1:100, R&D), Brachyury T (gIgG, 1:50, R&D), Cdx2 (mIgG, 1:200, Biogenex, Fremont, CA, USA), Sox1 (gIgG, 1:1000, R&D).
Luciferase reporter assay
Targeting sites of miR-96-182 were analyzed by the TargetScan program (http://www.targetscan.org) (Lewis et al., 2005) and the mirWIP program (http://sfold.wadsworth.org). All miRNA reporter constructs were built on the pmirGLO dual-luciferase miRNA target expression vector (Promega, Madison, WI, USA). HEK293T cells were transfected 24 hour after seeding by using the calcium phosphate method. Luciferase activity was assayed 48 hours later using the Dual Luciferase Assay System (Promega) and normalized to the co-expressed Renilla Luciferase. All luciferase assays were repeated at least three times and performed in triplicates each time. Luciferase assays for target validation in epidermal cells were performed by transfecting reporter constructs with FugeneHD reagent (Roche).
Statistical analyses
For quantifications, experiments were performed at least in triplicate. Statistical significance was assessed using Student's t-test. P<0.05 was considered significant. Data were presented as mean±s.e.m.
RESULTS
Identification of miRNAs unique to neuroectoderm specification of hESCs
Under a chemically defined culture system, hESCs differentiate to enriched PAX6-expressing neuroepithelia (>90%) with remaining cells being other ectodermal cells (Zhang et al., 2001; Pankratz et al., 2007) (Fig. 1A,B). We have shown that PAX6-expressing neuroepithelia begin to appear after 4 days of hESC differentiation and reach a peak at day 10 (Pankratz et al., 2007). We therefore compared the miRNA expression profiles between day 4 and day 10 cells by miRNA qPCR arrays, which will probably exclude miRNAs that are involved in ESC self-renewal and initial spontaneous differentiation. More than 50 miRNAs were differentially expressed between these two cell populations, which include miRNAs involved in ESC differentiation to other cell lineages (supplementary material Table S1).
Fig. 1.
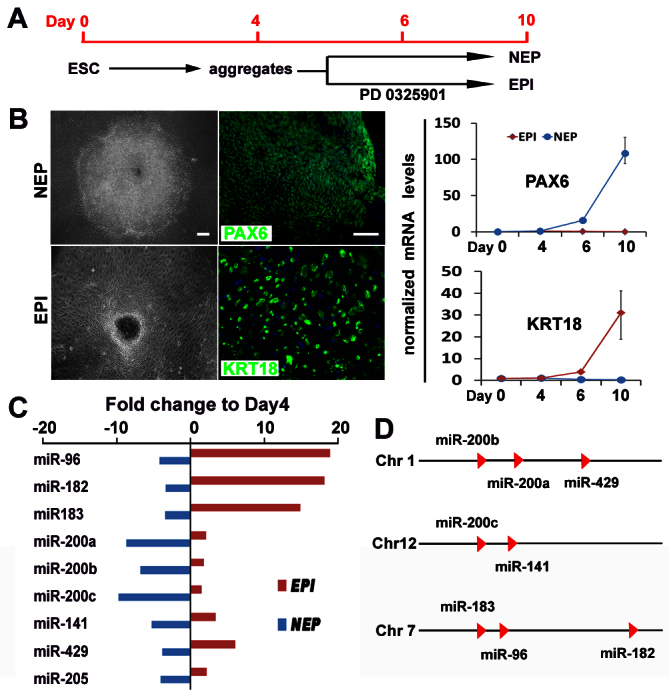
Identifying miRNAs specific to NEP differentiation from hESCs. (A) Time course and culture conditions for hESC differentiation into NEP or EPI cells. (B) Phase-contrast images (left column), expression of PAX6 and KRT18 by immunocytochemistry (middle column) and by qPCR analysis (right column) of NEP and EPI cells at day 10. Scale bars: 50 μm. The relative gene expression levels were normalized to day 4 cells. qPCR data are presented as mean±s.e.m. from triplicate samples (n=3). (C) Nine members of miR-200 and miR-96 families were differentially expressed in NEP and EPI cells. (D) The genomic location of the nine members of miR-200 and miR-96 families.
To further narrow down miRNAs that are specifically involved in NEP specification, we set up EPI as the second negative control group because of their close lineage relationship with the NEP. In our chemically defined condition, hESCs differentiate to primitive ectoderm cells that transiently express FGF5 at days 4-6 and these cells primarily differentiate to ectoderm cells (Pankratz et al., 2007). By treatment with an ERK inhibitor PD0325901 to block FGF signaling from day 4 to day 10, the hESCs were differentiated into EPI, as shown by the flat epithelial morphology and expression of epidermal progenitor marker keractin 18 (KRT18) (Fig. 1A,B). By comparing the miRNA profiles of day 10 NEP and EPI with day 4 cells, nine miRNAs were found to be downregulated by 4- to 12-fold in NEP, but were upregulated or not changed in EPI cells (Fig. 1C).
The differential expression pattern of these miRNAs was confirmed in another hESC line (H1 line), as well as under another neural induction method, the dual Nodal/BMP inhibition (Chambers et al., 2009; Li et al., 2011) in a monolayer culture (supplementary material Fig. S1). Among these miRNAs, miR-200a, miR-200b, miR-200c, miR-141, miR-429 belong to the miR-200 family, which are clustered on the chromosomes 1 and 12; miR-96, miR-182 and miR-183 are clustered at one locus of the chromosome 7 (Fig. 1D).
miR-200 family members repress neural differentiation by targeting ZEBs
The miR-200 family members target ZEB1 and ZEB2 (Gregory et al., 2008), and ZEB2 (also known as SMAD interacting protein 1, SIP1) is known to mediate cell fate decision between the neuroectoderm and meso-endoderm (Chng et al., 2010). qPCR analysis indicated that ZEB1 and ZEB2 were rarely expressed in hESCs and remained at a low level until day 4. During neural induction, both ZEB1 and ZEB2 were upregulated by 150- to 300-fold in day-10 NEP cells (Fig. 2A). By contrast, the expression of miR-200 family members was high in hESCs, halved on day 4, and was further downregulated by 10- to 20-fold to nearly loss of expression in day 10 NEP cells (Fig. 2A). The contrasting expression patterns suggest that ZEB transcription factors are targeted and repressed by miR-200 family members.
Fig. 2.
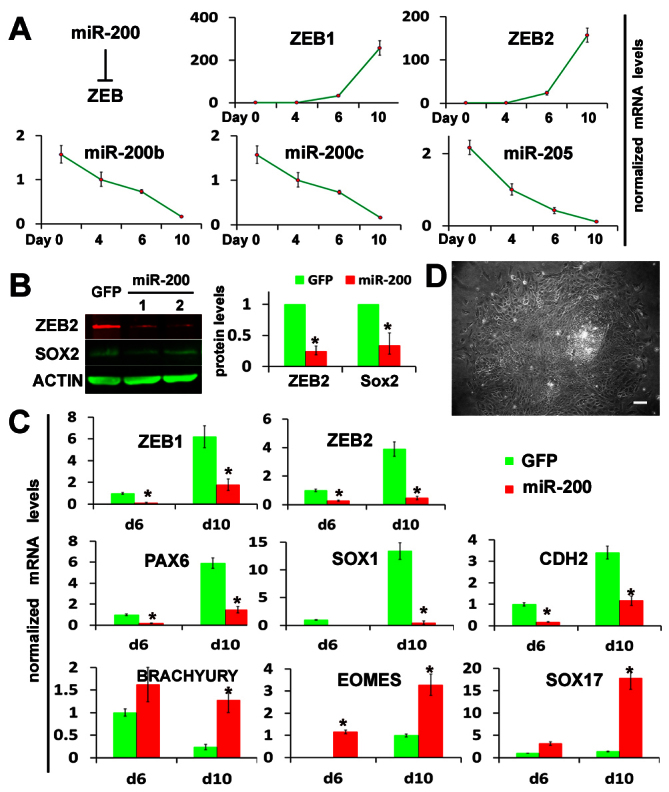
miR-200 affects ESC differentiation fate choice between neuroectoderm and meso-endoderm by regulating ZEBs. (A) Fold changes in expression of ZEBs and miR-200s during neuroepithelial specification from hESCs quantified by qPCR. Data are mean±s.e.m. (B) The protein levels of ZEB2 and SOX2 were quantified by western blot analysis at day 10 of differentiation of miR-200-overexpressing hESC lines. The GFP-hESC line (without miR-200 insertion) was used as a control. Quantification of protein content was normalized to actin content. (C) The expression of ZEB genes, neuroectodermal markers (PAX6, SOX1, CDH2) and meso-endodermal markers [brachyury (T), EOMES, SOX17] was quantified by qPCR during differentiation of miR-200-overexpressing and GFP control hESCs. The relative gene expression levels were normalized to day 6 GFP control cells. qPCR data are presented as mean±s.e.m. from triplicate samples (n=3, *P<0.05). (D) The phase-contrast image at day 10 differentiation of the miR-200-overexpressing hESC line, showing the reduced area of columnar NE cells. Scale bar: 50 μm.
To determine the effect of miR-200 family members on NEP specification, we stably expressed miR-200 family members in hESCs. A DNA fragment containing the miR-200b-200a-429 cluster sequence was placed after the GFP gene, which is driven by the CAG promoter in a lentiviral vector (supplementary material Fig. S2A). GFP was used to monitor the miRNA expression. The miR-200-hESC clones expressed 10 to 15-fold higher levels of miR-200b-200a-429 than the control GFP-hESCs (without miRNA insertion) at day 10 of differentiation, a level comparable with their expression in day 10 EPI (supplementary material Fig. S2A). Accompanying the overexpression of miR-200 was the downregulation of ZEBs, as indicated by western blotting and qPCR. The expression of ZEB2 protein showed about 75% reduction when compared with that in the control GFP-ESCs. Along with the changes in miR-200 and ZEBs, we observed a reduction in SOX2 protein (Fig. 2B) as well as mRNAs of other neuroectoderm genes PAX6, SOX1 and CDH2 (also known as N-cadherin) and upregulation of meso-endoderm genes brachyury (T), EOMES and SOX17 (Fig. 2C). At the cellular level, differentiating miR-200-hESCs presented a much reduced area of columnar NEP cells in the colony center but a much larger area of flat epithelial cells in the surrounding area, indicating the repression of neural induction (Fig. 2D). These results suggest that miR-200 family members affect hESC differentiation fate choices between the neuroectoderm and the meso-endoderm by regulating ZEB transcription factors.
miR-200 family members promote epidermal differentiation through regulating ZEB transcription factors and BMP signaling
ZEB transcription factors are crucial repressors of BMP signaling, and BMP signaling acts as an epidermal inducer during ectoderm development (Wilson and Hemmati-Brivanlou, 1995). We therefore asked whether miR-200 family members modulate the BMP signaling to promote epidermal differentiation. As overexpression of miR-200 family members at the hESC stage resulted in differentiation into the meso-endoderm fate, which prevents the delineation of neural versus epidermal ectoderm fate choice, we infected day 4 cell aggregates to overexpress miR-200 family members, right before the neural or epidermal fate is specified (Fig. 3A). This approach, however, prevents us from establishing a hESC line and the effect of miR-200 may only be assessed in the infected cells. Under our chemically defined neural differentiation condition, a few EPI cells, detected by KRT18 staining, were observed in the cells infected with GFP (no miRNA-200) (Fig. 3B), but most of cells were PAX6-expressing NEP (as shown above in Fig. 1B). Overexpression of miR-200 family members increased the production of KRT18+ EPI cells by sevenfold when compared with the control GFP group (Fig. 3B). To confirm that this effect is mediated through BMP signaling, we applied a chemical BMP inhibitor LDN193189 in the culture. LDN193189 completely blocked the production of KRT18+ EPI cells induced by overexpressing miR-200 (Fig. 3B), and promoted nearly all the cells to differentiate into NEP. Thus, miR-200 family members affect the differentiation fate choices between the NEP and the EPI via BMP signaling.
Fig. 3.
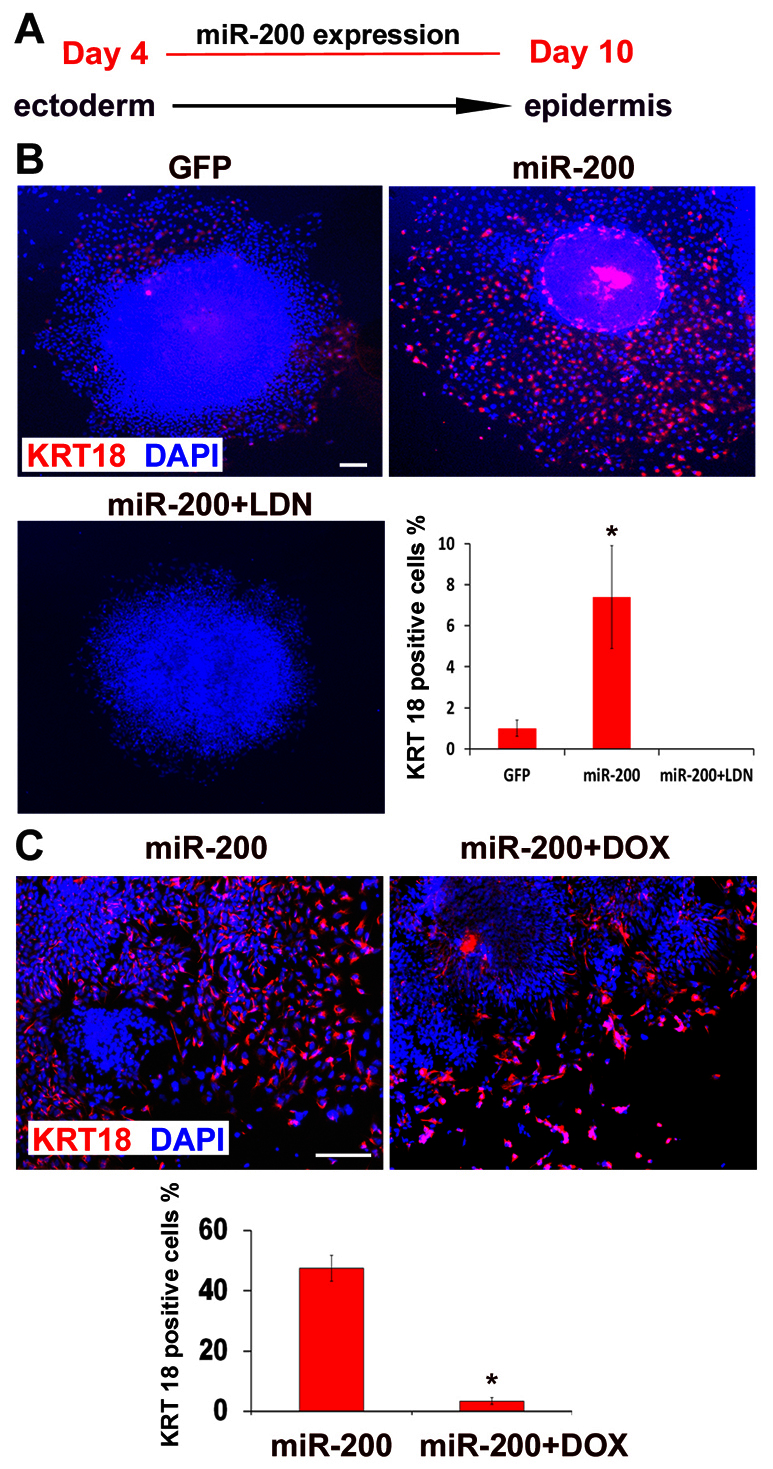
miR-200 promotes epidermal differentiation through BMP signaling. (A) Time course and condition for epidermal differentiation induced by overexpressing miR-200. (B) The expression and quantification of KRT18 (red) among total cells (DAPI stained, blue) under control (GFP), miR-200 overexpression (miR-200), and a combination of miR-200 overexpression and treatment with LDN (miR-200+LDN). Scale bar: 50 μm. Data are mean±s.e.m. *P<0.05. (C) The expression and quantification of KRT18 (red) among total cells (DAPI stained, blue) under the control (miR-200 overexpression) or the doxycycline-treated hESCs with inducible ZEB2 expression (miR-200 + DOX) at day 10. Scale bar: 50 μm. Data are mean±s.e.m. *P<0.05.
To further confirm that the effect of miR-200 on promoting the epidermal fate is mediated by repressing ZEB transcription factors, we established a hESC line with inducible ZEB2 expression by inserting the Tet-inducible expression cassette in the AAVS1 genomic locus. In this cell line, the transgene was homogeneously expressed in ESC-differentiated cells, including EPI cells upon induction by doxycycline, as indicated by GFP (supplementary material Fig. S2C). We then expressed miR-200 in the ZEB2-inducible hESCs. To increase the transfection efficiency, day 4 cell aggregates were dissociated and plated as a monolayer before they were transfected with the miR-200 lentivirus. These miR-200-expressing hESCs were divided into two groups and one of them was treated with doxycycline to induce ZEB2 expression. Without induction of ZEB2 (control group), miR-200 overexpression induced about 45% EPI cells at day 10, as detected by KRT18 staining. By contrast, induction of ZEB2 by doxycycline prevented the differentiation of KRT18+ EPI cells (to 4%) that were induced by miR-200 overexpression (Fig. 3C). This result confirms that miR-200 family members regulate the differentiation fate choices between the NEP and the EPI by regulating its target gene ZEBs.
miRNA-96 family members directly target the neural determinant gene PAX6
The miR-96-182-183 share a similar seed sequence (Fig. 4A) and are the most differentially expressed miRNAs between day 10 NEP and EPI cells. qPCR analysis confirmed that members of the miR-96 family exhibited the same expression pattern during hESC differentiation, i.e. a slight upregulation at day 4 followed by a fourfold downregulation in day 10 NEP cells but a 16-fold increase in day 10 EPI cells (Fig. 4B). This result suggests a crucial role for miR-96 family members in fate specification of neural versus epidermal ectoderm. By screening genes with sequences that match the seed sequence of miR-96 via the TargetScan program and comparing with the genes that are specifically expressed in day 10 NEP cells (Zhang et al., 2010), we identified a site at the 3′UTR 2783 bp of PAX6, the NEP determinant gene, to be a target of miR-96 and miR-182 (Fig. 4C, TS2 site). The 3′UTR typically contains several sites targeted by miRNAs, and some target sites do not perfectly match the miRNA seed sequence. We therefore applied the mirWIP program that predicts target sites without perfect match with miRNA seed sequences (Hammell et al., 2008) and identified another potential site for miR-96 and miR-182 at the 3′UTR 2425 bp (Fig. 4C, TS1 site). The nucleotide sequence of TS1 and TS2 sites are conserved during evolution.
Fig. 4.
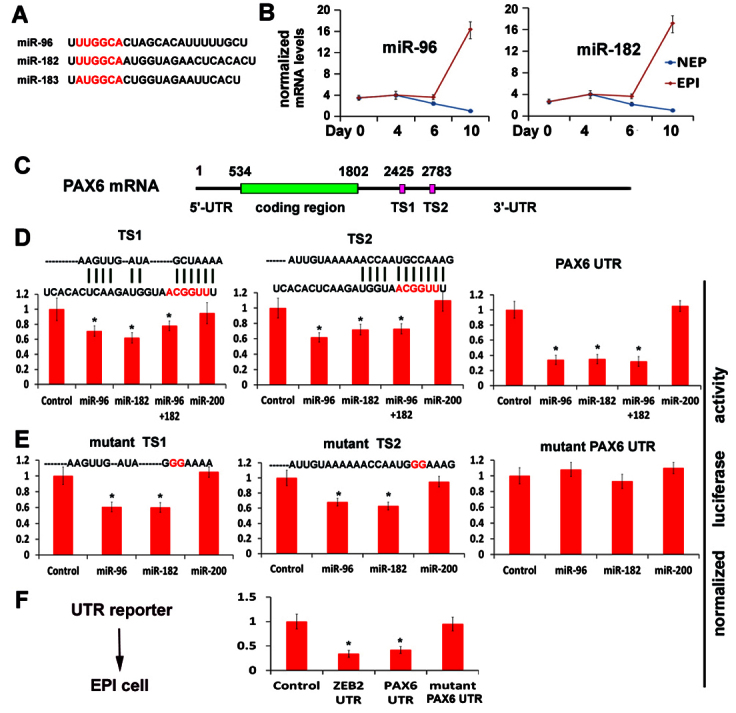
miR-96 directly targets the PAX6 gene. (A) The nucleotide sequences of miR-96 family members. The seed sequence is marked in red. (B) Fold changes in miR-96 and miR-182 expression during NEP and EPI differentiation from hESCs, quantified by qPCR. (C) The miR-96 targeting sites TS1 and TS2 are located in PAX6 3′UTR. (D) The luciferase reporter assays were performed on TS1 and TS2 sites, and the whole PAX6 UTR. The complementary sequences of TS1 or TS2 sites with miR-182 are shown. (E) The luciferase reporter analyses were performed on mutant TS1, mutant TS2 sites or mutant PAX6 UTR (both TS1 and TS2 mutated). The mutant nucleotides are shown in red. (F) The luciferase reporter analyses were performed on wild-type and mutant PAX6 UTR reporter in EPI cells, which endogenously express miR-96 and miR-200 family members. The reporter without the UTR is used as a negative control, and the reporter with ZEB2 UTR as a positive control. The results are presented as mean±s.e.m. of three experiments (n=3, *P<0.05, Student's t-test).
To investigate whether these two sites of PAX6 are targeted by miR-96 and miR-182, we constructed luciferase reporters that contain the TS1 site, the TS2 site, or a 2kb 3′UTR of the PAX6 gene that includes both the TS1 and TS2 sites. The luciferase reporters were co-transfected with a plasmid overexpressing miR-96 and/or miR-182 into 293T cells. A miR-200b-expressing plasmid was used as a negative control. As expected, miR-200 did not significantly affect the reporter activities. The miR-96 or miR-182 reduced the luciferase activities of TS1 and TS2 reporters by 30%, and reduced the luciferase activities of PAX6 UTR reporters by 70% (Fig. 4D). The co-transfection of both miR-96 and miR-182 did not result in further reduction, indicating that they target the same sites. To further confirm that these two sites are directly targeted, we made a mutation at the TS1 or TS2 site that is complementary to the seed sequence of miR-96-182. Mutation on both the TS1 and TS2 sites lost the repression of reporter activity induced by miR-96 or miR-182, but mutation on either the TS1 or TS2 site resulted in only minor reduction in the repression of reporter activity (Fig. 4E). Therefore, the miR-96-182 family targets the TS1 and TS2 sites of the PAX6 gene.
To verify the role of endogenous miR-96 family members in repressing the PAX6 gene, we transfected the day 10 EPI cells with the PAX6 UTR reporters. A luciferase reporter without the UTR was used as a negative control, and a luciferase reporter with the ZEB2 UTR was used as a positive control. At 48 hours after transfection, there was 60% reduction in the PAX6 and ZEB2 UTR reporter activities when compared with the non-UTR control (Fig. 4F). As an additional control, a reporter with a mutant PAX6 UTR (mutation on both TS1 and TS2 sites, shown above) was used. In this case, no change in reporter activity was observed (Fig. 4F). Taken together, these results indicate that miR-96 family members directly target the PAX6 gene.
Overexpression of miR-96 family members represses neural specification
As miR-96 represses PAX6 expression and PAX6 is a human neuroectoderm determinant (Zhang et al., 2010), we asked whether miR-96 family members inhibit neural induction from hESCs. Using lentiviral transfection, we selected hESC clones that expressed miR-96 or miR-182 around 15-fold higher than that in the control GFP-hESCs at day 10 of differentiation, a level comparable with their expression in day 10 EPI cells (supplementary material Fig. S2A). Overexpression of miR-96 or miR-182 in hESCs showed no effect on ESC self-renewal and proliferation. Under our neural differentiation conditions, miR-96 or miR-182-hESCs remained as round aggregates formed by stem-like cells, which was confirmed by positive staining for POU5F1, but not columnar NEP morphology (Fig. 5A). Consistent results were obtained with different miR-96 or miR-182-hESC lines, indicating that the phenotype was not due to different viral integration. Western blotting confirmed that the PAX6 expression was significantly decreased, whereas the expression of pluripotent marker POU5F1 was maintained at a high level at day 10 (Fig. 5B). qPCR analysis revealed a similar expression pattern for neuroectoderm genes (PAX6, SOX1 and CDH2) and pluripotent genes (POU5F1 and NANOG) (Fig. 5C). These phenotypes of miR-96 or miR-182-hESCs are reminiscent of those in PAX6 knockdowns (Zhang et al., 2010), in which the hESCs do not differentiate into the neuroectoderm.
Fig. 5.
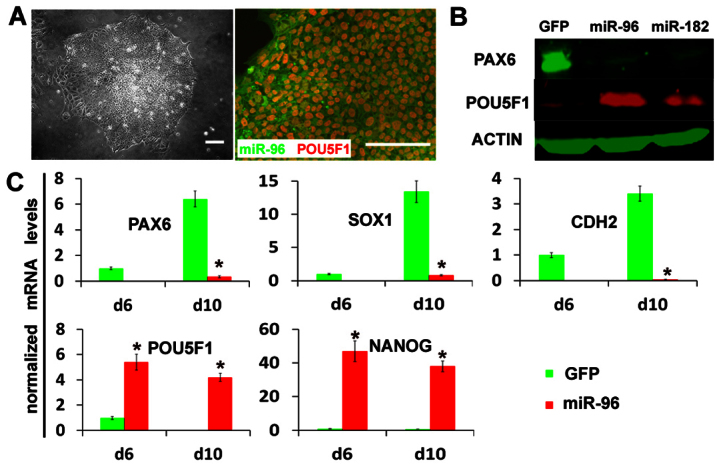
Overexpression of miR-96 family members represses neural specification. (A) hESCs with overexpression of miR-96 (indicated by GFP) exhibited a flat morphology, as shown by phase-contrast microscopy, and retained the expression of POU5F1 (red) after 10 days of neural differentiation. Scale bars: 50 μm. (B) The protein levels of PAX6 and POU5F1 were analyzed by western blotting at day 10 of differentiation of miR-96- or miR-182-overexpressing hESC lines. The GFP-hESC line (without miRNA insertion) was used as a control. (C) The fold changes in expression of neuroectodermal markers (PAX6, SOX1, CDH2) and pluripotent cell markers (POU5F1 and NANOG) were quantified by qPCR during differentiation of miR-96-overexpressing and GFP control hESCs. The relative gene expression levels were normalized to day 6 GFP control cells. qPCR data are presented as mean±s.e.m. from triplicate samples (n=3, *P<0.05).
To ascertain whether overexpression of miR-96-182 also interferes with the differentiation of hESCs into other cell lineages, we cultured the miR-96 or miR-182-hESCs in the meso-endoderm differentiation condition. The miR-96 or miR-182-hESCs differentiated into mesoderm and endoderm cell lineages at a similar efficiency to the control GFP cells, as evidenced by immunostaining for brachyury (T) and SOX17 (supplementary material Fig. S3). Therefore, overexpression of miR-96 family members prevents hESCs from differentiating to the NEP fate by repressing PAX6 expression without interfering with the meso-endoderm lineages.
Inhibition of miR-96 family members derepresses PAX6 expression to interfere with cell fates
Given the repressive role of miR-96-182 on PAX6 and neural induction from hESCs, we hypothesized that inhibition of miR-96-182 would promote hESC differentiation towards a neural fate. Using the miRNA sponge strategy, which contains multiple tandem binding sites to a miRNA of interest to compete with target genes for interacting with miRNA (Ebert et al., 2007), we constructed miR-96-182 sponge by inserting eight copies of the sponge sequence (Fig. 6A) into the 3′UTR of mcherry (red fluorescence reporter) gene in a lentiviral vector (supplementary material Fig. S2B). First, we intended to establish hESC lines that constitutively express the miR-96-182 sponge. Infection with miR-96-182 sponge lentivirus produced few ESC colonies, and these hESCs differentiated spontaneously (data not shown). This result indicates that inhibition of miR-96-182 drives differentiation and prevents hESC from self-renewal. Interestingly, miR-96-182 sponge hESCs did not form round aggregates and differentiate to columnar neuroepithelia under our neural differentiation condition. Instead, they formed irregular aggregates at day 4 and some aggregates possessed a cystic structure (Fig. 6B). When these cell aggregates were attached to laminin-coated coverslips, they formed large flat cells with trophoblast morphology at day 10. These cells were indeed positive for trophoblast marker CDX2 (Fig. 6B). qPCR analysis also showed upregulation of other extra-embryonic ectoderm markers GATA3 and SOX7, and downregulation of ESC pluripotent markers POU5F1 and NANOG (Fig. 6C).
Fig. 6.
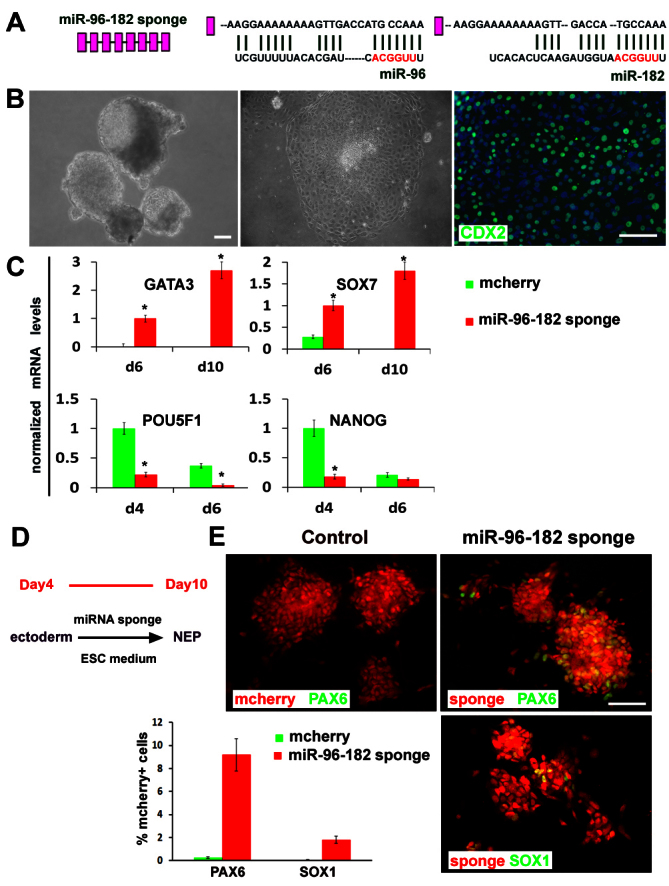
Inhibition of miR-96 family induces extra-embryonic differentiation at an early stage and promotes neural specification at a later stage. (A) Diagram of an eight-copy miR-96-182 sponge. The complementary nucleotide sequences of miR-96 or miR-182 with sponge sequence is shown. (B) The miR-96-182-sponge hESCs exhibit cystic structures at day 4 and flat trophoblast cells that are positively stained for CDX2 (green) at day 10 of differentiation. Scale bars: 50 μm. (C) The gene expression of trophoblast markers (GATA3 and SOX7) and pluripotent cell markers (POU5F1 and NANOG) was quantified by qPCR during differentiation of miR-96-182 sponge and mcherry control hESCs. The relative gene expression levels were normalized to day 6 miR-96-182 sponge cells for GATA3 and SOX7, and normalized to day 6 mcherry control cells for POU5F1 and NANOG. qPCR data are presented as mean±s.e.m. from triplicate samples (n=3, *P<0.05). (D) Treatment with miR-96-182 sponge at day 4-10 promotes neuroepithelial differentiation. (E) The neuroepithelial markers PAX6 and SOX1 (green) were stained and quantified in miR-96-182 sponge (red)-expressing cells. Scale bar: 50 μm. Data are mean±s.e.m.
Inhibition of miR-96-182 at the ESC stage possibly derepresses PAX6, which in turn represses pluripotent genes (Zhang et al., 2010), thus preventing us from studying its role in neuroectoderm specification. We therefore infected differentiating hESCs with miR-96-182 sponge lentivirus on day 4 with mcherry as an indicator of infected cells in the cell aggregates, and then cultured cells in the ESC growth medium that prevents neural differentiation (Fig. 6D). Under this condition, hardly any NEP cells were induced from the control mcherry-hESCs. By contrast, about 10% of the miR-96-182-sponge-expressing cells, as indicated by mcherry, were positive for PAX6, and about 2% were positive for SOX1, the neuroectodermal markers (Fig. 6E). Therefore, miR-96-182 sponge, by derepressing PAX6 expression, induces ESC differentiation into extra-embryonic lineage at an early stage and promotes neural specification at a later stage.
DISCUSSION
Dissection of the roles of miRNAs in early lineage choices requires distinguishing those for maintaining pluripotency and those for instructing a lineage fate. For this reason, we compared miRNA profiles during neuroepithelial differentiation with those during epidermal differentiation from day 4 to 10 to minimize miRNAs that are involved in pluripotency exit and spontaneous hESC differentiation. This strategy has allowed us to identify two miRNA families, miR-200 and miR-96, that are uniquely involved in neural induction from hESCs. We confirmed ZEB transcription factors as the known target of miR-200 family members, and identified PAX6 transcription factor as the new target of miR-96 family members. Not only does the expression pattern of miR-200 and miR-96 inversely correspond to that of ZEB and PAX6, transcription factors that are crucial for neural induction, but also do our genetic analyses confirm that miR-200 and miR-96 families target and regulate ZEBs and PAX6 genes, respectively, thus modulating neural induction (Fig. 7A).
Fig. 7.
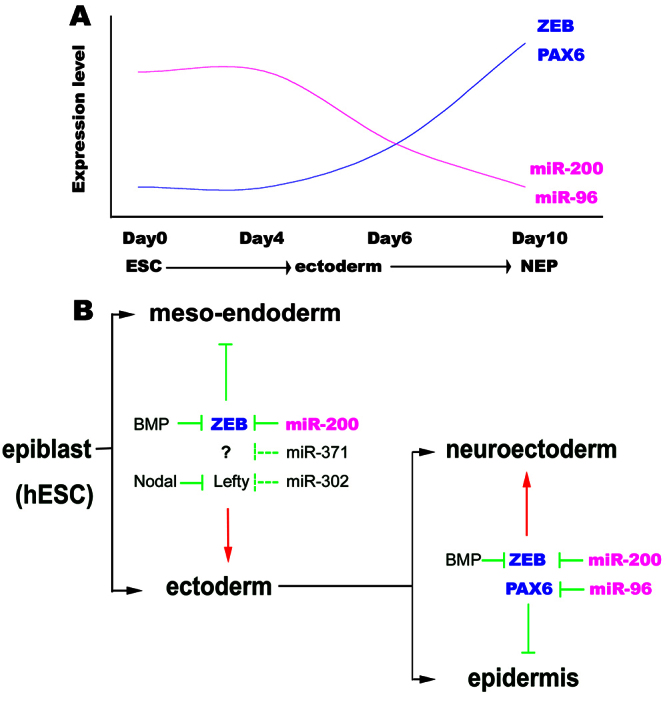
The miRNA network during neural induction from hESCs. (A) The expression change of miR-200 and miR-96 family members, and their target genes (ZEBs and PAX6) during neural induction from hESCs. (B) miR-200, miR-302 and miR-371 segregate the meso-endodermal fate from the ectodermal fate by regulating Nodal/BMP signaling in an early stage, and miR-200 and miR-96 segregate the neuroectodermal fate from the epidermal fate by regulating ZEBs and PAX6 factors at a later stage.
miRNAs are crucial for maintaining hESCs and they are largely downregulated upon differentiation (Laurent et al., 2008; Martinez and Gregory, 2010). A number of miRNAs were found to be downregulated during hESC differentiation. Similar to our identified miR-200 and miR-96 families, miR-302 and miR-371 families were downregulated during hESC differentiation to neuroepithelia, suggesting their potential roles in inhibiting neural fate specification from hESCs (Rosa et al., 2009; Kim et al., 2011). However, they are downregulated in not only NEP but also EPI cells (supplementary material Fig. S4; Table S1), suggesting that their roles are not necessarily specific to the neural fate. miR-302 targets Lefty factors, which are inhibitors of Activin/Nodal signaling, and the target of miR-371 is unknown. Activin/Nodal signaling can block the formation of neural tissues from animal cap cells and adopt a mesodermal fate, instead of the epidermal fate, but BMP signaling is a potent epidermal inducer and neural inhibitor (Wilson and Hemmati-Brivanlou, 1995). BMP inhibition is sufficient to induce the neural tissue within the ectoderm, but combined BMP and Nodal inhibition is required to induce neural tissue from the epiblast (Henry et al., 1996). Therefore, miR-302 and miR-371 families may play a role only in the early stage to segregate the meso-endodermal fate from the ectodermal fate (Fig. 7B). That explains why miR-302 and miR-371 are downregulated in both NEP and EPI.
miR-125 miRNAs are recently identified miRNA family that promotes neural specification from hESCs by inhibition of BMP/SMAD pathway (Boissart et al., 2012). However, the expression of miR-125a and miR-125b was upregulated 4- to 10-fold in both NEP and EPI cells when we compared with the day 4 differentiating hESCs (supplementary material Fig. S4; Table S1), suggesting that the role of the miR-125 family is not specific to neural specification. The upregulation of miR-125 family members is consistent with its role in the repression of self-renewal of ESCs (Rybak et al., 2008; O'Loghlen et al., 2012), and upregulation of miR-125 family members is indeed involved in the ESC differentiation into other cell linages, such as mesodermal cardiomyocytes (Wong et al., 2012). The robust induction of miR-125 is most likely due to the potent effect of small molecules of BMP inhibitors used in their study (Boissart et al., 2012). Whether the miR-125 family plays a role in NEP specification under normal development remains to be verified. Taken together, it is crucial to set up rigorous controls to segregate stem cell self-renewal from directed differentiation in order to identify miRNAs that are specific to lineage specification.
The miR-200 family was reported to regulate epithelial to mesenchymal transition by targeting ZEB transcription factors (Burk et al., 2008; Gregory et al., 2008). The miR-200 family inhibits expression of ZEBs at the post-transcriptional level by binding to highly conserved target sites in their 3′UTRs. Interestingly, ZEBs directly inhibit the transcription of miR-200 genes by binding to highly conserved sites in their common promoter (Burk et al., 2008). Thus, miR-200 members and ZEB factors reciprocally regulate each other in a negative-feedback loop. Consistent with their molecular interactions in other systems, we found that miR-200 targets and represses ZEBs during neuroepithelial specification from hESCs. ZEB factors are crucial regulators of TGFβ/BMP signaling through binding SMADs and the regulation of their downstream gene expression. ZEB2−/− or ZEB2−/−/ZEB1−/− embryos showed severe defects in neural plate/tube morphogenesis with a drastic reduction of neuroepithelial marker Sox2 (Miyoshi et al., 2006). Indeed, inhibition of BMP signaling by small molecules induces a quick increase in ZEB levels and decrease in miR-200 levels (supplementary material Fig. S1B), suggesting a direct role for the miR-200/ZEB network in inhibiting BMP signaling. This is further confirmed by the fact that the repression of neural induction by miR-200 is restored by simply expressing its target, ZEB (Fig. 3C). Thus, a new function of miR-200/ZEB network is to modulate neural induction.
The miR-96 family is expressed in retina, inner ear, nose, tongue and dorsal root ganglia, and is thus regarded as a sensory organ-specific paralogous miRNA cluster (Xu et al., 2007; Christodoulou et al., 2010). Consistent with its expression pattern, mutations in the seed sequence of human miR-96 are responsible for non-syndromic progressive hearing loss (Mencía et al., 2009) and miR-96-182 sponge transgenic mice display severe retinal degeneration after acute bright light exposure (Zhu et al., 2011). We found that the miR-96 family is perhaps the most differentially expressed miRNA between neuroectoderm and epidermis. The phenotypes of gain or loss of miR-96-182 function are strikingly similar to those of PAX6 knockdown or overexpression (Zhang et al., 2010). Importantly, overexpression of miR-96-182 results in repression of PAX6 and subsequent inhibition of neural induction from hESCs but without affecting the differentiation to other lineages, illustrating its specific effect on PAX6 and neuroectoderm specification (Fig. 7B). The phenotypes of miR-96-182 inhibition appear more complicated. Constitutive inhibition of miR-96-182 in hESCs results in the de-repression of PAX6, thus preventing the hESCs from self-renewal. Because both PAX6a and PAX6b (especially PAX6b, which is turned on earlier during differentiation) are significantly upregulated, which represses the expression of pluripotent genes, the outcome at an early stage is default differentiation to extra-embryonic trophectoderm. This is nearly identical to the phenotype of overexpression of PAX6b (Zhang et al., 2010). When the miR-96-182 sponge is expressed after day 4, during which the differentiating cells are biased towards the ectoderm fate, it promotes the neural fate. This phenomenon again illustrates the importance of delineating the stage-specific effects of miRNAs. Taken together, miR-96-182 targets and regulates PAX6, thus modulating neuroectoderm specification from hESCs.
Multiple pathways, including BMP-SMAD-ZEB (LaVaute et al., 2009; Postigo et al., 2003; Nitta et al., 2004) and FGF-ERK-PARP (Yoo et al., 2011), converge on neuroectoderm transcription factor PAX6 (Zhang et al., 2010) to regulate neuroectoderm specification from hESCs. In this study, we have revealed another level of the regulatory network by miRNAs. The miR-200 family, by targeting ZEBs, affects ESC differentiation choices between ectodermal fate and meso-endodermal fate at an early stage, but also distinguishes the neuroectodermal fate from the epidermal fate at a later stage. The miR-96 family, by targeting PAX6, may solidify the neuroectoderm fate at a later stage (Fig. 7B), as overexpression of miR-96 family does not interfere with the differentiation of hESCs into the meso-endodermal fate. The present finding adds complexity of the regulatory network but offers opportunities for modulating neural specification and/or reprogramming.
Supplementary Material
Acknowledgements
We thank Professor D. Trono for the plasmid pLVCT-tTRKRAB.
Footnotes
Funding
This study was supported by the National Institutes of Health (NIH)-National Institute of Neurological Disorders and Stroke (NINDS) [NS045926] and in part by a core grant to the Waisman Center from the National Institute of Child Health and Human Development [P30 HD03352]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.092809/-/DC1
References
- Boissart C., Nissan X., Giraud-Triboult K., Peschanski M., Benchoua A. (2012). miR-125 potentiates early neural specification of human embryonic stem cells. Development 139, 1247-1257 [DOI] [PubMed] [Google Scholar]
- Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., Studer L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. A., Huang Y. P., Mazzoni E. O., Tan G. C., Zavadil J., Wichterle H. (2011). Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron 69, 721-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng Z., Teo A., Pedersen R. A., Vallier L. (2010). SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell 6, 59-70 [DOI] [PubMed] [Google Scholar]
- Christodoulou F., Raible F., Tomer R., Simakov O., Trachana K., Klaus S., Snyman H., Hannon G. J., Bork P., Arendt D. (2010). Ancient animal microRNAs and the evolution of tissue identity. Nature 463, 1084-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J. J., Mandel G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. USA 103, 2422-2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. H., Cuellar T. L., Koch S. M., Barker A. J., Harfe B. D., McManus M. T., Ullian E. M. (2008). Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322-4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z. W., Zhang S. C. (2010). Lentiviral vector-mediated transgenesis in human embryonic stem cells. Methods Mol. Biol. 614, 127-134 [DOI] [PubMed] [Google Scholar]
- Du Z. W., Hu B. Y., Ayala M., Sauer B., Zhang S. C. (2009). Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells 27, 1032-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas J. C., Cuellar T. L., Scholze A., Ason B., Ibrahim A., Emery B., Zamanian J. L., Foo L. C., McManus M. T., Barres B. A. (2010). Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 65, 597-611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M. S., Neilson J. R., Sharp P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg S. K., Kosik K. S., Davidson B. L. (2009). MicroRNAs potentiate neural development. Neuron 64, 303-309 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593-601 [DOI] [PubMed] [Google Scholar]
- Hammell M., Long D., Zhang L., Lee A., Carmack C. S., Han M., Ding Y., Ambros V. (2008). mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods 5, 813-819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry G. L., Brivanlou I. H., Kessler D. S., Hemmati-Brivanlou A., Melton D. A. (1996). TGF-beta signals and a pattern in Xenopus laevis endodermal development. Development 122, 1007-1015 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Wang H., Kiani S., Lai C. S., Gao Q., Cassady J. P., Cost G. J., Zhang L., Santiago Y., Miller J. C., et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee G., Ganat Y., Papapetrou E. P., Lipchina I., Socci N. D., Sadelain M., Studer L. (2011). miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell 8, 695-706 [DOI] [PubMed] [Google Scholar]
- Laurent L. C., Chen J., Ulitsky I., Mueller F. J., Lu C., Shamir R., Fan J. B., Loring J. F. (2008). Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 26, 1506-1516 [DOI] [PubMed] [Google Scholar]
- LaVaute T. M., Yoo Y. D., Pankratz M. T., Weick J. P., Gerstner J. R., Zhang S. C. (2009). Regulation of neural specification from human embryonic stem cells by BMP and FGF. Stem Cells 27, 1741-1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15-20 [DOI] [PubMed] [Google Scholar]
- Li W., Sun W., Zhang Y., Wei W., Ambasudhan R., Xia P., Talantova M., Lin T., Kim J., Wang X., et al. (2011). Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl. Acad. Sci. USA 108, 8299-8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill S. T., Cambronne X. A., Luikart B. W., Lioy D. T., Leighton B. H., Westbrook G. L., Mandel G., Goodman R. H. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. USA 107, 20382-20387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. J., Gregory R. I. (2010). MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell 7, 31-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía A., Modamio-Høybjør S., Redshaw N., Morín M., Mayo-Merino F., Olavarrieta L., Aguirre L. A., del Castillo I., Steel K. P., Dalmay T., et al. (2009). Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 41, 609-613 [DOI] [PubMed] [Google Scholar]
- Miyoshi T., Maruhashi M., Van De Putte T., Kondoh H., Huylebroeck D., Higashi Y. (2006). Complementary expression pattern of Zfhx1 genes Sip1 and deltaEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev. Dyn. 235, 1941-1952 [DOI] [PubMed] [Google Scholar]
- Muñoz-Sanjuán I., Brivanlou A. H. (2002). Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3, 271-280 [DOI] [PubMed] [Google Scholar]
- Nitta K. R., Tanegashima K., Takahashi S., Asashima M. (2004). XSIP1 is essential for early neural gene expression and neural differentiation by suppression of BMP signaling. Dev. Biol. 275, 258-267 [DOI] [PubMed] [Google Scholar]
- O'Loghlen A., Muñoz-Cabello A. M., Gaspar-Maia A., Wu H. A., Banito A., Kunowska N., Racek T., Pemberton H. N., Beolchi P., Lavial F., et al. (2012). MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10, 33-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz M. T., Li X. J., Lavaute T. M., Lyons E. A., Chen X., Zhang S. C. (2007). Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells 25, 1511-1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A. A., Depp J. L., Taylor J. J., Kroll K. L. (2003). Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins. EMBO J. 22, 2453-2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Spagnoli F. M., Brivanlou A. H. (2009). The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517-527 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E. E., Nitsch R., Wulczyn F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987-993 [DOI] [PubMed] [Google Scholar]
- Shi Y. H., Zhao X. Y., Hsieh J., Wichterle H., Impey S., Banerjee S., Neveu P., Kosik K. S. (2010). MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 30, 14931-14936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc J., Wiznerowicz M., Sauvain M. O., Trono D., Aebischer P. (2006). A versatile tool for conditional gene expression and knockdown. Nat. Methods 3, 109-116 [DOI] [PubMed] [Google Scholar]
- Vo N., Klein M. E., Varlamova O., Keller D. M., Yamamoto T., Goodman R. H., Impey S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 102, 16426-16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P. A., Hemmati-Brivanlou A. (1995). Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 376, 331-333 [DOI] [PubMed] [Google Scholar]
- Wong S. S., Ritner C., Ramachandran S., Aurigui J., Pitt C., Chandra P., Ling V. B., Yabut O., Bernstein H. S. (2012). miR-125b promotes early germ layer specification through Lin28/let-7d and preferential differentiation of mesoderm in human embryonic stem cells. PLoS ONE 7, e36121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Witmer P. D., Lumayag S., Kovacs B., Valle D. (2007). MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 282, 25053-25066 [DOI] [PubMed] [Google Scholar]
- Yoo Y. D., Huang C. T., Zhang X., Lavaute T. M., Zhang S. C. (2011). Fibroblast growth factor regulates human neuroectoderm specification through ERK1/2-PARP-1 pathway. Stem Cells 29, 1975-1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. C. (2006). Neural subtype specification from embryonic stem cells. Brain Pathol. 16, 132-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. C., Wernig M., Duncan I. D., Brüstle O., Thomson J. A. (2001). In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 19, 1129-1133 [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang C. T., Chen J., Pankratz M. T., Xi J., Li J., Yang Y., Lavaute T. M., Li X. J., Ayala M., et al. (2010). Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G. Q., Li S. X., Shi Y. H. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. H., He X. L., Han X. L., Yu Y., Ye F., Chen Y., Hoang T., Xu X. M., Mi Q. S., Xin M., et al. (2010). MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 65, 612-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Li H., Huang H., Qiu M. S. (2012). MicroRNAs and glial cell development. Neuroscientist 18, 114-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Sun W., Okano K., Chen Y., Zhang N., Maeda T., Palczewski K. (2011). Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J. Biol. Chem. 286, 31749-31760 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


