Abstract
Alagille syndrome (ALGS, MIM 118450) is an autosomal dominant, multisystem disorder with high variability. Two genes have been described: JAG1 and NOTCH2. The population prevalence is 1:70,000 based on the presence of neonatal liver disease. The majority of cases (∼97%) are caused by haploinsufficiency of the JAG1 gene on 20p11.2p12, either due to mutations or deletions at the locus. Less than 1% of cases are caused by mutations in NOTCH2. The most widely used methods for mutational screening include denaturing high-performance liquid chromatography (DHPLC) and multiplex ligation-dependent probe amplification (MLPA). Very recently, whole-exome sequencing (WES) has become technically feasible due to the recent advances in next-generation sequencing technologies, therefore offering new opportunities for mutations/genes identification. A proband and its family, negative for the presence of mutations in JAG1 and NOTCH2 genes by neither DHPLC nor MLPA, were analyzed by WES. A missense mutation, not previously described, in JAG1 gene was identified. This result shows an improvement in the mutation detection rate due to novel sequencing technology suggesting the strong need to reanalyze all negative cases.
Key Words : Alagille syndrome, JAG1, Whole-exome sequencing
Alagille syndrome (ALGS, MIM 118450) is an autosomal dominant, multisystem disorder with high variability [Alagille et al., 1975]. Two genes were described as associated to ALGS: JAG1 [Li et al., 1997] and NOTCH2 [McDaniell et al., 2006]. Disease general population prevalence is 1:70,000 based on the presence of neonatal liver disease [Danks et al., 1977]. The majority of cases (∼97%) are caused by haploinsufficiency of the JAG1 gene [Li et al., 1997], located on 20p11.2p12, either due to mutations or deletions at the locus. Less than 1% of cases are caused by mutations in NOTCH2 [McDaniell et al., 2006]. The most widely used methods for mutational screening include denaturing high-performance liquid chromatography (DHPLC) and multiplex ligation-dependent probe amplification (MLPA). Recently, whole-exome sequencing (WES) has become technically feasible due to the current advances in sequencing technologies, therefore offering new opportunities for mutation/gene identification in Mendelian disorder studies.
Materials and Methods
Patients
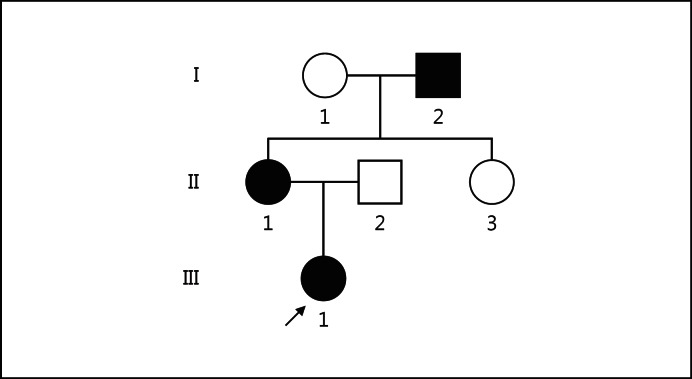
The proband (III-1) is the first female child of nonconsanguineous Italian parents. At birth, she presented neonatal jaundice, treated with phototherapy for 14 h. At 1 month, poor visual fixation was obvious, and the ophthalmological evaluation revealed retinal hemorrhage with abnormal latency of the visual evoked potentials. Serological examination showed high levels of hepatic enzyme values (AST, ALT and γGT), which remained abnormal during further evaluations. Abdominal ultrasound showed no abnormalities, while cardiac sonography indicated a thoracic aorta hypoplasia. ALGS was suspected, and a liver biopsy confirmed the paucity of intrahepatic bile ducts. At the age of 4, typical facial dysmorphisms of ALGS were clearly evident. Spinal X-rays and ophthalmological examination did not reveal any abnormalities. In the maternal proband's family, the mother (II-1) presented the same facial dysmorphisms and the coarctation of aorta but with normal liver function. The aunt (II-3) was referred with no clinical symptoms, and the grandfather (I-2), also with facial dysmorphisms, presented unspecified renal failure; however, no further clinical data are available (fig. 1).
Fig. 1.
Family tree. ○, □ = Nonaffected individuals; ●, ■ = affected individuals. The arrow indicates the proband.
Mutation Analysis
WES was performed on 3 family members (III-1, II-1 and II-3). Starting from as little as 3 μg of genomic DNA, the exome of each patient was enriched using SureSelect Human All Exon 38Mb Target Enrichment System (Agilent Technologies, Inc.); a whole-exome fragment library was constructed following the manufacturer's protocols (SureSelect Target Enrichment System for the Applied Biosystems SOLiD System-Version 2.0.1). Paired-end sequencing was performed on the SOLiD4 platform (Life Technologies), and the sequence reads were mapped against the human reference genome (hg19) using SOLiD bioscope software v1.3. Single nucleotide variants (SNVs) were called by SAMTools V0.1.8 [Li et al., 2009] and then filtered according to a dominant inheritance model. Among the SNVs identified, those present in NCBI dbSNP build v132 and in our own database were excluded. Results were confirmed by direct Sanger sequencing.
Results and Discussion
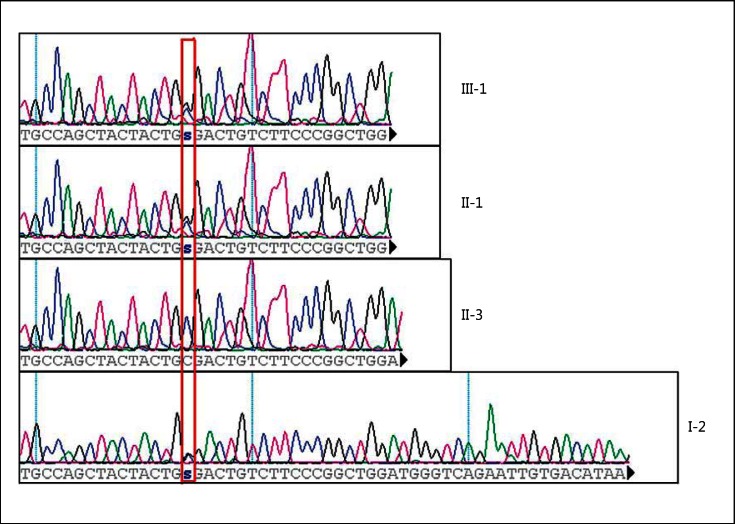
The proband (III-1) was referred to be negative at the mutation molecular screening of the entire coding region of JAG1 and NOTCH2 genes; the molecular analysis was done by DHPLC and MLPA. A comprehensive whole-exome sequencing was performed on subjects III-1, II-1 and II-3. The above-described data analysis pipeline allowed the detection of a missense mutation, never described before, in the JAG1 gene (c.1308C>G, p.C436W, NM_00124); this nucleotide variation was confirmed by Sanger sequencing (fig. 2) and has not yet been reported until today in any public databases, such as NCBI dbSNP or in 1000 Genomes Project. The proband (III-1), the mother (II-1) and the grandfather (I-2) were heterozygous (c.1308C>G), while the aunt (II-3) was normal (fig. 2). PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), Mutation Tester (http://www.mutationtaster.org/) and Condel (http://bg.upf.edu/condel/home) predict the mutation c.1308C>G, p.C436W, NM_00124 as pathogenic. This mutation affects the EGF-like6 jagged-1 protein region; the replaced cysteine is evolutionarily highly conserved and is involved in disulfide bonds within the EGF-like domain.
Fig. 2.
Sanger sequencing electropherogram; the variant [JAG-1, c.1308C>G, p.C436W, NM_00124] is highlighted by a red rectangle. s = Genotype C/G.
A mutation causing a cysteine substitution in the same JAG1 EGF-like6 domain (p.C438F), just 2 amino acids downstream from the one we are now discussing, was already described and its pathogenic role established [Crosnier et al., 1999].
Neither the mutation c.1308C>G nor others were revealed by DHPLC analysis of the entire coding region of JAG1 and NOTCH2 genes; DHPLC mutation discovery rate is lower than 100%, for different reasons, e.g. a large deletion spanning several exons or mutations localized within zones not covered by PCR amplified regions (such as promoters or introns) or, last, it could be due to technical failure [Samejima et al., 2007]. Mutation c.1308C>G was not detected by DHPLC because of technical failure.
Alagille syndrome is a genetic disorder with autosomal dominant inheritance; every affected individual could transmit the mutation and thus, the phenotype to future generations (regardless of the gender of the unborn) with a risk of 50%. Genetic counseling is very important to inform people of the risks, even though it may encounter difficulties in their ability to define the phenotype. Indeed, no genotype-phenotype correlations exist between clinical manifestations of ALGS and specific JAG1 mutation types or location within the gene [Crosnier et al., 1999]. However, many authors described a specific tissue sensitivity to JAG1 dosage, for example, the heart is more sensitive to JAG1 dosage than the liver [Eldadah et al., 2001; Lu et al., 2003]; this could explain why both, the mother (II-1) and the daughter (III-1), have cardiological involvement, while only the child (III-1) has liver affection.
The improvement of the mutation detection rate observed using WES strategy compared to DHPLC and MLPA approach strongly suggests reanalyzing all negative cases so far reported at mutational screenings (done with lower-resolution techniques). This example highlights the huge impact of new sequencing technologies, including targeted resequencing, in reducing false negative mutation detection rate and offering better clinical management of patients and their families, leading to remarkable improvements of counseling and of diagnostic algorithms.
Acknowledgement
This work was supported by the Italian Ministry of Health, Ricerca Finalizzata. We would like to acknowledge Angela D'Eustacchio for her technical support during laboratory work.
References
- Alagille D, Odièvre M, Gautier M, Dommergues JP. Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. J Pediatr. 1975;86:63–71. doi: 10.1016/s0022-3476(75)80706-2. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, et al. Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology. 1999;116:1141–1148. doi: 10.1016/s0016-5085(99)70017-x. [DOI] [PubMed] [Google Scholar]
- Danks DM, Campbell PE, Jack I, Rogers J, Smith AL. Studies of the aetiology of neonatal hepatitis and biliary atresia. Arch Dis Child. 1977;52:360–367. doi: 10.1136/adc.52.5.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, et al. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Lu F, Morrissette JJ, Spinner NB. Conditional JAG1 mutation shows the developing heart is more sensitive than developing liver to JAG1 dosage. Am J Hum Genet. 2003;72:1065–1070. doi: 10.1086/374386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the Notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima H, Torii C, Kosaki R, Kurosawa K, Yoshihashi H, et al. Screening for Alagille syndrome mutations in the JAG1 and NOTCH2 genes using denaturing high-performance liquid chromatography. Genet Test. 2007;11:216–227. doi: 10.1089/gte.2006.0519. [DOI] [PubMed] [Google Scholar]