Abstract
A decade ago, we identified a novel gene, glomulin (GLMN) in which mutations cause glomuvenous malformations (GVMs). GVMs are bluish-purple cutaneous vascular lesions with characteristic glomus cells in the walls of distended venous channels. The discovery of the genetic basis for GVMs allowed the definition of clinical features to distinguish GVMs from other venous anomalies. The variation in phenotype was also highlighted: from a single punctate blue dot to a large plaque-like lesion. In this study, we screened GLMN in a large cohort of patients to broaden the spectrum of mutations, define their frequency and search for possible genotype-phenotype correlations. Taking into account 6 families published by others, a mutation in GLMN has been found in 162 families. This represents 40 different mutations; the most frequent one being present in almost 45% of them. Expressivity varies largely, without a genotype/phenotype relationship. Among 381 individuals with a mutation, we discovered 37 unaffected carriers, implying a penetrance of 90%. As nonpenetrant individuals may transmit the disease to their descendants, knowledge on the mutational status is needed for appropriate genetic counseling.
Key Words : Anomaly, Gene, Glomulin, Glomuvenous malformation, Vascular
The classification of vascular anomalies was established following the guidelines of Mulliken and Glowacki [1982]. It differentiated vascular tumors from vascular malformations, the latter being subdivided depending on the type(s) of vessel(s) affected. Glomuvenous malformation (GVM, OMIM 138000) belongs to the subgroup of venous anomalies [Brouillard and Vikkula, 2003]. It is distinguished from sporadic venous malformation (VM) and inherited cutaneomucosal venous malformation (VMCM, OMIM 600195) [Soblet et al., in press]. A GVM is usually raised, nodular, multifocal, and hyperkeratotic (fig. 1A). Color varies from pink to purplish-dark blue [Boon et al., 2004]. Lesions are often present at birth and slowly expand during childhood. Plaque-like GVM, which is less frequent, is flat and purple in the newborn but darkens with time (fig. 1A, individual 810) [Mallory et al., 2006]. Most are located on the extremities. GVMs involve skin and subcutis, rarely mucosa or muscle, cannot be emptied by compression and are often painful on palpation. The lack of intestinal lesions and bleeding also distinguishes GVMs from Blue Rubber Bleb Nevus syndrome (BRBN) (OMIM 112200). The best therapy for GVM is surgical resection of the entire lesion; usually there is no infiltration of underlying tissues, although sometimes, only partial resection is possible. Sclerotherapy is another option in specific cases [Boon et al., 2004].
Fig. 1.
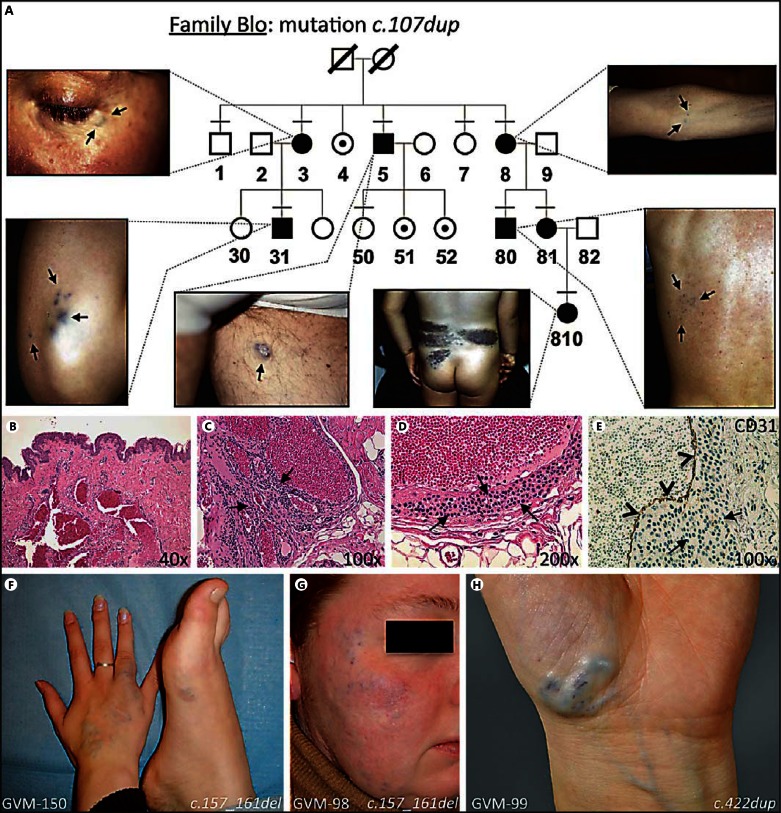
A Representative GVM family with mutation in glomulin (c.107dup), illustrating autosomal dominant inheritance with reduced penetrance (individuals 4, 51 and 52, unaffected carriers) and phenotypic variability. Most lesions are small and nodular (Blo-3, lower eyelid; Blo-5, left thigh; Blo-8, left arm; Blo-31, right elbow; and Blo-80, back). Blo-810 with large plaque-like lesion on back. Tested individuals numbered. Small horizontal bar = Clinically examined; black symbol = affected individual; dotted symbol = carrier. Adapted from Brouillard et al. [2008], with permission of Oxford University Press, USA. B-E Typical histology of GVM. B Hematoxylin-eosin-stained section at 40× magnification: dilated venous channels and normal overlying skin. C, D 100× and 200× magnifications demonstrate rounded glomus cells (arrows). E CD31 staining at 200×; normal endothelial cell layer (arrowheads). F-H Patients with GLMN mutation initially diagnosed as VM or VMCM.
Histologically, GVM is characterized by the presence of a variable number of mural glomus cells in distended venous channels (fig. 1B-E) [Gorlin et al., 1960; Goodman and Abele, 1971]. Similar cells are present in solitary glomus tumors. The generic term ‘glomangioma' has been used for these entities in the past. Solitary glomus tumors are subungual, painful lesions exclusively comprised of glomus cells without a major vascular component [Laymon and Peterson, 1965]. The glomus cells of GVM and subungual glomus tumors must not be confused with similarly named glomus cells of normal paraganglia (including glomus jugulare and glomus tympanicum) that are neuroendocrine cells. Paragangliomas are tumors derived from this second type of glomus cells, mainly found in the head and neck areas and caused by mutations in subunits of the succinate dehydrogenase complex (OMIM 115310, 168000, 601650, and 605373).
GVM is frequently inherited. It segregates as an autosomal dominant disease, with incomplete penetrance and variable expressivity (fig. 1A) [Brouillard et al., 2002]. Usually, most of the affected family members have small lesions and never seek treatment. Some individuals are more severely affected, depending on the size, number and location of the lesion(s), and seek medical attention [Boon et al., 2004]. We mapped the VMGLOM locus on the short arm of chromosome 1, in 1p21p22 [Boon et al., 1999; Brouillard et al., 2000; Irrthum et al., 2001], and identified the causative gene that we named glomulin (GLMN) [Brouillard et al., 2002]. We have reported a GLMN mutation in 87 families [Brouillard et al., 2002, 2005, 2008; Mallory et al., 2006; Goujon et al., 2011; Butler et al., 2012], and others reported 6 [O'Hagan et al., 2006; Ostberg et al., 2007; Borroni et al., 2011]. The mutations result in loss of function, either by causing a stop codon, by altering the reading frame or by changing the splice-site consensus sequences, leading to aberrant transcripts. The fact that expressivity is variable for the same mutation, from patient to patient, that penetrance is incomplete and that some affected individuals develop new lesions with time suggested paradominant inheritance (fig. 1A). We proved this by our discovery of a somatic second-hit, resulting in complete lack of GLMN in one GVM lesion [Brouillard et al., 2002] and, more recently, in 15 additional tissues [Amyere et al., 2013].
In this report, we expanded the panel of patients screened for mutations in GLMN to better define the scope of mutations and phenotypic characteristics and determine reliable mutational frequencies.
Materials and Methods
Subject Recruitment and Screening
Informed consent was obtained from all the patients prior to their participation in the study, as approved by the Ethical Committee of the Medical Faculty at the Université catholique de Louvain (Brussels, Belgium). For 58 families, a detailed clinical questionnaire was filled out, and for 66 additional families, a description was provided by the clinicians involved in this study. No further details were received for the other patients. In 37 families, pictures of the lesions were taken. Screening for mutations was performed on genomic DNA from 1 or 2 patients per family, and when possible, cosegregation was verified in the rest of the family. In brief, venous blood samples were taken from all participants, and DNA was extracted from whole blood (Wizard genomic DNA purification kit, Promega). Mutational screening was first performed by allele-specific PCR for the 3 most common mutations (c.157_161del, c.108C>A and c.1179_1181del), as described previously [Brouillard et al., 2005]. The negative samples were subsequently screened by high resolution melting analysis for all 19 exons of GLMN and surrounding splice-sites (Light Cycler 480, Roche). Fragments exhibiting an abnormal melting profile were reamplified and sequenced on an ABI 3130xl capillary sequencer (Applied Biosystems). For the analysis, sequences were aligned against the genomic sequence using CLCbio Main Workbench 6 software (CLCbio).
Results
We screened for the glomulin gene in 207 new samples clinically diagnosed as GVM or likely GVM (n = 80), VM without a mutation in TIE2 (n = 96) or BRBN syndrome (n = 31). A mutation was found in 60 GVM, 9 VM and none of the BRBN samples. Taking into account the 87 families that we reported previously [Brouillard et al., 2002, 2005, 2008; Mallory et al., 2006; Goujon et al., 2011; Butler et al., 2012], we identified a GLMN mutation in 156 index patients. If we include the 6 families reported by others [O'Hagan et al., 2006; Ostberg et al., 2007; Borroni et al., 2011], 40 different mutations have been discovered in GLMN altogether in 162 families (fig. 2A; table 1). All mutations were submitted to the Vascular Anomaly and Lymphedema Mutation Database (http://www.icp.ucl.ac.be/vikkula/VAdb/). The majority (143/162; 88%) of the patients in which a mutation was found had a familial history of the disorder; only 19 patients were considered sporadic. We were not able to confirm the de novo appearance of these mutations, as parental samples were not available for testing, except for the mother of one individual, who was negative for the mutation.
Fig. 2.
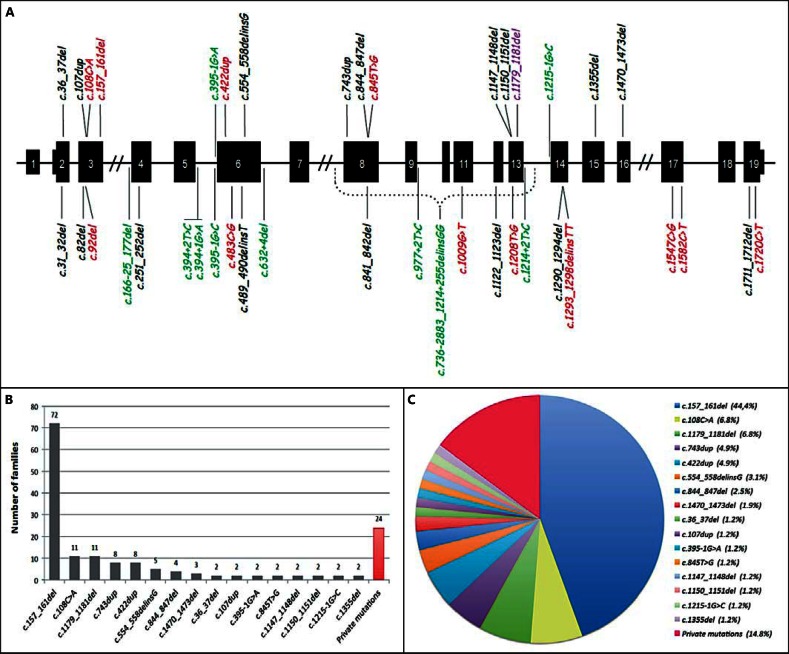
GLMN mutations. A Gene with shared mutations (top) and private ones (below). Red = Direct stop codon; black = frameshift leading to premature truncation; green = splice-site alteration; purple = deletion of asparagine 393. B Frequency of mutations in 162 families. Private mutations only identified in one index case or family. C Proportion of the different mutations.
Table 1.
The 40 mutations identified in glomulin
| No. | GLMN mutation (HGVS name) (refseq # NG_009796.1) | Exon | Protein | Freq. (index cases) | Mutation old nomenclature (= former published name) | First published in |
|---|---|---|---|---|---|---|
| 1. | c.31_32del | 2 | p.(Lys11Glufs*11) | 1 | 31delAA | Brouillard et al., 2002 |
| 2. | c.36_37del | 2 | p.(Cys13Serfs*9) | 2 | 36delAT | Brouillard et al., 2008 |
| 3. | c.82del | 3 | p.(Leu28Tyrfs*4) | 1 | 81delC | Brouillard et al., 2008 |
| 4. | c.92del | 3 | p.(Leu31*) | 1 | – | - |
| 5. | c.107dup | 3 | p.(Cys36Trpfs*17) | 2 | 107insG | Brouillard et al., 2002 |
| 6. | c.108C>A | 3 | p.(Cys36*) | 11 | 108C>A | Brouillard et al., 2002 |
| 7. | c.157_161del | 3 | p.(Lys53*) | 72 | 157delAAGAA | Brouillard et al., 2002 |
| 8. | c.166–25_177del | 4 | altered splicing | 1 | (IVS3–25)–177del37nt | Brouillard et al., 2008 |
| 9. | c.251_252del | 4 | p.(Lys84Serfs*7) | 1 | 251delAA | Brouillard et al., 2008 |
| 10. | c.394+1G>A | 5 | altered splicing | 1 | IVS5+1(G>A) | Brouillard et al., 2008 |
| 11. | c.394+2T>C | 5 | altered splicing | 1 | – | – |
| 12. | c.395–1G>A | 6 | altered splicing | 2 | IVS5–1(G>A) | Brouillard et al., 2008 |
| 13. | c.395–1G>C | 6 | altered splicing | 1 | IVS5–1(G>C) | Brouillard et al., 2008 |
| 14. | c.422dup | 6 | p.(Tyr141*) | 8 | 422insA (reported as 421insT) | Brouillard et al., 2002 |
| 15. | c.483C>G | 6 | p.(Tyr161*) | 1 | – | – |
| 16. | c.489_490delinsT | 6 | p.(Lys163Asnfs*18) | 1 | – | – |
| 17. | c.554_558delinsG | 6 | p.(Lys185Serfs*19) | 5 | 554delA+556delCCT | Brouillard et al., 2002 |
| 18. | c.632+4del | 6 | altered splicing | 1 | IVS6+4delA | Brouillard et al., 2002 |
| 19. | c.736–2883_1214+255delinsGG | 8–13 | altered splicing | 1 | (IVS7–2884)–(IVS13+255) del8.4kbp+insGG | Brouillard et al., 2002 |
| 20. | c.743dup | 8 | p.(Leu248Phefs*14) | 8 | 738insT | Brouillard et al., 2005 |
| 21. | c.841_842del | 8 | p.(Gln281Valfs*20) | 1 | – | – |
| 22. | c.844_847del | 8 | p.(Leu282Glnfs*10) | 4 | 842delAGTT | Brouillard et al., 2002 |
| 23. | c.845T>G | 8 | p.(Leu282*) | 2 | 845T>G | Brouillard et al., 2008 |
| 24. | c.977+2T>C | 9 | altered splicing | 1 | – | – |
| 25. | c.1009G>T | 11 | p.(Glu337*) | 1 | – | – |
| 26. | c.1122_1123del | 12 | p.(Cys375Profs*3) | 1 | c.1121delTT | Borroni et al., 2011 |
| 27. | c.1147_1148del | 13 | p.(Lys383Glufs*10) | 2 | – | – |
| 28. | c.1150_1151del | 13 | p.(ser384Phefs*9) | 2 | 1150delAG | Brouillard et al., 2005 |
| 29. | c.1179_1181del | 13 | p.(Asn393del) | 11 | 1179delCAA | Brouillard et al., 2002 |
| 30. | c.1208T>G | 13 | p.(Leu403*) | 1 | – | – |
| 31. | c.1214+2T>C | 13 | altered splicing | 1 | IVS13+2(T>C) reported as IVS13+1 | Brouillard et al., 2008 |
| 32. | c.1215–1G>C | 14 | altered splicing | 2 | IVS13–1(G>C) | Brouillard et al., 2008 |
| 33. | c.1290_1294del | 14 | p.(Met430Ilefs*6) | 1 | – | – |
| 34. | c.1293_1298delinsTT | 14 | p.(Leu432*) | 1 | 1293delA+1296delAAA | Brouillard et al., 2005 |
| 35. | c.1355del | 15 | p.(Leu452Trpfs*24) | 2 | 1355delT | Brouillard et al., 2002 |
| 36. | c.1470_1473del | 16 | p.(Asn490Lysfs*16) | 4 | 1470delTCAA | Brouillard et al., 2002 |
| 37. | c.1547C>G | 17 | p.(ser516*) | 1 | 1547C>G | Brouillard et al., 2002 |
| 38. | c.1582C>T | 17 | p.(Gln528*) | 1 | 1582C>T | Brouillard et al., 2008 |
| 39. | c.1711_1712del | 19 | p.(val571Serfs*9) | 1 | 1711delGT | Brouillard et al., 2002 |
| 40. | c.1720C>T | 19 | p.(Arg574*) | 1 | 1720C>T | Brouillard et al., 2008 |
Since our original reports, the Human Gene Variation Sequence nomenclature has evolved (http://www.hgvs.org/); the names in this report are given according to the new guidelines. The novel names were verified using Mutalyzer (https://www.mutalyzer.nl/); the former and corresponding novel names are shown in table 1. All mutations were heterozygous nonsense (n = 12), splice-site (n = 10) or frame-shift changes (n = 17), with the exception of one, predicted to result in premature truncation of the protein and, therefore, in loss-of-function of GLMN (fig. 2A). If the mutant proteins were produced, the longest form (c.1720C>T; p.Arg574*) would only miss the 21 last amino acids, yet the phenotype is indistinguishable from patients with mutations in exon 2. Alternatively, the RNAs carrying premature stop codons are likely to undergo nonsense-mediated mRNA decay. The only exception is c.1179_1181del, corresponding to the loss of one asparagine within the correct open reading frame. This mutant allele was shown to be stable [Brouillard et al., 2005].
A GLMN mutation was discovered in most GVM index cases, with the exception of 20 patients with such a clinical diagnosis. Of these negative patients, 5 were familial, 12 sporadic and 3 had an unknown mode of inheritance. Among the familial cases, only one was a probable (plaque-like) GVM. A posteriori, the other lesions turned out to be glomangiomyoma (n = 1), VMCM or sporadic multifocal VM (n = 2) and a fifth family comprised of 3 affected individuals, one of whom had a somatic TIE2 mutation. All 3 of these individuals had only small lesions. On the 12 negative sporadic cases, only 3 were probable GVM, whereas 3 others had only one small lesion, which could be a VM or a GVM. The other 6 were subsequently found to have a clinical diagnosis of one hemangioma, one dermoid cyst, one complex vascular anomaly, one malignant glomus tumor, one likely BRBN; and one had insufficient clinical information. Interestingly, in 6 of the GVM families with a mutation, at least one individual was diagnosed as a VM. Nine patients clinically diagnosed as ‘VM or GVM’ also had a GLMN mutation, and 3 of them were clinically considered to have multiple VM or VMCM (fig. 1F-H).
Some of the mutations were encountered in several families (fig. 2; table 1). The most frequent mutation, c.157_161del, was discovered in 72 of the 162 families (44.4%). Using polymorphic markers, we previously showed that there was a strong founder effect associated with this mutation for 21 families, indicating a common ancestor in whom the original mutation occurred [Brouillard et al., 2005]. It is likely that most of the 51 novel families with this mutation also share the same haplotype. A similar situation was evidenced for mutations c.108C>A, and c.1179_1181del, each of which was found in 11 families (6.8% each). Two other mutations, c.743dup and c.422dup were present in 8 families (5% each), c.554_558delinsG in 5 (3.1%), c.844_847del in 4 (2.5%), c.1470_1473del in 3 (1.8%) and 8 mutations were found in 2 families (1.2% each). Overall, these 16 mutations explain the disorder in 85% of GVM index patients. The remaining 15% are accounted for by 24 private mutations that are unique to the respective pedigree (table 1).
We assessed cosegregation for all index patients for which DNA was obtained from other family members. Within the 162 families, 381 individuals carried a GLMN mutation, while 84 unaffected ones did not: 184 were males and 197 were females. Among the samples with a mutation, we identified 37 unaffected carriers, i.e. healthy individuals at the time of examination (age range from 6 months to 76 years old; mean = 34 years old). We also found 3 phenocopies, i.e. patients who had been considered as affected (i.e. thought to have a GVM), but did not have the familial mutation. None of the 71 spouses harbored a mutation. Carriers were found for different mutations, in different pedigrees.
Some atypical clinical features were detected in the patients in whom we found a mutation in GLMN. The most frequent unusual presentation was a plaque-like GVM, which can be confused with a capillary malformation. We reported 10 such cases earlier, 3 of which had a proven GLMN mutation [Mallory et al., 2006]. Nine additional plaque-like GVMs were detected in this series. This phenotype was observed with different mutations. In 2 instances, they were associated with chylous ascites and pleural effusions [Tejedor et al., 2010; Goujon et al., 2011]. A third patient with a large GVM had been diagnosed with hydrops fetalis at 36 weeks of gestation. Another GVM patient had transposition of the great vessels [Chen et al., 2009].
GVMs were mostly cutaneous; however, one patient had an unusual localization on the palate. Mucosal lesions were also observed on the upper and lower lips in another patient. Other possible associations included a hemangioma of infancy in 2 children of the same family; pial arteriovenous malformation; varicose veins in 2 sisters; mitral valve prolapse; thromboembolism of the middle cerebral artery at 6 years of age; and a patient with macrocephaly, mental retardation, obesity, bone deformation, scoliosis and fibrous dysplasia of the frontal bone. One patient also had lymphedema distichiasis and microcephaly in addition to GVM, but these features are most likely due to a microdeletion in nearby FOXC2 [Butler et al., 2012].
Discussion
We have identified a loss-of-function glomulin mutation in 381 individuals, accounting for 40 different mutations in 162 distinct families or index cases (fig. 2; table 1). There were no differences between males and females. The only peculiar mutation is c.1179_1181del, resulting in the loss of one asparagine in the protein. This mutation probably leads to a defective GLMN protein, but the exact mechanism is unknown [Brouillard et al., 2002, 2005]. The phenotype of the carriers of this mutation is indistinguishable from the other GVM patients. The most frequent mutation, c.157_161del, was detected in almost 45% of the families (72 of the families, accounting for 153 mutation carriers). The presence of all shared glomulin mutations facilitates genetic diagnosis and counseling for up to 85% of the patients (fig. 2C). Indeed, when assessing a likely GVM patient, testing for this mutation first, by allele-specific PCR as well as for c.108C>A and c.1179_1181del [Brouillard et al., 2005], gives a 58% chance to unravel the genetic cause of the disease when the clinical diagnosis is correct.
The detection of 37 unaffected mutation carriers, independent of the mutation, underscores the high (90%), but not complete penetrance of GVM. Large lesions are usually present at birth, but some affected individuals develop new small lesions with time [Boon et al., 2004]. Moreover, there is a large phenotypic variability, even for the same mutation in the same family (fig. 1A) [Brouillard et al., 2005, 2008]. Based on the double-hit hypothesis, we have detected a series of somatic second-hit mutations altering the second intact allele locally [Brouillard et al., 2002; Amyere et al., 2013]. Thus, we think that the size and number of lesions relate to the rate of somatic second-hit mutation(s) and to the angiogenic activity that continues until the end of growth in young adulthood. Large GVM lesions likely arise from mutations occurring early in development, whereas small lesions are due to mutations appearing later and/or in areas of lower angiogenic activity.
As GVMs are almost exclusively familial (88%), the presence of other affected family members strongly suggests GVM or VMCM. When an individual with a glomulin mutation is identified, it is important to test the other family members, as they can be nonpenetrant. These apparently unaffected members have a 50% chance of transmitting the mutation to their children, who can be severely affected depending on the temporal and spatial occurrence of the second-hit mutations.
Since the description of GVMs and the discovery that they are caused by mutations in GLMN, there have been several reports of likely affected patients and/or families in which no genetic testing has been performed [Blume-Peytavi et al., 2000; Vercellino et al., 2006; Henning et al., 2007; Torchia et al., 2007; Hill and Rademaker, 2009; Al Dhaybi et al., 2010; Hoekzema et al., 2010; Yoruk et al., 2010; Brauer et al., 2011]. Based on our results, it is likely that most of these patients have a mutation in GLMN. Indeed, very few clinically diagnosed GVMs were negative for GLMN. The majority of the samples that were GLMN-negative had only 1 (small) lesion, rendering clinical diagnosis difficult. Even if the mutation could have been missed because of limitations inherent to the screening method, these rare lesions may have a different etiology. It could, however, be that they are caused by small intragenic deletions that would need complementary techniques such as MLPA or SNP-chips for detection. If needed, exploration at the RNA level would be another option, as for the detection of the large deletion (c.736-2883_1214 +255delinsGG) in family Ad [Brouillard et al., 2002]. Alternatively, these patients could be sporadic cases with a somatic change only, undetectable in the blood DNA, but detectable in tissue DNA. Overall, we have no strong data to suggest that there is locus heterogeneity in GVMs.
Clinical distinction of GVM from sporadic VM and VMCM can be difficult in patients with only a few small lesions and without familial history of the disorder (fig. 1F-H) [Boon et al., 2004]. Diagnosis can also be confounded with a capillary malformation and a plaque-like GVM [Mallory et al., 2006]. Several clinical criteria and a biomarker have been defined [Dompmartin et al., 2010]. In practice, this distinction is important because, unlike sporadic VMs, for which elastic stocking is helpful, GVMs are often painful on palpation, and compression is not recommended. A GVM lesion is usually raised, nodular, present at birth, and slowly expands during childhood. It is often multifocal and hyperkeratotic. Its color varies from pink to purplish-dark blue [Boon et al., 2004]. The clinical signs described by Boon et al. [2004] were confirmed in the large number of patients examined in this study. In addition, except for 2 instances, GVM was not encountered in mucosa, contrary to other VMs. Three patients with large thoracico-abdominal lesions had chylous ascites and/or pleural effusions. These findings occurred only in 1 patient, thus, it is unlikely that they are attributable to mutations in GLMN.
Patients with GVM are clinically distinct from those with BRBN syndrome. GVMs do not present in the gastrointestinal tract. D-dimer level is also normal, although it can be extremely high in BRBN and VMs [Dompmartin et al., 2009]. Biopsy or surgical resection of the usually well-delimited GVM, followed by histological analysis could confirm the diagnosis of GVM, especially if cuboidal glomus cells are present. In contrast, absence of the latter is not an exclusion criterion, since glomus cells are not present in all lesions with a GLMN mutation. Therefore, genetic testing is needed for proper diagnosis, genetic counseling and management of the GVM patients.
Acknowledgements
We are grateful to all patients and families for their participation in the present study. The authors' research was partially funded by the Interuniversity Attraction Poles initiated by the Belgian Federal Science Policy, network 7/43; Concerted Research Actions (A.R.C.) – Convention No 07/12-005 of the Belgian French Community Ministry; the National Institute of Health, Program Project P01 AR048564; the F.R.S.-FNRS (Fonds de la Recherche Scientifique), all to M.V.; and the Fédération Wallonie-Bruxelles and the Lotterie nationale, Belgium.
References
- Al Dhaybi R, Powell J, McCuaig C, Kokta V. Differentiation of vascular tumors from vascular malformations by expression of Wilms tumor 1 gene: evaluation of 126 cases. J Am Acad Dermatol. 2010;63:1052–1057. doi: 10.1016/j.jaad.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Amyere M, Aerts V, Brouillard P, McIntyre BAS, Duhoux FP, et al. Somatic Uniparental Isodisomy: an Explanation for Multifocality of Glomuvenous Malformations. Am J Hum Genet. 2013;92:188–196. doi: 10.1016/j.ajhg.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Peytavi U, Adler YD, Geilen CC, Ahmad W, Christiano A, et al. Multiple familial cutaneous glomangioma: a pedigree of 4 generations and critical analysis of histologic and genetic differences of glomus tumors. J Am Acad Dermatol. 2000;42:633–639. [PubMed] [Google Scholar]
- Boon LM, Brouillard P, Irrthum A, Karttunen L, Warman ML, et al. A gene for inherited cutaneous venous anomalies (‘glomangiomas') localizes to chromosome 1p21-22. Am J Hum Genet. 1999;65:125–133. doi: 10.1086/302450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon LM, Mulliken JB, Enjolras O, Vikkula M. Glomuvenous malformation (glomangioma) and venous malformation: distinct clinicopathologic and genetic entities. Arch Dermatol. 2004;140:971–976. doi: 10.1001/archderm.140.8.971. [DOI] [PubMed] [Google Scholar]
- Borroni RG, Narula N, Diegoli M, Grasso M, Concardi M, et al. A novel mutation of the glomulin gene in an Italian family with autosomal dominant cutaneous glomuvenous malformations. Exp Dermatol. 2011;20:1032–1034. doi: 10.1111/j.1600-0625.2011.01387.x. [DOI] [PubMed] [Google Scholar]
- Brauer JA, Anolik R, Tzu J, Meehan S, Lieber CD, Geronemus RG. Glomuvenous malformations (familial generalized multiple glomangiomas) Dermatol Online J. 2011;17:9. [PubMed] [Google Scholar]
- Brouillard P, Vikkula M. Vascular malformations: localized defects in vascular morphogenesis. Clin Genet. 2003;63:340–351. doi: 10.1034/j.1399-0004.2003.00092.x. [DOI] [PubMed] [Google Scholar]
- Brouillard P, Olsen BR, Vikkula M. High-resolution physical and transcript map of the locus for venous malformations with glomus cells (VMGLOM) on chromosome 1p21-p22. Genomics. 2000;67:96–101. doi: 10.1006/geno.2000.6232. [DOI] [PubMed] [Google Scholar]
- Brouillard P, Boon LM, Mulliken JB, Enjolras O, Ghassibé M, et al. Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (‘glomangiomas') Am J Hum Genet. 2002;70:866–874. doi: 10.1086/339492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, Ghassibé M, Penington A, Boon LM, Dompmartin A, et al. Four common glomulin mutations cause two thirds of glomuvenous malformations (‘familial glomangiomas'): evidence for a founder effect. J Med Genet. 2005;42:e13. doi: 10.1136/jmg.2004.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, Enjolras O, Boon LM, Vikkula M. Glomulin and glomuvenous malformation. In: Epstein CJ, Erickson RP, Wynshaw-Boris A, editors. Inborn Errors of Development. ed 2. New York: Oxford University Press; 2008. [Google Scholar]
- Butler MG, Dagenais SL, Garcia-Perez JL, Brouillard P, Vikkula M, et al. Microcephaly, intellectual impairment, bilateral vesicoureteral reflux, distichiasis, and glomuvenous malformations associated with a 16q24.3 contiguous gene deletion and a Glomulin mutation. Am J Med Genet A. 2012;158A:839–849. doi: 10.1002/ajmg.a.35229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Eide M, Shwayder T. Glomuvenous malformation in a boy with transposition of the great vessels: a case report and review of literature. Pediatr Dermatol. 2009;26:70–74. doi: 10.1111/j.1525-1470.2008.00826.x. [DOI] [PubMed] [Google Scholar]
- Dompmartin A, Ballieux F, Thibon P, Lequerrec A, Hermans C, et al. Elevated D-dimer level in the differential diagnosis of venous malformations. Arch Dermatol. 2009;145:1239–1244. doi: 10.1001/archdermatol.2009.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dompmartin A, Vikkula M, Boon LM. Venous malformation: update on aetiopathogenesis, diagnosis and management. Phlebology. 2010;25:224–235. doi: 10.1258/phleb.2009.009041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman TF, Abele DC. Multiple glomus tumors. A clinical and electron microscopic study. Arch Dermatol. 1971;103:11–23. doi: 10.1001/archderm.103.1.11. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Fusaro RM, Benton JW. Multiple glomus tumor of the pseudocavernous hemangioma type. Arch Dermatol. 1960;82:776–778. doi: 10.1001/archderm.1960.01580050118018. [DOI] [PubMed] [Google Scholar]
- Goujon E, Cordoro KM, Barat M, Rousseau T, Brouillard P, et al. Congenital plaque-type glomuvenous malformations associated with fetal pleural effusion and ascites. Pediatr Dermatol. 2011;28:528–531. doi: 10.1111/j.1525-1470.2010.01216.x. [DOI] [PubMed] [Google Scholar]
- Henning JS, Kovich OI, Schaffer JV. Glomuvenous malformations. Dermatol Online J. 2007;13:17. [PubMed] [Google Scholar]
- Hill S, Rademaker M. A collection of rare anomalies: multiple digital glomuvenous malformations, epidermal naevus, temporal alopecia, heterochromia and abdominal lipoblastoma. Clin Exp Dermatol. 2009;34:e862–864. doi: 10.1111/j.1365-2230.2009.03616.x. [DOI] [PubMed] [Google Scholar]
- Hoekzema R, Zonneveld IM, van der Wal AC. Type 2 segmental glomangiomas. Dermatol Online J. 2010;16:8. [PubMed] [Google Scholar]
- Irrthum A, Brouillard P, Enjolras O, Gibbs NF, Eichenfield LF, et al. Linkage disequilibrium narrows locus for venous malformation with glomus cells (VMGLOM) to a single 1.48 Mbp YAC. Eur J Hum Genet. 2001;9:34–38. doi: 10.1038/sj.ejhg.5200576. [DOI] [PubMed] [Google Scholar]
- Laymon CW, Peterson WC., Jr Glomangioma (glomus tumor). A clinicopathologic study with special reference to multiple lesions appearing during pregnancy. Arch Dermatol. 1965;92:509–514. doi: 10.1001/archderm.92.5.509. [DOI] [PubMed] [Google Scholar]
- Mallory SB, Enjolras O, Boon LM, Rogers E, Berk DR, et al. Congenital plaque-type glomuvenous malformations presenting in childhood. Arch Dermatol. 2006;142:892–896. doi: 10.1001/archderm.142.7.892. [DOI] [PubMed] [Google Scholar]
- Mulliken JB, Glowacki J. Hemangiomas and vascular malformations in infants and children: a classification based on endothelial characteristics. Plast Reconstr Surg. 1982;69:412–422. doi: 10.1097/00006534-198203000-00002. [DOI] [PubMed] [Google Scholar]
- O'Hagan AH, Maloney FJ, Buckley C, Bingham EA, Walsh MY, et al. Mutation analysis in Irish families with glomuvenous malformations. Br J Dermatol. 2006;154:450–452. doi: 10.1111/j.1365-2133.2005.07041.x. [DOI] [PubMed] [Google Scholar]
- Ostberg A, Moreno G, Su T, Trisnowati N, Marchuk D, Murrell DF. Genetic analysis of a family with hereditary glomuvenous malformations. Australas J Dermatol. 2007;48:170–173. doi: 10.1111/j.1440-0960.2007.00373.x. [DOI] [PubMed] [Google Scholar]
- Soblet J, Limaye N, Uebelhoer M, Boon LM, Vikkula M. Variable somatic TIE2 mutations in half or sporadic venous malformations. Mol Sydromol, DOI: 10.1159/000348327 (2013). [DOI] [PMC free article] [PubMed]
- Tejedor M, Martin-Santiago A, Gomez C, Fiol M, Benitez-Segura I. Congenital plaque-type glomuvenous malformation associated with chylous ascites. Pediatr Dermatol. 2010;27:673–675. doi: 10.1111/j.1525-1470.2010.01337.x. [DOI] [PubMed] [Google Scholar]
- Torchia D, Palleschi GM, Urso C. Unilateral glomuvenous malformations. J Eur Acad Dermatol Venereol. 2007;21:552–553. doi: 10.1111/j.1468-3083.2006.01950.x. [DOI] [PubMed] [Google Scholar]
- Vercellino N, Nozza P, Oddone M, Bava GL. Large plaque-like glomuvenous malformation (glomangioma) simulating venous malformation. Clin Exp Dermatol. 2006;31:538–541. doi: 10.1111/j.1365-2230.2006.02147.x. [DOI] [PubMed] [Google Scholar]
- Yoruk O, Ucuncu H, Aktan B, Calik M, Kilic K. Glomuvenous malformations in the buccal area. J Craniofac Surg. 2010;21:2001–2003. doi: 10.1097/SCS.0b013e3181f535a2. [DOI] [PubMed] [Google Scholar]