Abstract
Hereditary haemorrhagic telangiectasia (HHT) is a disease characterised by abnormal vascular structures, and most commonly caused by mutations in ENG, ACVRL1 or SMAD4 encoding endothelial cell-expressed proteins involved in TGF-β superfamily signalling. The majority of mutations reported on the HHT mutation database are predicted to lead to stop codons, either due to frameshifts or direct nonsense substitutions. The proportion is higher for ENG (67%) and SMAD4 (65%) than for ACVRL1 (42%), p < 0.0001. Here, by focussing on ENG, we report why conventional views of these mutations may need to be revised. Of the 111 stop codon-generating ENG mutations, on ExPASy translation, all except one were premature termination codons (PTCs), sited at least 50-55 bp upstream of the final exon-exon boundary of the main endoglin isoform, L-endoglin. This strongly suggests that the mutated RNA species will undergo nonsense-mediated decay. We provide new in vitro expression data to support dominant negative activity of stable truncated endoglin proteins but suggest these will not generate HHT: the single natural stop codon mutation in L-endoglin (sited within 50-55 nucleotides of the final exon-exon boundary) is unlikely to generate functional protein since it replaces the entire transmembrane domain, as would 8 further natural stop codon mutations, if the minor S-endoglin isoform were implicated in HHT pathogenesis. Finally, next-generation RNA sequencing data of 7 different RNA libraries from primary human endothelial cells demonstrate that multiple intronic regions of ENG are transcribed. The potential consequences of heterozygous deletions or duplications of such regions are discussed. These data support the haploinsufficiency model for HHT pathogenesis, explain why final exon mutations have not been detected to date in HHT, emphasise the potential need for functional examination of non-PTC-generating mutations, and lead to proposals for an alternate stratification system of mutational types for HHT genotype-phenotype correlations.
Key Words : Alternative splicing, Nonsense-mediated decay, Pervasive transcription, Premature termination codons
Hereditary haemorrhagic telangiectasia (HHT, also known as Osler Weber Rendu syndrome) is one of the most common disorders to be inherited as an autosomal dominant trait [Guttmacher et al., 1995; Shovlin, 2010]. Careful epidemiological studies reveal that it affects approximately 1 in 5,000 individuals [Bideau et al., 1992; Kjeldsen et al., 1999; Dakeishi et al., 2002]. Causative gene mutations (most commonly in ENG, ACVRL1 and SMAD4) lead to the development of abnormal vascular structures which cause the major complications of the disease. Nosebleeds and chronic gastrointestinal bleeding result from telangiectasia in the nasal and gastrointestinal tracts; complex organ-specific pathology results from the presence of larger arteriovenous malformations in the pulmonary, hepatic, cerebral, pancreatic, spinal and other circulations [Shovlin, 2010].
Currently, 5 types of HHT are recognised. The majority of HHT patients will have HHT1 due to mutations in ENG encoding endoglin [McAllister et al., 1994] or HHT2 due to mutations in ACVRL1 encoding activin receptor-like kinase (ALK1) [Johnson et al., 1996]. As summarised elsewhere [Govani and Shovlin, 2009; Shovlin, 2010], more than 500 different mutations have been found in ENG (HHT1) and ACVRL1 (HHT2) HHT families (www.hhtmutation.org at time of submission, now at www.arup.utah.edu/database/hht/). One to two percent of cases have mutations in SMAD4, mutations that also cause the gastrointestinal epithelial precancerous state of juvenile polyposis [Gallione et al., 2004, 2010]. There are at least 2 further unidentified genes that can cause pure HHT, HHT3 [Cole et al., 2005; Govani and Shovlin, 2010] and HHT4 [Bayrak-Toydemir et al., 2006a], to date, each identified in single families. While all cause a wide spectrum of potential phenotypic consequences, genotype-phenotype correlations indicate significant differences between HHT1, HHT2, and juvenile polyposis/HHT (JPHT), particularly with respect to the prevalence of pulmonary, cerebral and hepatic arteriovenous malformations [Kjeldsen et al., 2005; Bayrak-Toydemir et al., 2006b; Bossler et al., 2006; Letteboer et al., 2006; Lesca et al., 2007; Sabbà et al., 2007], and the JPHT-specific pathologies associated with juvenile polyposis [Gallione et al., 2004, 2010].
The genes mutated in HHT encode endothelial cell-expressed proteins that transmit or regulate signals by the transforming growth factor (TGF)-β superfamily of ligands. These ligands (including TGF-βs, activins, bone morphogenetic proteins (BMPs), growth/differentiation factors (GDFs) and inhibins) [Heldin et al., 1997; Massague and Chen, 2000; Miyazono et al., 2001] signal through heteromeric complexes comprised of type I and type II cell surface receptor serine-threonine kinases [Wrana et al., 1994]. Endoglin is a transmembrane glycoprotein, highly expressed on endothelial cells. L-endoglin, the predominant form in endothelial cells, has a cytoplasmic tail of 47 amino acids including 19 Ser/Thr residues; S-endoglin shares the identical extracellular and transmembrane domains, but due to retention of the penultimate intron, has a truncated cytoplasmic tail of 14 amino acids [Bellón et al., 1993]. Endoglin interacts with multiple superfamily receptor complexes [Abdalla and Letarte, 2006; Lebrin and Mummery, 2008]. ACVRL1 encodes ALK1, a relatively endothelial cell-specific TGF-β type I receptor that can associate with 2 type II receptors, TβRII and BMPR2 [Bertolino et al., 2005]. TβRII also associates with TβRI (also known as ALK5), with activation of the alternate ALK1/ALK5 pathways generating different, and mutually antagonistic downstream cellular responses [Goumans et al., 2003; Lebrin et al., 2004; Blanco et al., 2005, 2008]. The precise ligand(s) responsible for the HHT phenotype is/are not yet determined [Park et al., 2008; Upton et al., 2009], with models proposed for both TGF-β1 [Goumans et al., 2003; Lebrin et al., 2004, 2010; Blanco et al., 2005; Pece-Barbara et al., 2005] and the ALK1-specific ligand, BMP9 [David et al., 2007; Scharpfenecker et al., 2007; Bailly, 2008].
For endoglin, a body of evidence indicates that the HHT gene mutations lead to reduced endoglin expression at ∼50% of normal in activated monocytes, human umbilical vein endothelial cells [Pece et al., 1997; Pece-Barbara et al., 1999; Bourdeau et al., 2000; Cymerman et al., 2000; Paquet et al., 2001] and blood outgrowth endothelial cells [Fernandez et al., 2006]. There are individual mutations which cannot generate a mutated protein, most obviously the single base change mutations disrupting the ENG start codon [Gallione et al., 1998] and nonsense mutations or large deletion mutations in which only the wild type allele could be detected at an RNA level [Shovlin et al., 1997; Gallione et al., 1998]. A high proportion of HHT-causing mutations generate stop codons either directly by nonsense mutations, or as a result of out-of-frame deletions and/or insertions. An early model proposed was that translation of truncated proteins from the mutated ENG allele would lead to interference with the normal function of the protein from the wild type allele in a dominant negative manner [McAllister et al., 1995]. Naturally occurring truncated endoglin protein moeities do exist, for example the rare S-endoglin isoform which is increased in senescent endothelial cells [Blanco et al., 2008], and a soluble form of endoglin protein [Venkatesha et al., 2006]. Each of these exhibits functions that can be interpreted as dominant negative activity when co-expressed with L-endoglin [Venkatesha et al., 2006; Blanco et al., 2008]. The HHT mutational relevance of these data, and similar evidence for dominant negative activity of endoglin derived from recombinant intron-less endoglin cDNA expression constructs in vitro [Lux et al., 2000] is, however, not clear: when endogenous mutated endoglin protein species are examined by metabolic labelling of cells from HHT1 patients, it is rare for any mutant species to be detected [Pece et al., 1997; Pece-Barbara et al., 1999; Cymerman et al., 2000]. For only 2 of 16 ENG mutations studied could the mutated endoglin protein be detected in human umbilical vein endothelial cells or activated monocytes. These were splice site mutation c360+1G>A, resulting in an in-frame exon 3 skip [Pece et al., 1997; Cymerman et al., 2000], and missense substitution T157C [Cys53Arg] [Pece-Barbara et al., 1999; Cymerman et al., 2000]. In each case, the mutant proteins were retained intracellularly at low levels [Pece et al., 1997; Pece-Barbara et al., 1999; Cymerman et al., 2000].
To summarise, currently it is believed that in most if not all cases, HHT results from endoglin or ALK1 haploinsufficiency, that is the mutational inactivation of one allele, such that the remaining wild type allele is unable to contribute sufficient protein for normal function [Shovlin, 2010]. Recent attention has focused on the cellular mechanisms underlying the generation of HHT telangiectasia and arteriovenous malformations, invoking defective responses to angiogenic stimuli in particular settings [Park et al., 2009; Lebrin et al., 2010; Mahmoud et al., 2010]. In contrast, the molecular mechanisms involved in ENG HHT mutations have not been subjected to detailed recent scrutiny. In this article, we present new data that prompt a re-evaluation of the concepts regarding the most common HHT mutations.
Methods
HHT Gene Analyses
HHT causing mutations were downloaded from the HHT mutation database (www.hhtmutation.org at time of submission, now at www.arup.utah.edu/database/hht/) and classified into each of the conventional mutational types. Figures were generated using Microsoft Excel. The number of times each mutation in ENG and ACVRL1 had been reported in different families was recorded as a categorical variable. Graphs and one phase exponential decay models were constructed using GraphPad Prism 5.00 (GraphPad Software, San Diego, Calif., USA).
Premature Termination Codon Analyses
The alternate frame reads for the L-endoglin and S-endoglin were determined by ExPASy translation of FASTA RefSeq sequences NM_001114753.1 and NM_000118.2, and stop codons assigned to the endoglin exon map.
ENG mutations predicted to generate frameshift mutations were analysed by ExPASy (www.expasy.org) to translate and determine the exact site of the first stop codon generated. These were correlated to the known positions of stop codons in alternate reading frames. For stop codons generated directly by single nucleotide nonsense substitutions, exon position was determined by eye. The relative position of each stop codon to the final 3′ exon-exon boundary of L-endoglin and S-endoglin was determined using RefSeq sequences NM_001114753.1 and NM_000118.2, to classify the stop codons as either premature termination codons (PTCs; more than 50-55 nucleotides upstream of the final exon-exon boundary) or natural stop codons within 50-55 nucleotides of the final exon-exon boundary.
Endoglin Expression Data
The coding sequences for naturally occurring L-endoglin and S-endoglin were excised from the previously described pCEXV-EndoL and pCEXV-EndoS vectors [Bellón et al., 1993] and ligated into pcDNA3.1Zeo+ (Invitrogen). A construct corresponding to an exon 13 stop codon was created by PCR-based site-directed mutagenesis of the pCEXV-EndoL template to introduce a stop codon in place of the second cytoplasmic amino acid of endoglin (I588X). To provide an additional control for transient transfection (and to explore a role for the 2 cytoplasmic serine residues conserved between L- and S-endoglin), a full length L-endoglin construct in which Ser590 and Thr592 were mutated to alanines was also created by PCR-based site-directed mutagenesis of the pCEXV-EndoL template, employing a PCR overlap extension method (details available on request). Both constructs were amplified by PCR, ligated into pGEM®-T easy (Promega) for sequencing validation, prior to ligation into pcDNA3.1Zeo+. Insert orientation and sequence was confirmed at all stages and the final products resequenced prior to use. All transfections were performed using Polyfect reagent (Qiagen) according to the manufacturer's instructions. Luciferase activity was measured using a Luclite kit and TopCount luminometer, both from Packard Biosciences. Luciferase activity was measured 48 h after initial transfection.
L6E9 rat myoblast cells that had been mock-transfected, or stably expressing L-endoglin [Letamandía et al., 1998], were a kind gift of Dr. Carmelo Bernabeu, and maintained in DMEM/10% FBS/G418/L-glutamine, and penicillin/streptomycin. L6E9 cells stably expressing L-endoglin were transfected with empty vector (mock transfections), S-endoglin, or one of the endoglin mutants, and analysed within 48 h. For Western blots, total cell proteins were extracted from transiently and stably transfected cells using a Triton X-100/Tris/NaCl/EDTA solution, electrophoresed under non-reducing conditions on a 7.5% acrylamide gel (Bio-Rad), transferred to a nitrocellulose membrane, and blocked overnight in 5% skim milk powder in PBS-Tween. Endoglin protein was detected using the monoclonal mouse anti-human endoglin antibody SN6h (Dako) and an anti-mouse IgG-horse radish peroxidase conjugate (Sigma)/chemiluminescence (Renaissance, Dupont-NEN) strategy. For luciferase assays, mock and L-endoglin-expressing L6E9 rat myoblast cells were transfected with empty vector or one of the endoglin variants, and the p800PAI-1-Luc plasmid, a generous gift of Dr. D. Rifkin. Luciferase activity was measured 48 h after initial transfection, and raw data of 5 replicates entered into GraphPad Prism. For statistical analyses, group data were compared using the Kruskal Wallis test using a post estimation multiple comparison test (Dunn's) to derive pairwise p value statistics for the 3 transient transfectants compared to the parental L-transfected line. For Northern blots, mock and L-endoglin-expressing L6E9 rat myoblast cells were transiently transfected with empty vector or one of the endoglin variants, and incubated in the absence or presence of 3 μg/ml TGF-β1 (Sigma) for 24 h, before extraction of total RNA using TriReagent (Sigma). Approximately equal amounts of RNA were size-separated on a 1.5% agarose/formaldehyde gel, and transferred to a GeneScreen Plus membrane and fixed. A 32P-dATP-labelled fragment of exon 2 of the rat c-myc gene amplified using primers S 5′-CGACGAGGAAGAGAATTTGTATCA and AS 5′-GAATGGACGAGGTACAGGATT was hybridised to the membrane for subsequent autoradiographic exposure. Following appropriate decay, the membrane was then hybridised to a 32P-dATP-labelled fragment of rat β-actin (amplified using S 5′-AAAACTGGAACGGTGAAGGC, AS 5′-GCCTCAACACCTCAAACCAC), to allow for subsequent normalisation of loading differences. Image densities for each band were quantified using Adobe Photoshop to calculate the mean, median and standard deviation across the selected number of pixels.
Pervasive Transcription across Endoglin Locus – Evidence in Endothelial Cells
Total RNA was extracted from 7 different confluent cultures of proliferating primary human microvascular endothelial cells, as described in Shovlin et al. [2010]. Using RNA sequencing methods described in detail elsewhere (Mollet et al., manuscript in preparation, methodological details supplied), Illumina small RNA sample prep kit was used to prepare directional whole transcriptome libraries for RNAseq. For each of the 7 libraries, 8 pM was used for cluster generation and sequencing on separate lanes on Genome Analyser II, following the standard protocol for single 76-base reads. Image processing and base-calling was performed with RTA version 1.6.47.1. To generate the endoglin transcript data, the demultiplexed RNAseq reads were aligned to the human endoglin gene region from assembly hg18 (chr9: 129616500-129660000) using Seqmap [Jiang and Wong, 2008]. Base calls with phred scores below 20 were converted to Ns and alignments were performed allowing 2 mismatches per read and a minimum read length of 25 nucleotides. An average read depth of 2 was obtained outside repeat masked regions. Absolute read depth plots were created with R (www.r-project.org), highlighting regions for repeat masking in black on the negative axis.
Results
Breakdown of HHT Mutational Types
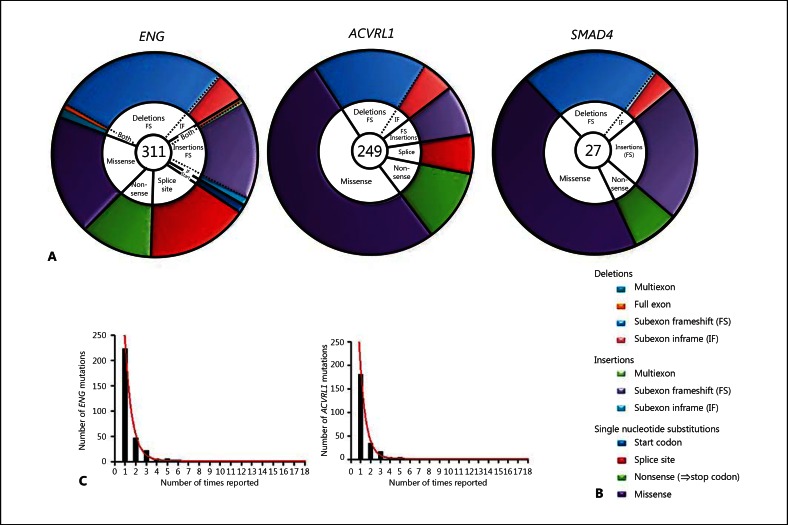
As illustrated in figure 1A, many more HHT mutations are reported for ENG and ACVRL1 than for SMAD4. In all 3 genes, HHT disease-causing mutations include insertions and deletions and single nucleotide substitutions. No individual mutation is particularly common: For ENG and ACVRL1, the number of times each individual mutation was reported corresponded to first-order exponential decay (ENG r2 0.997; ACVRL1 r2 0.997), suggesting that the pattern of mutations was random (fig. 1C).
Fig. 1.
Mutational types in HHT. A, B Comparison for all known HHT genes with pie chart representation of the different mutational types. The number in the centre of each pie refers to the total number of mutations reported at the time of analysis. The colour-coding explained in the schematic key in B is used to illustrate the relative prevalence of the different mutational types and to emphasise different genomic DNA mutational types with similar transcript consequences: single nucleotide substitutions are sub-classified conventionally as missense, nonsense, splice site and start codon mutations; deletions and insertions are sub-classified to emphasise the proportion generating nonsense codons and include complex rearrangements. Nonsense codons are generated by all direct nonsense single nucleotide substitutions (green); all subexon frameshift (FS) deletions (pale blue) and insertions (lilac) but no subexon inframe (IF) insertions or deletions; approximately two-thirds of multiexon and full exon deletions, insertions and rearrangements (aqua, orange and pale green), and approximately two-thirds of splice site mutations (red). The formal assignment of splice site mutations and multiexon deletions to frameshift or non-frameshift groupings depends upon evidence of the mutant endoglin RNA species generated – activation of cryptic splice sites may generate an inframe, detectable RNA species [Shovlin et al., 1997] instead of a more obvious exon skip that would lead to a frameshift. C The distribution of the number of times each individual ENG or ACVRL1 mutation was reported (illustrated as columns), with the respective best fit one phase exponential decay model illustrated.
As shown in figure 1A, B, single nucleotide substitutions in all 3 genes include nonsense mutations (which generate 1 of the 3 triplet stop codons), missense substitutions (particularly common in ACVRL1) and splice site mutations. Mutations of the start site have been reported only for ENG to date.
For subexon and multiexon insertion and deletion mutations, in-frame mutations, where the reading frame is restored after deletion/insertion of one or more groups of 3 nucleotides, are less common than frameshift mutations. Frameshift mutations generate stop codons because the 2 alternate reading frames contain frequent stop codons, with one usually reached within 1 or 2 exons of the frameshift. For example, for the open reading frame of ENG, the 2 alternate 5′ to 3′ reading frames contain 18 and 16 stop codons, respectively.
Stop codon generation is also predicted for approximately two-thirds of multiexon deletions/insertions, and splice site mutations, because of frameshifts in the RNA transcript. Using these calculations, 57% (95% confidence interval 53%, 61%) of HHT mutations reported on the HHT database will lead to stop codons, either as a result of frameshifts or direct nonsense substitutions. The proportion is higher for ENG (67% [95% confidence interval 62%, 73%]) and SMAD4 (65% [55%, 75%]) than for ACVRL1 (42% [35%, 47%]) (p < 0.0001).
Premature Termination Codons and Nonsense-Mediated Decay
Nonsense-mediated decay (NMD) has previously been implicated in the failure to identify RNA species for individual ENG nonsense mutations [Shovlin et al., 1997]. Examining the mechanisms of NMD allows a more global approach to HHT mutations, since in mammals, the PTC required to trigger NMD [Doma and Parker, 2007] is required to be a stop codon sited more than 50-55 nucleotides upstream of the final exon-exon junction [Maquat, 2005]. The distribution of stop codons generated by HHT mutations was therefore examined.
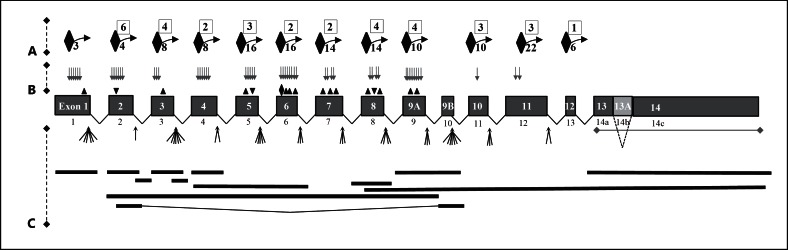
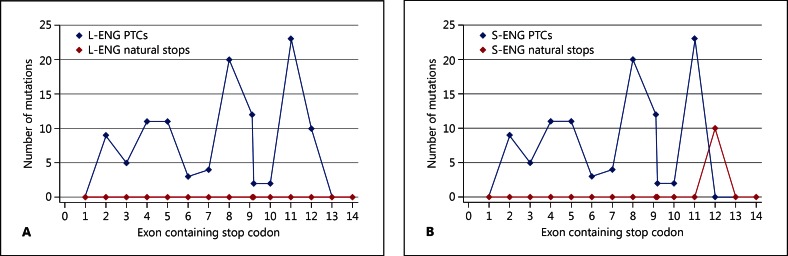
First, each mutation was assigned to 1 of 3 groups according to whether these could or could not generate a stop codon (fig. 2). Focussing on the 111 mutations which definitely generate stop codons directly or by frameshift mutations (fig. 2, group A), the sites of predicted stop codons were analysed. As shown in table 1 and figure 3, these stop codons were sited in exons 2 to 12. Only one was within 55 nucleotides of the final exon-exon boundary for L-endoglin. The remainder fulfil the requirements to act as PTCs, suggesting that the mutants will fail to generate an RNA product from which translation of truncated proteins could occur. For the minor alternatively spliced isoform, S-endoglin, 8 mutations were predicted to result in natural stop codons in exon 12.
Fig. 2.
Schematic of ENG HHT mutations mapped to exons in L-endoglin, classified into 3 groups. A Site of stop-generating mutations: nonsense (number in boxes) and frameshift from indels (number below curved arrow, indicating stop codons generated downstream by frameshift to an alternate reading frame). B Non-stop codon-generating mutations: missense (grey arrows), in-frame insertions (▼), and in-frame deletions (▲). C Mutations where 2 out of 3 are predicted to generate stop codons: splice site mutations (clustered arrows), and large deletions, insertions and rearrangements (black horizontal bars). White numbers in exons: exon numbering as in HHT database (1-9A, 9B-14, denoting S-endoglin retained intron as exon 13A). Black numerals below exons: RefSeq numbering (1-14c, denoting S-endoglin retained intron as exon 14b).
Table 1.
Detailed breakdown of ENG stop codon-generating mutations, by exon in which stop codon occurs (site of origin is indicated in fig. 2)
| Stop codon exon | Mutations |
|---|---|
| 1 | – |
| 2 | c.63delC (Thr22fs); c.90T>A (Cys30X); c.97C>T (Gln33X); c.121G>T (Glu41X); c.142C>T (Gln48X); c.159C>A (Cys53X); c.166C>T (Gln56X); c.107delG (Gly36fs); c.115delA (Arg39fsX42) |
| 3 | c.229C>T (Gln77X); c.247C>T (Gln83X); c.277C>T (Arg93X); c.360C>A (Tyr120X); c.277delC (Arg93fs) |
| 4 | c.214_215delCC (Pro72AsnfsX76); c.298_299delAG (Ser100CysfsX48); c.305delA (Asn102ThrfsX61); c.326_327delTC (Leu109fs); |
| c.332_338delCCCTGGG (Ala111fs); c.344delC (Pro115fs); c.447G>A (Trp149X); c.511C>T (Arg171X); c.392delC (Pro131fs); c.461delG (Gly154fsX162); c.472_472delCT (Ser158fsX160) | |
| 5 | c.496delC (Gln166fsX221); c.526C>T (Glu176X); c.587G>A (Trp196X); c.588G>A (Trp196X); c.574delC (ArgfsX221); c.617delG (Gly206Alafs); c.640_644delGGCC (Gly214fs); c.644delA (His215fs); c.647delA (Lys216fs); c.604_607delinsCCC (Ala202ProfsX20); c.586_604delTGGCGGCC (p.Trp196fs …) |
| 6 | c.715G>T (Glu239X); c.782G>A (Trp261X); c.704delC (Thr235fs) |
| 7 | c.537_538delGT (Ser180fs); c.591_619del29 (Pro198fsX125); c.831C>G (Tyr277X); c.889C>T (Gln297X) |
| 8 | c.653insC (His219fs); c.657_658delCA (Ile220fsX332); c.682_686delTCGGC (Ser228fs); c.733delG (Gly245fs); c.736delG (Asp246fs); c.740delT (Asp248fs); c.771delC (Pro257fsX358); c.772delT (Tyr258fs); c.775delG (Val259fs); c.785_789delTCATC (Leu262fsX331); c.834_837delCTTC (Phe279fs); c.893delG (Gly298fsX358); c.967_968delGT (Val323fs); c.820_826delACTGGAGinsTTGAAGGTCTTTCCAGAGAAAAAC (Thr274fsX338); c.1010C>G (Ser337X); c.1050T>A (Cys350X); c.1078C>T (Gln360X); c.1123G>T (Glu375X); c.995delG(Gly332fsX358); c.1085delA (Lys362fs) |
| 9A | c.1089_1090delTG (Ala363fs394X); c.1097_1119del23(Asp366GlufsX22); c.1111_1133del23(Val371fs); c.1120_1123delAAAG (Lys374fs); c.1124_1125delAG (Leu375fs); c.1243C>T (Gln415X); c.1186delG (Ala396fsX420); c.1195delA (Arg399fs); c.1199delG (Gly400fsX420); c.1205delA (Lys402SerfsX19); c.1206delG (Leu402fs); c.1255delA (Ser419fs) |
| 9B | c.1292C>A (ser431X); c.1306C>T (Gln436X) |
| 10 | c.1365C>A (Tyr455X); c.1414C>T (Gln472X) |
| 11 | c.1268delA (Asn423fsX490); c.1310delG c.1334delT (Met445Argfs); c.1342_1343delCT (Ser449fsX490); c.1346_1347delCT (Ser449fsX499); c.1347_1350delTTTC (Phe450fs); c.1410delG (Gly470fsX490); c.1415_1417delAGA insGT (Gln472fsX490); c.1513G>T (Glu505X) c.1611C>A (Tyr537X) c.1678C>T (Gln560X); c.1684C>T (Gln562X) c.1432_1433delAG (Arg478fs); c.1472_1475delACAG (Asp491-AlafsX26); c.1478delG (Ser492fsX516); c.1479delC (His494fsX517); c.1526delG (Gly509fs); c.1550_1551delTG (Val517fsX525); c.1553_1554delGC (Ser518fs); c.1554_1555delCC (Leu519fs); c.1605_1627del23 (535fs); c.1609delT (Tyr537fsX551); c.1630delA(Thr544fs) |
| 12 | c.1655delC (Ala552fs); c.1657delC (Leu553fs); c.1672_1684delGGGTCT (Gln558fsX568); c.1689_1699delAGTCCA (Glu563fsX574); |
| 13 | c.1687delG (Glu563LysfsX10); c.1698delG (Arg566fs); c.1712delG (Arg571fs); c.1715T>A (Leu572X) |
| 14 | c.1733_1737delACCTG (Asp578fs) |
Note that the table excludes 2 reported frameshift mutations where the genomic DNA mutation translated inframe (c.993_1134del; c.887_918del32, 919_920insCAAGCTCCCAG); 2 reported intronic deletions in introns 4 and 7 (c.524_689del; c.992-25_1120del152); the 5'UTR-exon 1 deletion deleting the start codon; and c.1-?_67+?del; c.896del186; and c1350C>T which translate as no amino acid change.
Fig. 3.
Summary of group A ENG mutations generating stop codons. Results are presented for L-endoglin transcript (the major transcript) (A) and for the S-endoglin transcript (B) for which, due to retention of the final intron, the final exon-exon junction is between exons 12 and 13 and not exons 13 and 14 (see fig. 4), with implications for stop codon mutations in exon 12.
To define whether these distributions were likely to have occurred by chance, the pattern of stop codons within the 3 alternate reading frames for ENG was examined further. Throughout all 3 reading frames, the stop codon prevalence between the ATG start and true stop codons was 34/658 codons. In any of the 3 reading frames, L-endoglin has only a single stop codon throughout the last 55 nucleotides of exon 13 and the 124 coding nucleotides of exon 14. The probability that no stop codons would be present in any frame for these 59 codons is ([658–34]/658)59, that is p < 0.05.
Natural Stop Codon Considerations
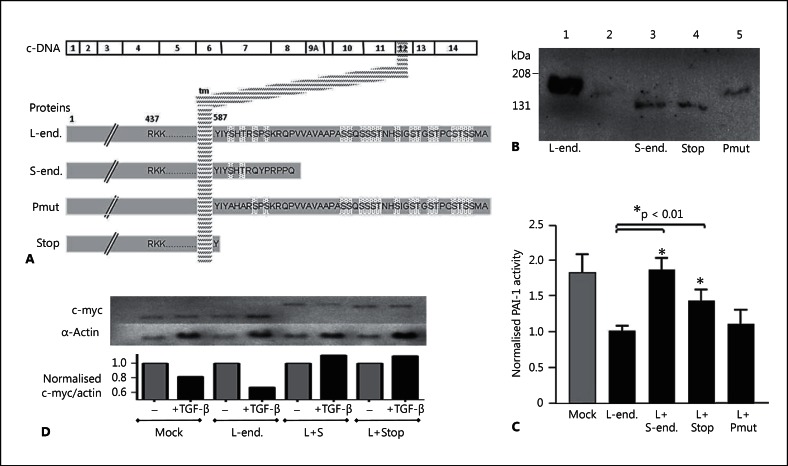
It would be predicted that were HHT nonsense mutations to generate a natural stop codon within the last 55 nucleotides of exon 13 and the 124 coding nucleotides of exon 14, the mutated L-endoglin protein moiety (with the transmembrane domain encoded by exon 12) might be stably expressed. We therefore re-examined unpublished data that we had performed as part of a previous project, generating expression constructs encoding a highly relevant moiety, an endoglin species with the shortest possible cytoplasmic tail equivalent to a stop codon in the second cytoplasmic codon (I588X, fig. 4A). Under non-reducing conditions, this, and a control construct encoding a species in which the 2 cytoplasmic residues conserved between L- and S-endoglin (Ser590 and Thr592) were mutated to alanines, were expressed as dimers, at similar expression levels to transiently transfected S-endoglin (fig. 4B).
Fig. 4.
Natural and recombinant endoglin variants. A Illustration of the variants with respect to the transmembrane domain and cytoplasmic tail (end. = endoglin). L-endoglin and S-endoglin are naturally occurring endoglin species; the artificial mutants have the 2 cytoplasmic serine residues conserved between L- and S-endoglin mutated to alanines (Pmut), or a stop codon in place of the second cytoplasmic amino acid of endoglin (I588X; Stop). B Western blot data of total cell protein extracted from L6E9 cells stably transfected with L-endoglin (lane 1), or the mock-transfected L6E9 cells transiently transfected with expression vectors for S-endoglin (lane 3), the truncated L-endoglin I588X stop mutant (lane 4) or the phosphorylation mutant (lane 5). C Modulation of PAI-1 promoter activity by stable expression of L-endoglin in L6E9 rat myoblasts (lanes 2-5) and subsequent modulation by transient transfection of empty vector (lanes 1 and 2) or other endoglin species (lanes 3-5): Basal PAI-1 activity was significantly (p < 0.0001) downregulated by stable transfection of L-endoglin. Transient transfection of S-endoglin, the truncation mutant, but not the phosphorylation mutant resulted in PAI-1 activities not different to the mock stable transfectants, L6E9 rat myoblasts not expressing L-endoglin. D Modulation of c-myc and α-actin expression in mock-transfected L6E9 or L6E9 stably expressing L-endoglin following transient transfection with an empty vector (mock; L-endoglin); S-endoglin or the truncated endoglin construct, before 24 h treatment with normal media or media supplemented with 3 μg/ml TGF-β1.
Such expressed moieties would have the potential to have functional consequences, potentially demonstrating the dominant negative activities, as observed for S-endoglin [Blanco et al., 2008] and soluble endoglin [Venkatesha et al., 2006]. This was examined by testing the effects of these constructs in established functional assays. As previously demonstrated [Letamandía et al., 1998; Blanco et al., 2008], activity of the plasminogen activator inhibitor (PAI-1) promoter was downregulated in L6E9 myoblasts stably expressing L-endoglin, and this response was abrogated in cells transiently overexpressing S-endoglin (fig. 4C). Transiently transfecting the truncated endoglin species into L6E9 myoblasts stably expressing L-endoglin also appeared to partially restore the activity of the PAI-1 promoter, whereas the full-length cytoplasmic tail phosphorylation mutant had no effect (fig. 4C). S-endoglin and the truncation mutant also demonstrated similar responses in a c-myc assay where TGF-β1 downregulated c-myc [Letamandía et al., 1998; Blanco et al., 2008]: Once corrected for differential loading, the fall in c-myc in response to TGF-β1, a response enhanced in cells stably transfected with L-endoglin, was not observed when L6E9 cells stably expressing L-endoglin were transiently transfected with S-endoglin or the truncated mutant (fig. 4D). Furthermore, both S-endoglin and the truncation mutant also altered the predominant c-myc transcript expressed [Bentley and Groudine, 1986, 1988] (http://genome.ucsc.edu). These data demonstrate that an endoglin species containing a post-transmembrane domain stop codon, is not only likely to be translated, but also may have similar activities to S-endoglin, the production of which is highly regulated [Blanco et al., 2008]. Such moieties would not be predicted to act as endoglin nulls, as for the PTC-containing mutants.
Additional Complexity for Multiexon Deletions
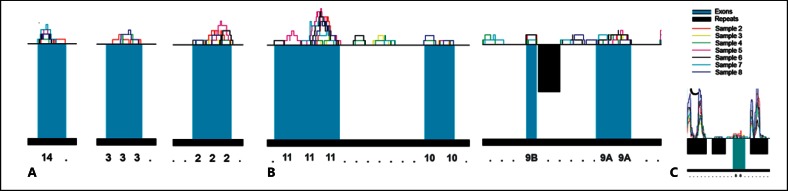
Multiexon deletions and insertions are frequently reported in HHT (fig. 1). To assess if the loss of intronic sequences spanned by the deletions were likely to be important, we examined data from our directional RNAseq programme of RNA libraries constructed from 7 different primary human microvascular endothelial cell cultures. As illustrated in figure 5A, there was a quasinormal distribution of transcripts spanning ENG exons, with the expected sharp delineation of exon-intron borders usually observed. Of importance to the large multiexon deletions of ENG in HHT patients, however, numerous transcribed fragments from the intronic regions were consistently detected in these endothelial-specific libraries (fig. 5B). In some cases, these were recognised to align to repetitive regions in the genome, and origin from the ENG locus could not be assumed (inset, fig. 5C), but many represented unique alignments to the ENG locus (fig. 5B). Importantly, as a directional sequencing strategy had been employed, we could assign the strand of origin of transcripts. In all libraries, the majority of intronic transcripts were from the complementary strand to ENG. In these healthy low passage number endothelial cells, there were no alignments to the final intron retained in S-endoglin.
Fig. 5.
Transcribed fragments across regions of the ENG locus deleted in HHT mutations. Colour-coded R-plot data for 7 endothelial cell libraries, with exons represented by blue boxes, repeat regions by black boxes, and individual library alignments by the coloured lines (key, samples 2-8). Note ENG is on the minus strand, so 5′ to 3′ is represented right to left. A Replicate alignments to exons 2, 3 and 14. B Alignments to regions spanning exon 9A-11, demonstrating replicate alignments to exons and to non-repetitive intronic regions. C Illustration of non-unique alignments to repetitive regions (including the Alu elements responsible for a 2-kb ENG deletion spanning exon 8 [Shovlin et al., 1997]). Note that the transcripts responsible for these very high alignments to repetitive regions will have originated throughout the genome and are not necessarily from the ENG locus.
Discussion
Modern molecular data and insights need to be incorporated into our understanding of mutational mechanisms of disease. The data presented in this article highlight the relevance of NMD for PTC-generating mutations, and potential implications of intronic transcribed fragments from loci deleted or duplicated by large genomic deletions, insertions or rearrangements.
For PTC-generating endoglin mutations, current molecular perspectives suggest that the majority will fail to generate an RNA product from which translation of truncated proteins could occur. For ENG and SMAD4, the most commonly reported mutations generate stop codons either directly or indirectly by out-of-frame insertions or deletions. These are often assumed to have the potential to generate truncated proteins. Where endogenous mutated endoglin protein species are examined by metabolic labelling of cells, however, it is rare for any mutant species to be detected [Pece et al., 1997; Pece-Barbara et al., 1999; Cymerman et al., 2000]. The data presented here highlight that such mutations are most likely to result in NMD of the mutated mRNA, as discussed further below.
One of the puzzles in HHT circles has been the absence of detected mutations in the final coding exon. At first sight, this may suggest that S-endoglin, which utilises only 7 amino acids of the L-endoglin cytoplasmic tail (fig. 2), could be the moiety responsible for HHT pathogenesis. However, the presented data provide alternate explanations for the lack of HHT genomic mutations at these sites. The absence of frameshift mutations leading to exon 13 and 14 stop codons is explained by the absence of any alternate reading frame stop codons. For missense substitutions, the absence of an effect of substituting the conserved serine residues (fig. 4) implies that there is no single cytoplasmic amino acid residue where modifications have been shown to generate a null allele. The absence of nonsense mutations requiring a specific nucleotide change may not be surprising given that only 97 single nucleotide substitutions were reported as HHT mutations for the entire ENG coding sequence (www.hhtmutation.org at time of analysis, now at www.arup.utah.edu/database/hht/), but a further explanation is detailed below. Overall, the data begin to explain the missing mutations, and neither support or refute the possibility that mutated S-endoglin causes HHT.
The detection of transcribed transcripts within intronic regions of the ENG locus deleted in HHT disease-causing mutations should not be surprising. Cellular RNA is normally quoted as comprising mRNAs (∼2-4%), rRNAs (∼80%), tRNAs (∼15%) and small RNAs (∼1%) [Lindberg and Lundeberg, 2010]. However, the development of high-throughput technologies such as tiling arrays and next-generation sequencing have made it evident that a large proportion of the eukaryotic intronic and intergenic regions of the genome are transcribed, in what is known as ‘pervasive transcription’. The biological consequences are still being elucidated and although most of these RNA fragments are not protein coding, some are very stable, highly conserved or regulated under different conditions, suggesting that they are functional with potential regulatory roles [Kapranov et al., 2002; Berretta and Morillon, 2009]. The detection of such transcripts within intronic regions of the ENG locus deleted in HHT disease-causing mutations highlights additional transcript changes that will be incurred by these multiexon deletion, insertion or rearrangement ENG mutations in HHT.
Overall, the data suggest that any phenotypic differences ascribable to different ENG mutation types are unlikely to segregate by the conventional mutational subgroupings illustrated in figure 1. The key distinction is the generation or non-generation of PTCs, with multiexon deletions, insertions or rearrangements predicted to have a more complex phenotype because affected patients will have a different copy number of other intronic transcribed fragments. Such predictions can and should be tested experimentally by the laboratories that have derived endothelial cells from HHT patients harbouring different ENG mutations. For example, loss of an exon due to a splice-site mutation and exon skip may differ functionally to loss of the same exon due to a large deletion spanning the exon and neighbouring intronic regions.
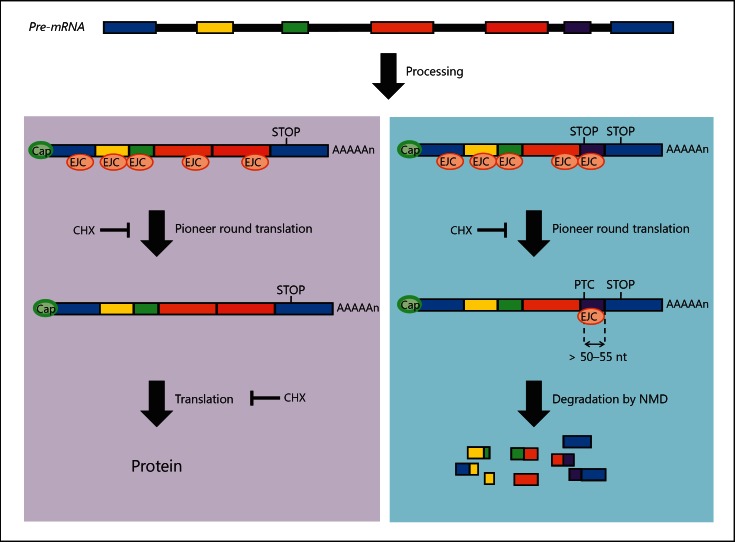
To understand why mRNAs containing stop codon mutations will usually be degraded, it is helpful to consider NMD in a more general context. NMD is one of a group of highly conserved quality control surveillance mechanisms operating in normal cells to recognise and degrade defective mRNAs, which if translated could produce harmful aberrant proteins. Such RNAs can be generated independently of any genomic DNA mutations, as a result of errors during transcription, RNA processing or RNA packaging. NMD is known to be translation-dependent in eukaryotes, as it can be reversed by protein synthesis inhibitors such as anisomycin and cycloheximide [Carter et al., 1996]. NMD also depends upon normal RNA splicing mechanisms: PTC-containing mRNAs derived from intronless genes (where no splicing occurs) do not undergo NMD [Brocke et al., 2002]. As illustrated in figure 6, during RNA splicing, exon-exon junctions are marked by exon junction complexes (EJCs) which are deposited ∼20-24 nucleotides upstream of each exon-exon junction. The current NMD model in mammals suggests that during the initial (pioneer) round of translation, the scanning ribosome displaces all the EJCs on an mRNA which would normally be upstream of the stop codon. Upon recognition of the stop codon, a termination complex is formed. In PTC-containing mRNAs, however, EJCs downstream of the PTC are not displaced by scanning the ribosome. This allows the termination complex to interact with the EJC, triggering NMD [Maquat, 2005]. Despite the frequency with which they are generated, PTC-containing mRNAs are low in abundance in vivo, unless unmasked by treating cells with protein synthesis inhibitors such as CHX [Carter et al., 1996], or RNA interference depletion of essential NMD factors [McGlincy and Smith, 2008].
Fig. 6.
Nonsense-mediated decay (NMD). A single gene locus is transcribed into a pre-mRNA containing all exons and introns. The left panel illustrates normal splicing when proteins of the exon junction complex (EJC) are deposited ∼20-24 nucleotides upstream of exon-exon boundaries. EJCs are displaced by the scanning ribosome in the ‘pioneer’ round of translation. The right panel illustrates an RNA with a premature termination codon (PTC), that is a stop codon more than 50-55 nucleotides upstream of the final exon-exon boundary. During the pioneer round of translation, EJCs downstream of PTCs are not displaced, triggering degradation by NMD, an mRNA surveillance system. Cap = 7-Methylguanosine; CHX = cycloheximide, a naturally occurring antibiotic that inhibits translation and therefore inhibits NMD; STOP = termination codon.
Historically, PTC-containing mRNAs were thought to be translated into truncated proteins based on experimental evidence of gain of function or dominant negative activities. Such studies used in vitro-generated cDNA constructs [Gong et al., 2007]. As for intronless genes [Brocke et al. 2002], cDNA constructs do not undergo splicing, do not contain EJCs, and therefore do not trigger NMD in mammalian cell lines [Gong et al., 2007]. Thus, many diseases thought to be due to gain of function or dominant negative effects may be more likely to be due to loss of function or haploinsufficiency. However, NMD is not generally appreciated as a concept relevant to DNA mutations. In September 2011, Web of Science search terms ‘nonsense, decay, mutation’ identified only 972 publications (including 50-90 per year since 2006), compared to 32,112 referring to ‘truncated, protein, mutation’.
The absence of ENG exon 13 and 14 mutations in HHT point to other generic processes. The widespread prevalence of insertions and deletions (indels, estimated to affect 30% of the human genome [Zhang et al., 2009]) highlights the importance of genomic sequence toleration of out-of-frame indels. Genome-wide analyses by others have addressed mechanisms relating to codon nucleotide use [Cusack et al., 2011; Wilke et al., 2011], but for ENG, we examined from a different perspective. For indels generating stop codons more than 55 nucleotides upstream of the final exon-exon boundary, the resultant PTCs will trigger NMD. For coding sequences beyond this safety net, it would be predicted that alternate strategies need to be employed. The absence of stop codons in any of the 3 reading frames throughout the last 55 nucleotides of exon 13, and the 124 coding nucleotides of exon 14 suggest that for L-endoglin, the most prevalent endoglin species, selective pressures may have limited the opportunity for indels to generate a natural stop codon which would generate functional truncated species (fig. 4) [Lux et al., 2000]. There is no particular reason why the phenotypic result would resemble that of a null allele and cause HHT. Yet for endoglin, there is 1 HHT-generating frameshift mutation predicted to generate a natural stop codon in exon 14 (fig. 3). ENG mutation c.1733_1737delACCTG changes 170 amino acids before a novel natural stop codon in exon 14. Similarly, were S-endoglin to be implicated in HHT pathogenesis, there are 8 natural stop codons generated in exon 12 by frameshift HHT mutations in exons 11 and 12 (fig. 3). None of these should undergo NMD. However, all 9 disrupt the transmembrane domain encoded by exon 12: functional nulls at a protein level might therefore explain the phenotypic similarity with other ENG null alleles.
In conclusion, using modern molecular perspectives to re-examine HHT mutations suggests that any phenotypic differences are unlikely to segregate by the conventional mutational subgroupings. Not only may additional data be required before a pathogenic role can be ascribed to potential missense ENG mutations [McDonald et al., 2009], but similar cautions may also need to be applied to small inframe indels to prevent provision of incorrect genotypic predictions. From the data presented in this article, we predict that instead of nonsense or missense mutations being more severe (as predicted by earlier models), it should be the multiexon deletions that have a more complex phenotype. The data also lead to the prediction that while nonsense mutations may be identified in the final exons of endoglin, when these occur, the phenotype will not be that of HHT.
Acknowledgements
This work was supported by the British Heart Foundation. C.L.S. is also grateful for support from the NIHR Biomedical Research Centre Funding Scheme. The authors thank Ellen Thomas for advice on SPRI purification, Sonal Shah and Nathalie Lambie for technical support, and Illumina TechSupport for helpful discussions. The endoglin expression work was supported by the Wellcome Trust. The L6E9 rat myoblast cells and pCEXV-EndoL and pCEXV-EndoS vectors were a kind gift of Dr. Carmelo Bernabeu. The p800PAI-1-Luc plasmid was a generous gift of Dr. D. Rifkin.
References
- Abdalla S, Letarte M. Hereditary haemorrhagic telangiectasia: current views on genetics and mechanisms of disease. J Med Genet. 2006;43:97–110. doi: 10.1136/jmg.2005.030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly S. HHT is not a TGFbeta disease. Blood. 2008;111:478. [Google Scholar]
- Bayrak-Toydemir P, McDonald J, Akarsu N, Toydemir RM, Calderon F, et al. A fourth locus for hereditary hemorrhagic telangiectasia maps to chromosome 7. Am J Med Genet. 2006a;140:2155–2162. doi: 10.1002/ajmg.a.31450. [DOI] [PubMed] [Google Scholar]
- Bayrak-Toydemir P, McDonald J, Markewitz B, Lewin S, Miller F, et al. Genotype-phenotype correlation in hereditary hemorrhagic telangiectasia. Am J Med Genet A. 2006b;140:463–470. doi: 10.1002/ajmg.a.31101. [DOI] [PubMed] [Google Scholar]
- Bellón T, Corbi A, Lastres P, Calés C, Cebrián M, et al. Identification and expression of two forms of the human transforming growth factor-beta-binding protein endoglin with distinct cytoplasmic domains. Eur J Immunol. 1993;23:2340–2345. doi: 10.1002/eji.1830230943. [DOI] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. Novel promoter upstream of the human c-myc gene and regulation of c-myc expression in B-cell lymphomas. Mol Cell Biol. 1986;6:3481–3489. doi: 10.1128/mcb.6.10.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL, Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988;53:245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino P, Deckers M, Lebrin F, ten Dijke P. Transforming growth factor-beta signal transduction in angiogenesis and vascular disorders. Chest. 2005;128(6 Suppl):585S–590S. doi: 10.1378/chest.128.6_suppl.585S. [DOI] [PubMed] [Google Scholar]
- Bideau A, Brunet G, Heyer E, Plauchu H, Robert J-M. An abnormal concentration of cases of Rendu-Osler disease in the Valserine valley of the French Jura: a geneological and demographic study. Ann Hum Biol. 1992;19:233–247. doi: 10.1080/03014469200002112. [DOI] [PubMed] [Google Scholar]
- Blanco FJ, Santibanez JF, Guerrro-Esteo M, Langa C, Vary CP, Bernabeu C. Interactions and functional interplay between endoglin and ALK-1, two components of the endothelial transforming growth factor-beta receptor complex. J Cell Physiol. 2005;204:574–584. doi: 10.1002/jcp.20311. [DOI] [PubMed] [Google Scholar]
- Blanco FJ, Grande MT, Langa C, Oujo B, Velasco S, et al. S-endoglin expression is induced in senescent endothelial cells and contributes to vascular pathology. Circ Res. 2008;103:1383–1392. doi: 10.1161/CIRCRESAHA.108.176552. [DOI] [PubMed] [Google Scholar]
- Bossler AD, Richards J, George C, Godmilow L, Ganguly A. Novel mutations in ENG and ACVRL1 identified in a series of 200 individuals undergoing clinical genetic testing for hereditary hemorrhagic telangiectasia (HHT): correlation of genotype with phenotype. Hum Mutat. 2006;27:667–675. doi: 10.1002/humu.20342. [DOI] [PubMed] [Google Scholar]
- Bourdeau A, Cymerman U, Paquet ME, Meschino W, McKinnon WC, et al. Endoglin expression is reduced in normal vessels but still detectable in arteriovenous malformations of patients with hereditary hemorrhagic telangiectasia type 1. Am J Pathol. 2000;156:911–923. doi: 10.1016/S0002-9440(10)64960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocke KS, Neu-Yilik G, Gehring NH, Hentze MW, Kulozik AE. The human intronless melanocortin 4-receptor gene is NMD insensitive. Hum Mol Genet. 2002;11:331–335. doi: 10.1093/hmg/11.3.331. [DOI] [PubMed] [Google Scholar]
- Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Cole SG, Begbie ME, Wallace GMF, Shovlin CL. A new locus for hereditary haemorrhagic telangiactasia (HHT3) maps to chromosome 5. J Med Genet. 2005;42:577–582. doi: 10.1136/jmg.2004.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack BP, Arndt PF, Duret L, Roest Crollius H. Preventing dangerous nonsense: selection for robustness to transcriptional error in human genes. PLoS Genet. 2011;7:e1002276. doi: 10.1371/journal.pgen.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymerman U, Vera S, Pece-Barbara N, Bourdeau A, White RI, Jr, et al. Identification of hereditary hemorrhagic telangiectasia type I in newborns by protein expression and mutation analysis of endoglin. Pediatr Res. 2000;47:24–35. doi: 10.1203/00006450-200001000-00008. [DOI] [PubMed] [Google Scholar]
- Dakeishi M, Shioya T, Wada Y, Shindo T, Otaka K, et al. Genetic epidemiology of hereditary hemorrhagic telangiectasia in a local community in the northern part of Japan. Hum Mutat. 2002;19:140–148. doi: 10.1002/humu.10026. [DOI] [PubMed] [Google Scholar]
- David L, Mallet C, Mazerbourg S, Feige JJ, Bailly S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood. 2007;109:1953–1961. doi: 10.1182/blood-2006-07-034124. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Sanz-Rodriguez F, Zarrabeitia R, Perez-Molino A, Morales C, et al. Mutation study of Spanish patients with hereditary hemorrhagic telangiectasia and expression analysis of endoglin and ALK1. Hum Mutat. 2006;27:295. doi: 10.1002/humu.9413. [DOI] [PubMed] [Google Scholar]
- Gallione CJ, Klaus DJ, Yeh EY, Stenzel T, Xue Y, et al. Mutation and expression analysis of the endoglin gene in hereditary hemorrhagic telangiectasia reveals null alleles. Hum Mutat. 1998;11:286–294. doi: 10.1002/(SICI)1098-1004(1998)11:4<286::AID-HUMU6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Gallione C, Repetto GM, Legius E, Rustgi AK, Schelley SL, et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia is associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- Gallione C, Aylsworth AS, Beis J, Berk T, Bernhardt B, et al. Overlapping spectra of SMAD4 mutations in juvenile polyposis (JP) and JP-HHT syndrome. Am J Med Genet A. 2010;152A:333–339. doi: 10.1002/ajmg.a.33206. [DOI] [PubMed] [Google Scholar]
- Gong Q, Zhang L, Vincent GM, Horne BD, Zhou Z. Nonsense mutations in hERG cause a decrease in mutant mRNA transcripts by nonsense-mediated mRNA decay in human long-QT syndrome. Circulation. 2007;116:17–24. doi: 10.1161/CIRCULATIONAHA.107.708818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumans MJ, Lebrin F, Valdimarsdottir G. Controlling the angiogenic switch: a balance between two distinct TGF-b receptor signaling pathways. Trends Cardiovasc Med. 2003;13:301–307. doi: 10.1016/s1050-1738(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Govani FS, Shovlin CL. Hereditary haemorrhagic telangiectasia: a clinical and scientific review. Eur J Hum Genet. 2009;17:860–871. doi: 10.1038/ejhg.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govani FS, Shovlin CL. Fine mapping of the hereditary haemorrhagic telangiectasia (HHT)3 locus on chromosome 5 excludes Sprouty 4, VE-cadherin 2 and other interval genes. J Angiogen Res. 2010;11:15. doi: 10.1186/2040-2384-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmacher AE, Marchuk DA, White RI. Hereditary hemorrhagic telangiectasia. New Engl J Med. 1995;333:918–924. doi: 10.1056/NEJM199510053331407. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, tenDijke P. TGF-b signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kjeldsen AD, Vase P, Green A. Hereditary haemorrhagic telangiectasia: a population-based study of prevalence and mortality in Danish patients. J Intern Med. 1999;245:31–39. doi: 10.1046/j.1365-2796.1999.00398.x. [DOI] [PubMed] [Google Scholar]
- Kjeldsen AD, Møller TR, Brusgaard K, Vase P, Andersen PE. Clinical symptoms according to genotype amongst patients with hereditary haemorrhagic telangiectasia. J Int Med. 2005;258:349–355. doi: 10.1111/j.1365-2796.2005.01555.x. [DOI] [PubMed] [Google Scholar]
- Lebrin F, Mummery C. Endoglin-mediated vascular remodeling: mechanisms underlying hereditary hemorrhagic telangiectasia. Trends Cardiovasc Med. 2008;18:25–32. doi: 10.1016/j.tcm.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Lebrin F, Goumans MJ, Jonker L, Carvalho RL, Valdimarsdottir G, et al. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 2004;23:4018–4028. doi: 10.1038/sj.emboj.7600386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrin F, Srun S, Raymond K, Martin S, van den Brink S, et al. Thalidomide stimulates vessel maturation and reduces epistaxis in individuals with hereditary hemorrhagic telangiectasia. Nat Med. 2010;16:420–428. doi: 10.1038/nm.2131. [DOI] [PubMed] [Google Scholar]
- Lesca G, Olivieri C, Burnichon N, Pagella F, Carette MF, et al. Genotype-phenotype correlations in hereditary hemorrhagic telangiectasia: data from the French-Italian HHT network. Genet Med. 2007;9:14–22. doi: 10.1097/gim.0b013e31802d8373. [DOI] [PubMed] [Google Scholar]
- Letamandía A, Lastres P, Botella L, Raab U, Langa C, et al. Role of endoglin in cellular responses to transforming growth factor-b. A comparative study with betaglycan. J Biol Chem. 1998;273:33011–33019. doi: 10.1074/jbc.273.49.33011. [DOI] [PubMed] [Google Scholar]
- Letteboer TGW, Mager JJ, Snijder RJ, Koeleman BP, Lindhout D, et al. Genotype-phenotype relationship in hereditary haemorrhagic telangiectasia. J Med Genet. 2006;43:371–377. doi: 10.1136/jmg.2005.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg J, Lundeberg J. The plasticity of the mammalian transcriptome. Genomics. 2010;95:1–6. doi: 10.1016/j.ygeno.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Lux A, Gallione CJ, Marchuk DA. Expression analysis of endoglin missense and truncation mutations: insights into protein structure and disease mechanisms. Hum Mol Genet. 2000;9:745–755. doi: 10.1093/hmg/9.5.745. [DOI] [PubMed] [Google Scholar]
- Mahmoud M, Allinson K, Zhai Z, Oakenfull R, Ghandi P, et al. Pathogenesis of arteriovenous malformations in the absence of endoglin. Circ Res. 2010;106:1425–1433. doi: 10.1161/CIRCRESAHA.109.211037. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen Y-G. Controlling TGF-b signalling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, et al. Endoglin, a TGF-b binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- McAllister KA, Baldwin MA, Thukkani AK, Gallione CJ, Berg JN, et al. Six novel mutations in the endoglin gene in hereditary hemorrhagic telangiectasia type I suggest a dominant-negative effect of receptor function. Hum Mol Genet. 1995;4:1983–1985. doi: 10.1093/hmg/4.10.1983. [DOI] [PubMed] [Google Scholar]
- McDonald J, Gedge F, Burdette A, Carlisle J, Bukjiok C, et al. Multiple sequence variants in hereditary hemorrhagic telangiectasia cases: illustration of complexity in molecular diagnostic interpretation. J Mol Diagnostics. 2009;11:569–575. doi: 10.2353/jmoldx.2009.080148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Kusanagi K, Inoue H. Divergence and convergence of TGF-b/BMP signaling. J Cell Physiol. 2001;187:265–276. doi: 10.1002/jcp.1080. [DOI] [PubMed] [Google Scholar]
- Paquet ME, Pece-Barbara N, Vera S, Cymerman U, Karabegovic A, et al. Analysis of several endoglin mutants reveals no endogenous mature or secreted protein capable of interfering with normal endoglin function. Hum Mol Genet. 2001;10:1347–1357. doi: 10.1093/hmg/10.13.1347. [DOI] [PubMed] [Google Scholar]
- Park SO, Lee YJ, Seki T, Hong KH, Fliess N, et al. ALK5- and TGFBR2-independent role of ALK1 in the pathogenesis of hereditary hemorrhagic telangiectasia type 2. Blood. 2008;111:633–642. doi: 10.1182/blood-2007-08-107359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Wankhede M, Lee Y, Choi EJ, Fliess N, et al. Real-time imaging of de novo arteriovenous malformation in a mouse model of hereditary hemorrhagic telangiectasia. J Clin Invest. 2009;119:3487–3496. doi: 10.1172/JCI39482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece N, Vera S, Cymerman U, White R, Wrana J, Letarte M. Mutant endoglin in hereditary hemorrhagic telangiectasia type I is transiently expressed intracellularly and is not a dominant negative. J Clin Invest. 1997;100:2568–2579. doi: 10.1172/JCI119800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece-Barbara N, Cymerman U, Vera S, Marchuk D, Letarte M. Expression analysis of four endoglin missense mutations suggests haploinsufficiency is the predominant mechanism for hereditary hemorrhagic telangiectasia type I. Hum Mol Genet. 1999;8:2171–2181. doi: 10.1093/hmg/8.12.2171. [DOI] [PubMed] [Google Scholar]
- Pece-Barbara N, Vera S, Kathirkamathamby K, Liebner S, Di Guglielmo GM, et al. Endoglin null endothelial cells proliferate faster and are more responsive to transforming growth factor beta1 with higher affinity receptors and an activated Alk1 pathway. J Biol Chem. 2005;280:27800–27808. doi: 10.1074/jbc.M503471200. [DOI] [PubMed] [Google Scholar]
- Sabbà C, Pasculli G, Lenato GM, Suppressa P, Lastella P, et al. Hereditary hemorrhagic telangiectasia: clinical features in ENG and ALK1 mutation carriers. J Thromb Haemost. 2007;5:1149–1157. doi: 10.1111/j.1538-7836.2007.02531.x. [DOI] [PubMed] [Google Scholar]
- Scharpfenecker M, van Dinther M, Liu Z, van Bezooijen RL, Zhao Q, et al. BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J Cell Sci. 2007;120:964–972. doi: 10.1242/jcs.002949. [DOI] [PubMed] [Google Scholar]
- Shovlin CL. Hereditary haemorrhagic telangiectasia: pathophysiology, diagnosis and treatment. Blood Rev. 2010;24:203–219. doi: 10.1016/j.blre.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Shovlin CL, Hughes JM, Scott J, Seidman CE, Seidman JG. Characterization of endoglin and identification of novel mutations in hereditary hemorrhagic telangiectasia. Am J Hum Genet. 1997;61:68–79. doi: 10.1086/513906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shovlin CL, Angus G, Manning RA, Okoli GN, Govani FS, et al. Endothelial cell processing and alternatively spliced transcripts of Factor VIII: potential implications for coagulation cascades and pulmonary hypertension. PLOS One. 2010;5:e9154. doi: 10.1371/journal.pone.0009154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton P, Davies R, Trembath R, Morrell N. BMP and activin type-II receptors balance BMP9 signals mediated by activin receptor-like kinase-1 in human pulmonary artery endothelial cells. J Biol Chem. 2009;284:15794–15804. doi: 10.1074/jbc.M109.002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesha S, Toporsian M, Lam C, Hanai J, Mammoto T, et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Natur Med. 2006;12:642–649. doi: 10.1038/nm1429. [DOI] [PubMed] [Google Scholar]
- Wilke CO. Transcriptional robustness complements nonsense-mediated decay in humans. PLoS Genet. 2011;7:e1002296. doi: 10.1371/journal.pgen.1002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-b receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease and evolution. Ann Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]