Summary
One of the most striking physiological features of skeletal muscle tissues are their enormous capacity to adapt to changed functional demands. Muscle plasticity has been extensively studied by histological, biochemical, physiological and genetic methods over the last few decades. With the recent emergence of high-throughput and large-scale proteomic techniques, mass spectrometry-based surveys have also been applied to the global analysis of the skeletal muscle protein complement during physiological modifications and pathophysiological alterations. This review outlines and discusses the impact of recent proteomic profiling studies of skeletal muscle transitions, including the effects of chronic electro-stimulation, physical exercise, denervation, disuse atrophy, hypoxia, myotonia, motor neuron disease and age-related fibre type shifting. This includes studies on the human skeletal muscle proteome, animal models of muscle plasticity and major neuromuscular pathologies. The biomedical importance of establishing reliable biomarker signatures for the various molecular and cellular transition phases involved in muscle transformation is critically examined.
Keywords: mass spectrometry, muscle plasticity, muscle proteomics, muscle transformation, muscle transition, skeletal muscle proteome
Introduction
The cellular constellation of skeletal muscles represents one of the most abundant tissue systems in the mammalian organism. Skeletal muscles are highly complex biological structures that are crucial for a variety of body functions, such as voluntary movements, posture, heat homeostasis and metabolic integration. Motor units consist largely of multi-nucleated fibres, represented by a diverse population of contractile cells, as well as a variety of other cell types including motor neurons, layers of connective tissue, capillaries, satellite cells and muscle spindles. The contractile fibres within individual muscles are characterized by molecular heterogeneity and the mature neuromuscular system exhibits a high degree of plasticity. This adaptive potential and functional heterogeneity of motor units is the physiological basis of highly variable and adaptable motor tasks in mammals (1). Healthy adult skeletal muscles usually contain a mixture of three main fibre types and a variety of hybrid fibres. The specific proportions of fibre types characterize an individual skeletal muscle and are a functional reflection of its various physiological, biochemical and metabolic tasks. Fibre types can be divided into slow-red oxidative type I fibres, fast-reddish oxidative-glycolytic type IIa fibres, and fast-white glycolytic type IIb/IIx fibres (2). Different whole skeletal muscles differ considerably in their proportions of fibre types, depending on the main functional involvement of an individual muscle with respect to contraction-relaxation cycles, movement patterns, postural control mechanisms or metabolic integration. A wide range of histological, physiological and biochemical differences between fast- versus slow-twitching fibres reflects the biological properties of the main contractile units. Over the last few decades, the cellular changes and underlying mechanisms of skeletal muscle plasticity have been investigated by conventional cell biological, biochemical and physiological techniques (3). More recently, mass spectrometry-based proteomics has been employed to study global changes in skeletal muscle proteins during fibre type shifting. This article reviews how high-throughput and large-scale proteomic surveys have influenced our molecular and cellular understanding of skeletal muscle transitions.
Proteomics as a new analytical approach for assessing muscle plasticity
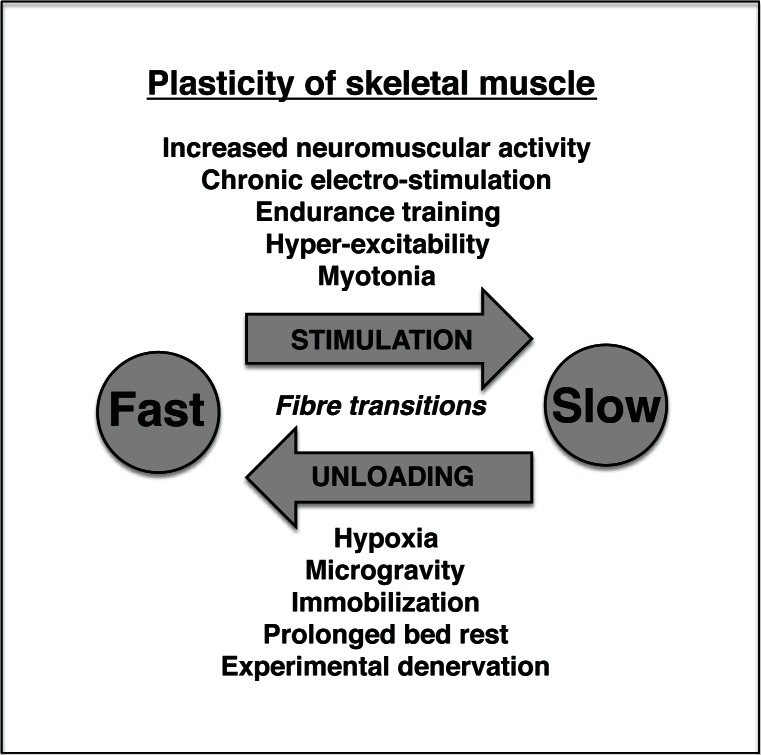
Muscle plasticity is an important topic in basic and applied myology. The proportions of fibre types in skeletal muscles are not static, but fibre type shifting is a frequent occurrence that can be observed as a natural physiological response during functional adaptations to altered neuromuscular activity levels. In addition, muscle changes occur as a consequence of experimental variations of neuronal input patterns, in association with many pathological processes that affect contractile tissues, and during the natural decline of bodily functions at old age. Figure 1 summarizes major aspects of skeletal muscle plasticity. Traditionally, muscle physiology and biochemistry has focused on individual or select groups of genes, proteins or metabolites. This analytical approach has drastically changed with the emergence of genomic, proteomic and metabolomic methodology. Mass spectrometry-based proteomics is concerned with the global survey of distinct protein complements from defined biological entities such as subcellular fractions, cells, tissues, organs or body liquids. Following separation of a given proteome by gel electrophoresis or liquid chromatography, protein species are usually identified by the mass spectrometric analysis of peptide populations that have been generated by controlled digestion (Fig. 2). Proteomics has become the method of choice for unbiased investigations into cell biological phenomena and represents now a core bioanalytical technique for discovery-oriented protein studies. Importantly, proteomics has been widely applied to studying skeletal muscle tissues in health and disease. However, since muscle fibres represent a tough tissue for homogenization and protein fractionation, as well as contain a large portion of high-molecular-mass proteins, integral membrane proteins and components with a low copy number that are difficult to isolate, muscle proteomics is complicated by a variety of technical challenges (4).
Figure 1.
Overview of fibre type transitions in skeletal muscle tissues. The diagram outlines major physiological factors or pathological insults that are involved in fast-to-slow or slow-to-fast fibre type transition processes in contractile tissues.
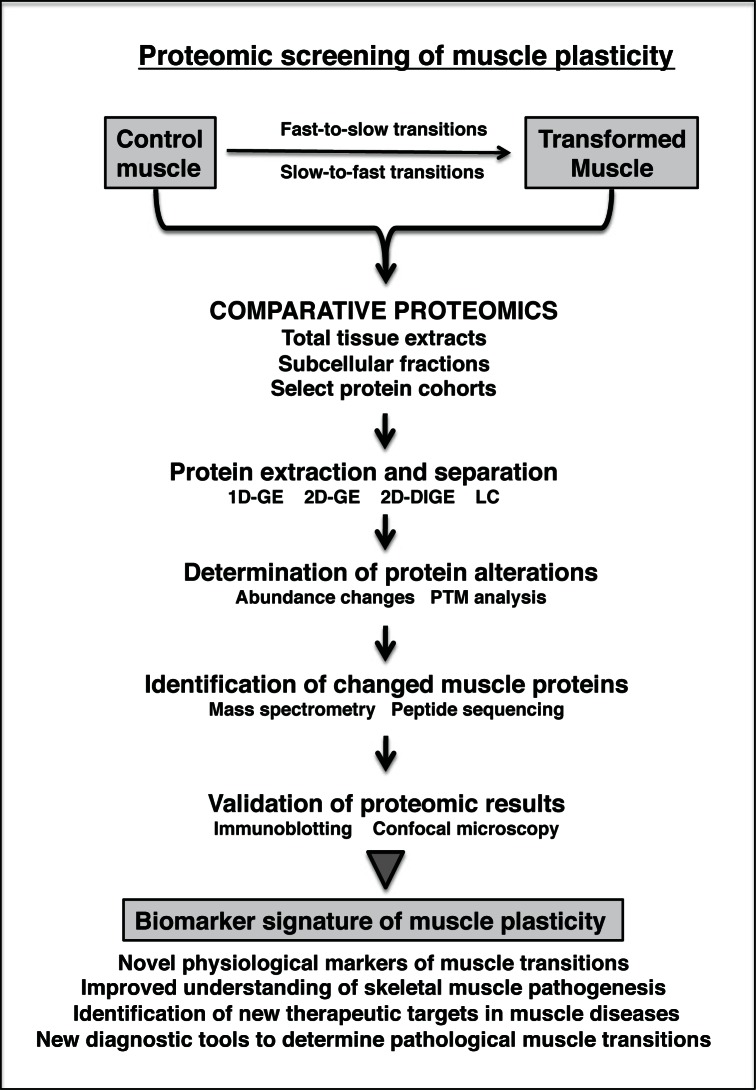
Figure 2.
Mass spectrometry-based proteomics for analysing fibre type transitions in skeletal muscle tissues. The flow diagram lists the various extraction, separation and identification steps for analysing transformed muscle samples.
With respect to the field of muscle plasticity, initial proteomic studies have evaluated global differences in the skeletal muscle proteome of fast versus slow animal and human muscles (5–7). The proteomic profiles clearly emphasize dissimilarity between predominantly fast- versus slow-twitching skeletal muscles. The differential expression patterns of distinct muscle protein isoforms were shown to form the underlying structural and functional template for key histological, physiological and biochemical characteristics of distinct skeletal muscle types. Histological hallmarks are differences in motor unit size, fibre diameter, muscle tissue color, mitochondrial content and capillary density. Physiological divergence is presented by differences in contraction speed, relaxation speed, ion homeostasis, regulation of excitation-contraction coupling, recruitment frequency, fatigability and maximum power output. Biochemical indicators of muscle specificity are the degree of metabolic integration, extent of oxidative enzyme activity, dependence on glycolytic enzyme activity, myoglobin content, glycogen levels, triglyceride concentration and phosphocreatine content. Gel electrophoresis-based proteomics of rat gastrocnemius versus soleus muscle (7) and the mass spectrometric analysis of human deltoideus versus vastus lateralis muscle (6) revealed distinct differences in the abundance and isoform expression pattern of key metabolic, ion handling, contrac-tile and signaling proteins in different fibre types.
Proteomic studies on the distribution of muscle enzymes belonging to anaerobic versus aerobic metabolic pathways have demonstrated the differential reliance of distinct muscle types on glycolysis, the creatine kinase shuttle, the citric acid cycle and oxidative phosphorylation (5–7). The energy supply of slow-twitching type I fibres is clearly based on oxidative metabolism, making them relatively resistant to fatigue with a low recruitment frequency and a low maximum power output. In contrast, fast-twitching type II fibres are fatigable, their supply of energy depends predominantly on the glycolytic pathway, and they exhibit a high recruitment frequency and show a high maximum power output (2). In general, the application of proteomics was highly successful for confirming many of the key protein factors involved in muscle specificity. However, even more importantly, mass spectrometry-based approaches have revealed many new markers of fibre alterations in health and disease that had previously not been identified by traditional biochemical or physiological analyses. An excellent example of the unparalleled analytical power of proteomics for global protein profiling surveys is the identification of a plethora of novel biomarkers of x-linked muscular dystrophy. Proteomic studies have shown that the primary deficiency in the membrane cytoskeletal protein dystrophin results in hitherto unrecognized alterations in diverse muscle proteins, including the enzyme adenylate kinase, the Ca2+-binding protein calsequestrin and the molecular chaperone cvHsp (8). Potential changes in various markers of fibre type shifting were also shown to occur in muscular dystrophy, including myosin heavy chains, glycolytic enzymes, mitochondrial enzymes and metabolic transporters (8). The establishment of proteomic maps of fast versus slow muscles has demonstrated that this technique is highly suitable to study muscle development, differentiation and plasticity, and has set the scene for detailed comparative studies of proteome-wide changes during muscle fibre transitions. This review focuses on the biochemical analysis of tissue plasticity in mature skeletal muscles, and discusses the impact of recent studies on skeletal muscle transitions that have employed proteomics.
Proteomic profiling of physical exercise
The determination of proteome-wide alterations due to physical exercise is of considerable interest to sports science (9). In addition, since regular exercise and a balanced diet have shown to be effective in the prevention of chronic diseases such as diabetes mellitus, and also appear to counter-act some of the symptoms of age-dependent muscle wasting, exercise biology is of considerable interest to biomedicine. The analysis of proteome-wide adaptations following physical exercise might improve our understanding of the molecular and cellular mechanisms that determine fibre specificity and explain how fibre populations adapt and shift in response to training. Genetic factors clearly play a central role in the establishment of muscle fibre type patterns in individual humans, but muscles also exhibit an extraordinary potential to adapt to changed functional demands (10). It is well established that the morphological and physiological effects of regular exercise are reflected by distinct molecular changes in skeletal muscles (11). Endurance training drastically increases levels of fatty acid oxidation and improves the aerobic capacity of muscles (12). In contrast, sprint training promotes carbohydrate utilization and is clearly associated with increased glycolytic and phosphocreatine metabolism in muscle fibres (13). The metabolic benefits of resistance training and accompanied advantages of increased fast-glycolytic skeletal muscles have also been recognized in the field of preventative medicine (14). Modern approaches in exercise biology have included proteomic analyses of endurance exercise and short-burst activities, as recently reviewed by Burniston and Hoffman (15). The physiological response to endurance exercise was shown to result in moderate changes in the skeletal muscle proteome (16–18). The proteomic survey of an intensity-controlled rat model of exercise training demonstrated elevated levels of trisosephosphate isomerase and the molecular chaperone alpha-B-crystallin, and a concomitant reduction in enolase, phospho-glucomutase and lacatate dehydrogenase (17). The slow isoform of myosin light chain MLC2 was shown too be increased in exercise-trained muscle, indicating a shift to a more fatigue-resistant and slower-contracting muscle phenotype due to moderate-intensity exercise (17). For the analysis of protein changes in human vastus lateralis muscle in response to interval-exercise training, a stable isotope-labeling technique named iTRAQ (isobaric tags for relative and absolute quantification) was successfully applied (18). Interval-exercise training appears to trigger distinct post-translational modifications in creatine kinase and enolase (18).
Proteomic profiling of chronic electro-stimulation
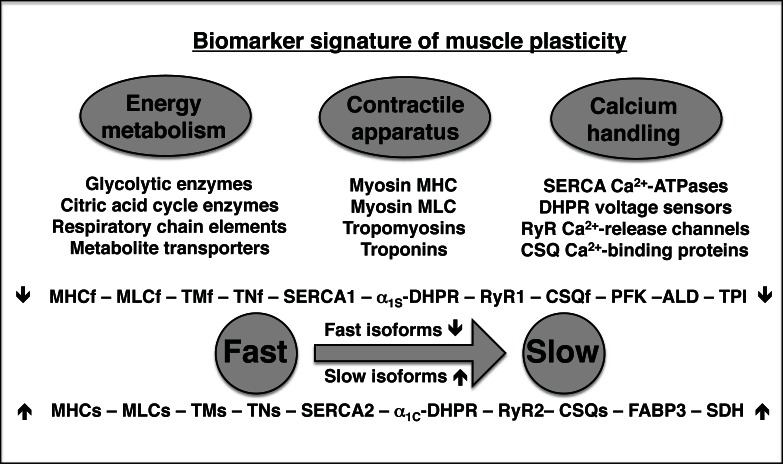
While endurance training triggers a graded physiological response of skeletal muscles (19), artificial non-physiological systems can be employed that activate all affected motor units to a maximum extent (20). For example, chronic low-frequency stimulation of fast muscle causes transitions to a slower contractile phenotype (1). Fast-to-slow transformations are characterized by an increase in the time-to-peak twitch tension and half-relaxation time of electro-stimulated muscles. Conditioned skeletal muscles exhibit a decreased fibre caliber, an elevation of aerobic-oxidative capacity and an improved resistance to fatigue (21). Extensive biochemical studies have established distinct changes in the density and/or isoform expression pattern of myosin heavy chains, myosin light chains, troponins, the transverse-tubular voltage-sensing dihydropyridine receptor, the relaxation-mediating Ca2+-ATPases of the sarcoplasmic reticulum, the triadic ryanodine receptor Ca2+-release channel, the Ca2+-binding protein calsequestrin of the terminal cisternae, the acetylcholine esterase of the basal lamina and the nicotinic acetylcholine receptor at the neuromuscular junction in stimulated fast muscle (1, 3, 20–22). Building on these findings, mass spectrometry-based proteomics was applied to the analysis of rabbit tibialis anterior muscle using a 42-days chronic stimulation protocol at 10Hz (23, 24). The biochemical analysis of proteome-wide changes confirmed stimulation-related alterations in elements involved in oxygen transportation, the efficient transfer and utilization of fatty acids, bioenergetic processes, the creatine kinase shuttle system, the contractile actomyosin apparatus, ion-handling, excitation-contraction coupling, muscle relaxation, and the cellular stress response (23, 24). Chronic electro-stimulation was shown to increase muscle proteins involved in aerobic substrate oxidation, which included key metabolic transporters such as myoglobin and the fatty acid binding protein FABP3, and enzymes of the citric acid cycle, fatty acid oxidation and the respiratory chain (23). In stark contrast, many enzymes belonging to the glycolytic pathway were reduced in fast muscles following low-frequency stimulation (24). As outlined in Figure 3, the proteomic profiling of skeletal muscle transformation supports the idea that adaptations to chronic electro-stimulation are characterized by a glycolytic-to-oxidative shift in muscle metabolism (24).
Figure 3.
Overview of the effect of chronic electro-stimulation on fast muscle. Shown are the main categories of muscle proteins involved in energy metabolism, the contraction-relaxation cycle and the ion-dependent regulation of excitation-contraction coupling, which are affected by chronic low-frequency stimulation. Proteins listed in the biomarker signature of fast-to-slow transition are named by standard abbreviations (22–24).
Proteomics of disuse atrophy
Disuse of skeletal muscle tissue or traumatic injury to the neurotransmission system leads inevitably to muscular atrophy and has severe adverse functional consequences for the entire body (25). Skeletal muscle atrophy attributable to extended periods of skeletal muscle inactivity occurs after prolonged bed rest, due to immobilization, during exposure to microgravity and as a result of traumatic nerve damage, as well as a consequence of experimental denervation or hindlimb unloading.
Skeletal muscles undergoing atrophy exhibit decreased contractile force, reduced protein content, a decreased muscle fiber cross-sectional area, insulin resistance, and slow-to-fast fiber type transitions (26). It is believed that the main degenerative mechanism of muscular atrophy is based on the early and transient decreases in muscle protein synthesis and a concomitant elevation of muscle protein degradation rates, which then result in a rapid decline in skeletal muscle mass (27). Proteomic profiling of muscular disuse has included the analysis of the cytosolic fraction from atrophying mouse skeletal muscle by ICAT (isotope-coded affinity tag) MS/MS technology (28), the fluorescence difference in-gel electrophoretic survey of rat soleus muscle changes during unloading (29), the analysis of protein alterations in murine skeletal muscle submitted to 1 week of hindlimb suspension (30) and the evaluation of long-term bed rest in humans with and without vibration exercise counter-measures (31). In general, the proteomics of skeletal muscle unloading has revealed considerable alterations in structural and contractile proteins, such as the various isoforms of myosins, actins and troponins, as well as numerous molecular chaperones. Importantly, changes in the concentration of enzymes such as enolase, triosephosphate isomerase, lactate dehydrogenase and isocitrate dehydrogenase clearly support a gradual slow-to-fast fibre type transformation process due to disuse atrophy (28–31).
Proteomic profiling of motor neuron disease
As outlined above, disuse of skeletal muscles due to physical inactivity or traumatic denervation has profound effects on the molecular and cellular composition of contractile tissue. In analogy, certain neuromuscular pathologies can also lead to slow-to-fast muscle transformations. A disease-induced form of muscular atrophy is represented by a heterogeneous group of fatal neurological diseases called motor neuron disease (32). In adult patients, the most common form of motor neuron disease is amyotrophic lateral sclerosis, which is characterized by highly progressive paralysis. This disorder can be sporadic or be of genetic origin, and motor neuron wasting triggers contractile weakness of limb, bulbar and respiratory muscles. Detailed biochemical studies into the underlying mechanisms of muscle paralysis often employ animal models that exhibit the progressive symptoms of neurodegeneration and muscular atrophy (33). The wobbler mouse model of motor neuron disease shows many cellular abnormalities seen in patients. A recessive missense mutation in the ubiquitously expressed gene Vps54 is the primary genetic defect in the wobbler mouse (34). In normal muscle, the expressed vesicular protein-sorting factor VPS54 was shown to maintain retrograde vesicle transport to the Golgi apparatus. Fluorescence difference in-gel electrophoretic analysis of the skeletal muscle proteome from wobbler mice revealed concentration changes in 24 proteins. The mass spectrometric identification of these muscle-associated proteins showed abnormalities in the actomyosin apparatus, metabolic pathways, intermediate filaments and the cellular stress response (35). The idea that an oxidative-to-glycolytic shift exists in disease-related muscular atrophy was confirmed by the proteomic finding that the concentration of the glycolytic marker enzyme glyceraldehyde-3-phosphate dehydrogenase was greatly increased in wobbler muscle. However, density of the cytosolic Ca2+-binding protein parvalbumin was shown to be decreased (35), which is usually observed during the opposite metabolic adaptations (36). Alterations in the Ca2+-handling apparatus of wobbler muscle and metabolic changes do not appear to occur in synchrony (35). Hence, pathobiochemical changes in highly progressive disease-associated muscular atrophy appear to differ in their molecular mechanisms from disuse atrophy.
Proteomic profiling of oxygen deprivation
The deprivation of an adequate oxygen supply may result in generalized hypoxia in the entire organism or regional hypoxia in select types of tissue. If the cellular demand of contractile fibres for oxygen is not met by the available supply of this crucial metabolite, muscle hypoxia occurs and causes extensive functional adjustments. The biological effects of muscle hypoxia are important factors in certain muscle diseases, in the course of strenuous exercise and during chronic exposure at altitude (37). Mass spectrometry-based proteomics has been applied to study hypoxia-related alterations in muscle proteins. This has included the evaluation of two well-established animal models of oxygen deprivation, zebrafish in hypoxic tanks (38) and rats in hypoxic chambers (39), and the examination of human vastus lateralis muscle during adaptations to hypobaric hypoxia (40). The proteomic analysis of both animal and human muscle samples exposed to chronic hypoxia have confirmed the increase in key regulatory enzymes of glycolysis, such as pyruvate kinase, and a decrease in components involved in the citric acid cycle and oxidative phosphorylation (38–40). This demonstrates an oxidative-to-glycolytic shift in skeletal muscle metabolism due to experimentally induced hypoxia or during physical activity at altitude. With respect to physiological adaptations of humans to altitude, these proteomic findings support the concept that the altitude-adapted muscle phenotype preserves energy balance at a drastically reduced aerobic capacity by supporting an anaerobic substrate flux.
Proteomics of myotonia-associated fibre type shifting
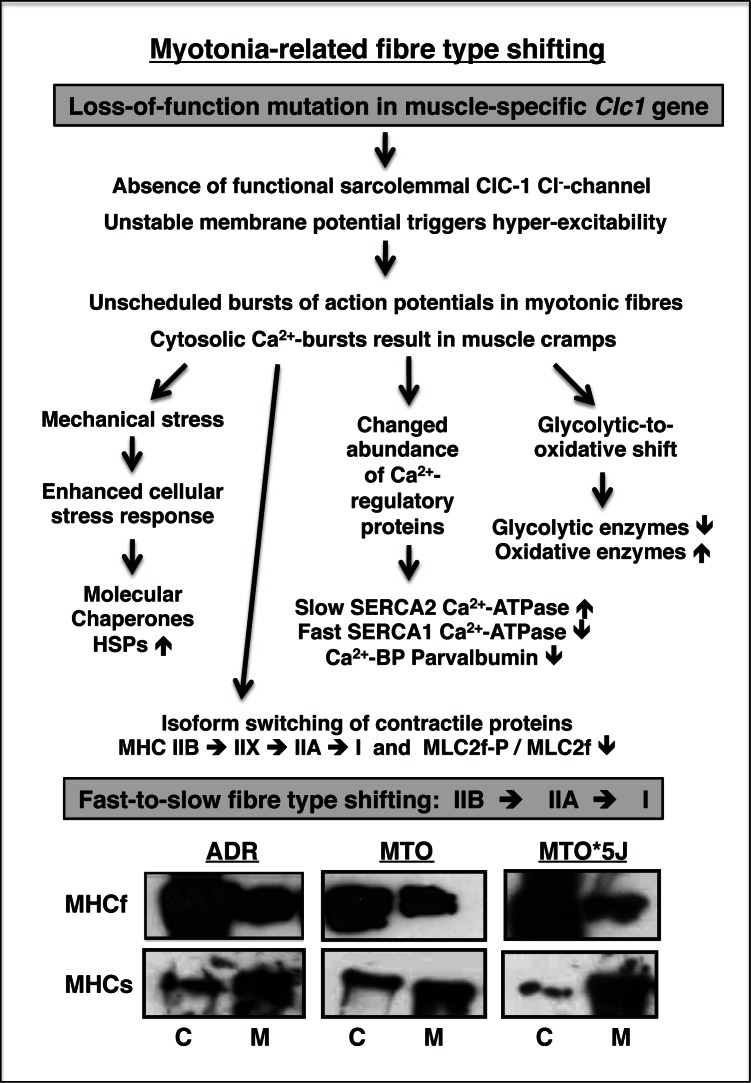
Channelopathies are neuromuscular disorders that exhibit recurrent patterns of genetic abnormalities, such as mutations in Na+-, K+-, Ca2+- or Cl−-channels. Myotonias and paramyotonias are characterized by involuntary contractions due to hyper-excitability of the sarcolemma (41). Research into the molecular mechanisms underlying hyper-excitability due to lowered Cl−-conductance in myotonias has been conducted for many years using genetic mouse models with distinct mutations at the gene locus Clc1 for the muscle-specific chloride channel ClC-1. Interestingly, the secondary effects of hyper-excitability cause biochemical changes in myotonic muscle that resemble the fast-to-slow transformation process observed in chronic low-frequency stimulated fast muscle (21). In myotonic skeletal muscles, switches in the fast and slow isoforms of myosin light and heavy chains occur during fibre transitons. Sarcolemmal hyper-excitability results in a reduced density of myosin heavy chain isoform MHC-IIB and lower phosphorylation level of fast myosin light chain MLC2f in myotonic fast muscles (42). These biochemical findings were confirmed by a recent proteomic survey of three mouse models of myotonia. The comparative proteomic survey of the severely myotonic ADR and MTO mutants with the milder phenotype of MTO*5J mice revealed a generally perturbed protein expression pattern in ADR and MTO muscle, but less pronounced alterations in mildly diseased MTO*5J tissue (36). The myotonic gastrocnemius muscle showed changes in the contractile machinery, various metabolic pathways, ion-regulatory proteins and the cellular stress response. The immunoblot analysis of fast versus slow myosin heavy chains in ADR versus MTO versus MTO*5J muscle confirms the switch of MHC-IIB to MHC-IIX to MHC-IIA to MHC-I in myotonia (Fig. 4). The biochemical and proteomic profiling of severely myotonic muscle revealed furthermore a drastic decrease in the phosphorylatable fast MLC2f isoform of myosin light chain, the cytosolic Ca2+-binding element parvalbumin, the fast SERCA1 isoform of the sarcoplasmic reticulum Ca2+-ATPase, and glycolytic enzymes such as triosephosphate isomerase and aldolase. In stark contrast, myoglobin, the slow/cardiac SERCA2 isoform of the sarcoplasmic reticulum Ca2+-ATPase, molecular chaperones and a variety of mitochondrial enzymes including NADH dehydrogenase and succinate dehydrogenase were found to be increased in myotonia (36). These proteomic findings support the concept of a glycolytic-to-oxidative transformation process in myotonic fast muscle. The establishment of a global bio-marker signature of myotonia using proteomics technology will be extremely helpful in improving our understanding of how a primary abnormality in a muscle-specific Cl−-channel triggers downstream alterations in the expression of a large number of genes in skeletal muscle.
Figure 4.
Effect of myotonia on the expression of skeletal muscle proteins. The flow diagram lists the various steps involved in the molecular pathogenesis of myotonia as revealed by biochemical and proteomic analyses. The immunoblot illustrates expression changes in the fast MHCf and slow MHCs myosin heavy chains in control muscle (C) versus myotonic muscle (M) from mouse mutants ADR, MTO and MTO*5J (36). Immuno-decoration was carried out by standard procedures as previously described by our laboratory (46). Although antibodies to MHCf and MHCs cannot differentiate between the many subspecies of myosin heavy chains, the immunoblot analysis illustrates the general trend of a fast-to-slow switch in isoforms of the contractile apparatus.
Proteomics of sarcopenia of old age
Progressive skeletal muscle wasting is an important feature of the overall aging process. Multiple factors are believed to trigger age-dependent loss in muscle mass and contractile strength, including denervation-associated muscular atrophy, excitation-contraction uncoupling, a drastically decreased capacity for fibre regeneration, disturbances in key metabolic pathways, deficiency in mitochondrial functions, an increased susceptibility to apoptosis, disturbed ion handling, impaired muscle protein synthesis, an altered equilibrium of growth factors and hormones involved in fibre maintenance, and an impaired response by molecular chaperones (43). Histological hallmarks of structural changes in aged muscle are the frequent occurrence of centrally located nuclei, grouped atrophying fibres and an increased variability in fibre diameter. Severe forms of age-related muscle degeneration in the elderly have been termed sarcopenia. Because the drastic decline in contractile efficiency associated with sarcopenia of old age is such an important clinical topic that affects a large proportion of society, a considerable number of proteomic studies have been devoted to the large-scale analysis of aged human and animal muscle, as reviewed by Doran et al. (44). Muscle aging is associated with a changed density in a variety of contractile and metabolic proteins, and has also a considerable effect on post-translational modifications. Glycosylation, phosphorylation and tyrosine nitration is altered in many metabolic enzymes (44).
With respect to muscle transformation, the mass spectrometric survey of senescent skeletal muscles has established a shift to a more aerobic oxidative metabolism in a slower-twitching aged fibre population (45–49). A glycolytic-to-oxidative shift during muscle aging is exemplified by the proteomic finding that enolase and phosphofructokinase are reduced in aged muscle while myoglobin, the fatty acid-binding protein FABP3, succinate dehydrogenase and mitochondrial ATP synthase are increased in senescent muscle preparations (46). These results were confirmed by a comprehensive survey of mitochondrial proteins using a fluorescence difference in-gel electrophoretic analysis of the mitochondria-enriched fraction from aged rat skeletal muscle (48). Proteomics of senescent gastrocnemius muscle showed an age-related increase in many mitochondrial marker proteins, such as mitofilin, peroxiredoxin, NADH dehydrogenase, ATPase synthase, succinate dehydrogenase, prohibitin, porin isoform VDAC2 and ubiquinol-cytochrome c reductase. The idea that pathological cycles of age-dependent denervation and faulty reinnervation processes trigger complex alterations in aged motor units that cause a drastic slowing of muscle contractile properties, was confirmed by a subproteomic analysis of the myofibrillar fraction (49). The slow-type myosin light chain isoform MLC2 was shown to be the contractile protein that exhibits the most dramatic alteration in expression and phosphorylation levels during muscle aging. The drastic increase of slow MLC-2 in aged muscle strongly indicates an extensive fast-to-slow fibre transition process in sarcopenia (49). Interestingly, a pathophysiological link between Ca2+-dependent proteolysis and abnormal functioning of the mitochondrial ATP synthase was recently established by a proteomic study on the role of calpain-binding proteins during muscle aging (50). It appears that mitochondrial function is impaired in aged fibres, but that the relative content of this organelle is increased in senescent muscle. The shift to more oxidative fibres is most likely due to the preferential degradation of fast-glycoloytic fibers during aging. Muscle fibre type shifting and metabolic changes are probably not causative factors of muscle wasting in the aged organism, but probably a pathophysiological consequence of age-related inflammation, reduced regenerative capacity and alterations in hormonal homeostasis.
Conclusion
Skeletal muscle fibres belong to the class of highly plastic tissues that can quickly and efficiently adapt to changed physiological or functional demands. In order to study global changes in the muscle proteome due to metabolic changes or fibre type shifting, it is crucial to study the entire protein complement of muscle tissue using a relatively rapid and reliable analytical approach. Mass spectrometry-based proteomics represents such a technique that combines a variety of traditional biochemical and protein chemical methods for a streamlined analytical workflow that can swiftly identify novel protein biomarker candidates. Over the last few years, proteomics has been successfully applied to studying skeletal muscle transitions during physiological and pathophysiological processes. This has included the mass spectrometric identification of novel protein factors involved in exercise-related fibre changes, stimulation-dependent adaptations, disuse-triggered muscular atrophy, hypoxia-dependent changes, myotonia-related fibre type shifting, disease-associated atrophy and age-dependent muscle transformation. However, most studies have focused on the urea-soluble fraction of the muscle protein complement and employed gel electrophoretic methodology for the separation of proteins. Since muscle proteins are expressed over a wide dynamic range and because different protein families exhibit greatly differing biochemical properties, the presence of a variety of muscle-associated proteins has been underestimated by traditional gel electrophoresis-based studies. In the future, it will be crucial to also include integral membrane proteins and proteins with a low copy number in proteomic surveys of muscle plasticity. More comprehensive studies of the entire skeletal muscle proteome promise to establish reliable biomarker signatures of important physiological and pathological processes and will thus be helpful for finding new therapeutic targets to treat skeletal muscle pathologies and improve diagnostic procedures of major neuromuscular diseases.
Acknowledgments
This work was supported by project grants from the Irish Health Research Board, Science Foundation Ireland, the Higher Education Authority and Muscular Dystrophy Ireland. The author would like to thank Dr. Lisa Staunton for excellent technical help with immunoblotting.
References
- 1.Pette D. Historical Perspectives: plasticity of mammalian skeletal muscle. J Appl Physiol. 2001;90:1119–1124. doi: 10.1152/jappl.2001.90.3.1119. [DOI] [PubMed] [Google Scholar]
- 2.Canepari M, Pellegrino MA, D’Antona G, Bottinelli R. Skeletal muscle fibre diversity and the underlying mechanisms. Acta Physiol (Oxf) 2010;199:465–476. doi: 10.1111/j.1748-1716.2010.02118.x. [DOI] [PubMed] [Google Scholar]
- 3.Matsakas A, Patel K. Skeletal muscle fibre plasticity in response to selected environmental and physiological stimuli. Histol Histopathol. 2009;24:611–629. doi: 10.14670/HH-24.611. [DOI] [PubMed] [Google Scholar]
- 4.Ohlendieck K. Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skelet Muscle. 2011;1(1):6. doi: 10.1186/2044-5040-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okumura N, Hashida-Okumura A, Kita K, Matsubae M, Matsubara T, Takao T, Nagai K. Proteomic analysis of slow-and fast-twitch skeletal muscles. Proteomics. 2005;5:2896–2906. doi: 10.1002/pmic.200401181. [DOI] [PubMed] [Google Scholar]
- 6.Capitanio D, Vigano A, Ricci E, Cerretelli P, Wait R, Gelfi C. Comparison of protein expression in human deltoideus and vastus lateralis muscles using two-dimensional gel electrophoresis. Proteomics. 2005;5:2577–2586. doi: 10.1002/pmic.200401183. [DOI] [PubMed] [Google Scholar]
- 7.Gelfi C, Vigano A, De Palma S, Ripamonti M, Begum S, Cerretelli P, Wait R. 2-D protein maps of rat gastrocnemius and soleus muscles: a tool for muscle plasticity assessment. Proteomics. 2006;6:321–340. doi: 10.1002/pmic.200501337. [DOI] [PubMed] [Google Scholar]
- 8.Lewis C, Carberry S, Ohlendieck K. Proteomic profiling of x-linked muscular dystrophy. J Muscle Res Cell Motil. 2009;30:267–279. doi: 10.1007/s10974-009-9197-6. [DOI] [PubMed] [Google Scholar]
- 9.Hittel DS, Hathout Y, Hoffman EP. Proteomics and systems biology in exercise and sport sciences research. Exerc Sport Sci Rev. 2007;35:5–11. doi: 10.1097/jes.0b013e31802d744a. [DOI] [PubMed] [Google Scholar]
- 10.Flueck M. Functional, structural and molecular plasticity of mammalian skeletal muscle in response to exercise stimuli. J Exp Biol. 2006;209:2239–2248. doi: 10.1242/jeb.02149. [DOI] [PubMed] [Google Scholar]
- 11.Booth FW, Thomason DB. Molecular and cellular adap-tation of muscle in response to exercise: perspectives of various models. Physiol Rev. 1991;71:541–585. doi: 10.1152/physrev.1991.71.2.541. [DOI] [PubMed] [Google Scholar]
- 12.Hawley JA. Adaptations of skeletal muscle to prolonged, intense endurance training. Clin Exp Pharmacol Physiol. 2002;29:218–222. doi: 10.1046/j.1440-1681.2002.03623.x. [DOI] [PubMed] [Google Scholar]
- 13.Ross A, Leveritt M. Long-term metabolic and skeletal muscle adaptations to short-sprint training: implications for sprint training and tapering. Sports Med. 2001;31:1063–1082. doi: 10.2165/00007256-200131150-00003. [DOI] [PubMed] [Google Scholar]
- 14.LeBrasseur NK, Walsh K, Arany Z. Metabolic benefits of resistance training and fast glycolytic skeletal muscle. Am J Physiol Endocrinol Metab. 2011;300:E3–E10. doi: 10.1152/ajpendo.00512.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burniston JG, Hoffman EP. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev Proteomics. 2011;8:361–367. doi: 10.1586/epr.11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guelfi KJ, Casey TM, Giles JJ, Fournier PA, Arthur PG. A proteomic analysis of the acute effects of high-intensity exercise on skeletal muscle proteins in fasted rats. Clin Exp Pharmacol Physiol. 2006;33:952–957. doi: 10.1111/j.1440-1681.2006.04470.x. [DOI] [PubMed] [Google Scholar]
- 17.Burniston JG. Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim Biophys Acta. 2008;1784:1077–1086. doi: 10.1016/j.bbapap.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Holloway KV, O’Gorman M, Woods P, Morton JP, Evans L, Cable NT, Goldspink DF, Burniston JG. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics. 2009;9:5155–5174. doi: 10.1002/pmic.200900068. [DOI] [PubMed] [Google Scholar]
- 19.Yan Z, Okutsu M, Akhtar YN, Lira VA. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J Appl Physiol. 2011;110:264–274. doi: 10.1152/japplphysiol.00993.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ljubicic V, Adhihetty PJ, Hood DA. Application of animal models: chronic electrical stimulation-induced contractile activity. Can J Appl Physiol. 2005;30:625–643. doi: 10.1139/h05-144. [DOI] [PubMed] [Google Scholar]
- 21.Hicks A, Ohlendieck K, Gopel SO, Pette D. Early functional and biochemical adaptations to low-frequency stimulation of rabbit fast-twitch muscle. Am J Physiol. 1997;273:C297–C305. doi: 10.1152/ajpcell.1997.273.1.C297. [DOI] [PubMed] [Google Scholar]
- 22.Froemming GR, Murray BE, Harmon S, Pette D, Ohlendieck K. Comparative analysis of the isoform expression pattern of Ca2+-regulatory membrane proteins in fast-twitch, slow-twitch, cardiac, neonatal and chronic low-frequency stimulated muscle fibers. Biochim Biophys Acta. 2000;1466:151–168. doi: 10.1016/s0005-2736(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 23.Donoghue P, Doran P, Wynne K, Pedersen K, Dunn MJ, Ohlendieck K. Proteomic profiling of chronic low-frequency stimulated fast muscle. Proteomics. 2007;7:3417–3430. doi: 10.1002/pmic.200700262. [DOI] [PubMed] [Google Scholar]
- 24.Donoghue P, Doran P, Dowling P, Ohlendieck K. Differential expression of the fast skeletal muscle proteome following chronic low-frequency stimulation. Biochim Biophys Acta. 2005;1752:166–176. doi: 10.1016/j.bbapap.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Chen X, Fan M. Signaling mechanisms involved in disuse muscle atrophy. Med Hypotheses. 2007;69:310–321. doi: 10.1016/j.mehy.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 27.Marimuthu K, Murton AJ, Greenhaff PL. Mechanisms regulating muscle mass during disuse atrophy and rehabilitation in humans. J Appl Physiol. 2011;110:555–560. doi: 10.1152/japplphysiol.00962.2010. [DOI] [PubMed] [Google Scholar]
- 28.Toigo M, Donohoe S, Sperrazzo G, Jarrold B, Wang F, Hinkle R, Dolan E, Isfort RJ, Aebersold R. ICAT-MS-MS time course analysis of atrophying mouse skeletal muscle cytosolic subproteome. Mol Biosyst. 2005;1:229–241. doi: 10.1039/b507839c. [DOI] [PubMed] [Google Scholar]
- 29.Moriggi M, Cassano P, Vasso M, Capitanio D, Fania C, Musicco C, Pesce V, Gadaleta MN, Gelfi CA. DIGE approach for the assessment of rat soleus muscle changes during unloading: effect of acetyl-L-carnitine supplementation. Proteomics. 2008;8:3588–3604. doi: 10.1002/pmic.200701176. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira R, Vitorino R, Neuparth MJ, Appell HJ, Duarte JA, Amado F. Proteolysis activation and proteome alterations in murine skeletal muscle submitted to 1 week of hindlimb suspension. Eur J Appl Physiol. 2009;107:553–556. doi: 10.1007/s00421-009-1151-1. [DOI] [PubMed] [Google Scholar]
- 31.Moriggi M, Vasso M, Fania C, Capitanio D, Bonifacio G, Salanova M, Blottner D, Rittweger J, Felsenberg D, Cerretelli P, Gelfi C. Long term bed rest with and without vibration exercise countermeasures: Effects on human muscle protein dysregulation. Proteomics. 2010;10:3756–3774. doi: 10.1002/pmic.200900817. [DOI] [PubMed] [Google Scholar]
- 32.Wood-Allum C, Shaw PJ. Motor neurone disease: a practical update on diagnosis and management. Clin Med. 2010;10:252–258. doi: 10.7861/clinmedicine.10-3-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boillee S, Peschanski M, Junier MP. The wobbler mouse: a neurodegeneration jigsaw puzzle. Mol Neurobiol. 2003;28:65–106. doi: 10.1385/MN:28:1:65. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt-John T, Drepper C, Mussmann A, Hahn P, Kuhlmann M, Thiel C, Hafner M, Lengeling A, Heimann P, Jones JM, Meisler MH, Jockusch H. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet. 2005;37:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- 35.Staunton L, Jockusch H, Ohlendieck K. Proteomic analy-sis of muscle affected by motor neuron degeneration: the wobbler mouse model of amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2011;406:595–600. doi: 10.1016/j.bbrc.2011.02.099. [DOI] [PubMed] [Google Scholar]
- 36.Staunton L, Jockusch H, Wiegand C, Albrecht T, Ohlendieck K. Identification of secondary effects of hyperexcitability by proteomic profiling of myotonic mouse muscle. Mol Biosyst. 2011;7:2480–2489. doi: 10.1039/c1mb05043e. [DOI] [PubMed] [Google Scholar]
- 37.Flueck M. Plasticity of the muscle proteome to exercise at altitude. High Alt Med Biol. 2009;10:183–193. doi: 10.1089/ham.2008.1104. [DOI] [PubMed] [Google Scholar]
- 38.Bosworth CA, Chou CW, Cole RB, Rees BB. Protein ex-pression patterns in zebrafish skeletal muscle: initial characterization and the effects of hypoxic exposure. Proteomics. 2005;5:1362–1371. doi: 10.1002/pmic.200401002. [DOI] [PubMed] [Google Scholar]
- 39.De Palma S, Ripamonti M, Vigano A, Moriggi M, Capitanio D, Samaja M, Milano G, Cerretelli P, Wait R, Gelfi C. Metabolic modulation induced by chronic hypoxia in rats using a comparative proteomic analysis of skeletal muscle tissue. J Proteome Res. 2007;6:1974–1984. doi: 10.1021/pr060614o. [DOI] [PubMed] [Google Scholar]
- 40.Vigano A, Ripamonti M, De Palma S, Capitanio D, Vasso M, Wait R, Lundby C, Cerretelli P, Gelfi C. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics. 2008;8:4668–4679. doi: 10.1002/pmic.200800232. [DOI] [PubMed] [Google Scholar]
- 41.Jurkat-Rott K, Lerche H, Lehmann-Horn F. Skeletal muscle channelopathies. J Neurol. 2002;249:1493–1502. doi: 10.1007/s00415-002-0871-5. [DOI] [PubMed] [Google Scholar]
- 42.Jockusch H, Reininghaus J, Stuhlfauth I, Zippel M. Reduction of myosin-light-chain phosphorylation and of parvalbumin content in myotonic mouse muscle and its reversal by tocainide. Eur J Biochem. 1988;171:101–105. doi: 10.1111/j.1432-1033.1988.tb13764.x. [DOI] [PubMed] [Google Scholar]
- 43.Edstrom E, Altun M, Bergman E, Johnson H, Kullberg S, Ramirez-Leon V, Ulfhake B. Factors contributing to neuromuscular impairment and sarcopenia during aging. Physiol Behav. 2007;92:129–135. doi: 10.1016/j.physbeh.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 44.Doran P, Donoghue P, O’Connell K, Gannon J, Ohlendieck K. Proteomics of skeletal muscle aging. Proteomics. 2009;9:989–1003. doi: 10.1002/pmic.200800365. [DOI] [PubMed] [Google Scholar]
- 45.Gelfi C, Vigano A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, Grassi B, Bottinelli R, Wait R, Cerretelli P. The human muscle proteome in aging. J Proteome Res. 2006;5:1344–1353. doi: 10.1021/pr050414x. [DOI] [PubMed] [Google Scholar]
- 46.Doran P, O’Connell K, Gannon J, Kavanagh M, Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8:364–377. doi: 10.1002/pmic.200700475. [DOI] [PubMed] [Google Scholar]
- 47.Capitanio D, Vasso M, Fania C, Moriggi M, Viganò A, Procacci P, Magnaghi V, Gelfi C. Comparative proteomic profile of rat sciatic nerve and gastrocnemius muscle tissues in ageing by 2-D DIGE. Proteomics. 2009;9:2004–2020. doi: 10.1002/pmic.200701162. [DOI] [PubMed] [Google Scholar]
- 48.O’Connell K, Ohlendieck K. Proteomic DIGE analysis of the mitochondria-enriched fraction from aged rat skeletal muscle. Proteomics. 2009;9:5509–5512. doi: 10.1002/pmic.200900472. [DOI] [PubMed] [Google Scholar]
- 49.Gannon J, Doran P, Kirwan A, Ohlendieck K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur J Cell Biol. 2009;88:685–700. doi: 10.1016/j.ejcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Brule C, Dargelos E, Diallo R, Listrat A, Bechet D, Cottin P, Poussard S. Proteomic study of calpain interacting proteins during skeletal muscle aging. Biochimie. 2010;92:1923–1933. doi: 10.1016/j.biochi.2010.09.003. [DOI] [PubMed] [Google Scholar]