Abstract
To develop a detailed double belt model for discoidal HDL, we previously scored inter-helical salt bridges between all possible registries of two stacked antiparallel amphipathic helical rings of apolipoprotein (apo) A-I. The top score was the antiparallel apposition of helix 5 with 5 followed closely by appositions of helix 5 with 4 and helix 5 with 6. The rationale for the current study is that, for each of the optimal scores, a pair of identical residues can be identified in juxtaposition directly on the contact edge between the two antiparallel helical belts of apoA-I. Further, these residues are always in the ‘9th position’ in one of the eighteen 11-mer repeats that make up the lipid-associating domain of apoA-I. To illustrate our terminology, 129j (LL5/5) refers to the juxtaposition of the Ca atoms of G129 (in a ‘9th position’) in the pairwise helix 5 domains. We reasoned that if identical residues in the double belt juxtapositions were mutated to a cysteine and kept under reducing conditions during disc formation, we would have a precise method for determining registration in discoidal HDL by formation of a disulfide-linked apoA-I homodimer. Using this approach, we conclude that 129j (LL5/5) is the major rotamer orientation for double belt HDL and propose that the small ubiquitous gap between the pairwise helix 5 portions of the double belt in larger HDL discoidal particles is significantly dynamic to hinge off the disc edge under certain conditions, e.g., in smaller particles or perhaps following binding of the enzyme LCAT.
Keywords: ApoA-I, site-directed mutagenesis, crosslinking, molecular dynamics simulation, molecular modeling, LCAT
1. Introduction
High density lipoprotein (HDL) represents a heterogeneous population of particles with apoA-I as the major protein [1]. A detailed understanding of the atheroprotective role of apoA-I/HDL requires knowledge of the structure and dynamics of its heterogeneous population of particles. Whether apoA-I/HDL plays a direct role in cardiovascular disease prevention (e.g., removal of cholesterol from clogged arteries) or an indirect one (e.g., acts as a platform for the clustering of protective molecules, such as anti-inflammatory or antioxidant proteins), detailed knowledge of HDL structure and dynamics is a key to understanding HDL function. Since the conformation of apoA-I on HDL is highly plastic [1], understanding apo-I/HDL structure and dynamics is not straight forward.
Both negative stain [2] and cryo-electron [3] microscopy of apoA-I reconstituted with phospholipid (apoA-I/HDL) reveal discoidal particles the thickness of a phospholipid bilayer that are similar to nascent HDL particles [4]. X-ray and neutron scattering studies support an annular protein-bilayer disc structure for apoA-I/HDL [5, 6].
The common lipid-associating motif in apoA-I is the amphipathic α helix [7, 8]. The first tangible experimental evidence for the conformation of apoA-I on the disc edge was determination of a 4-Å resolution solution phase x-ray structure for residues 44-243 of lipid-free apoA-I reported by Borhani, et al. in 1997 [9]. The structure determined for this fragment of apoA-I was that of an almost continuous amphipathic α helical non-planar horseshoe-shaped 4-helix bundle, a structure suggested by the authors to support the double belt model for discoidal HDL [9]. However, its relevance to lipid-associated apoA-I was not clear.
In the first experimental test of the belt model, Axelsen, et. al. [10] used polarized attenuated total internal reflection Fourier-transform infrared spectroscopy (PATIR-FTIR) to characterize the relative orientation of apoA-I and lipid components in reconstituted HDL (apoA-I/HDL) adsorbed to supported monolayers. The authors concluded that the result unambiguously supported the double belt model.
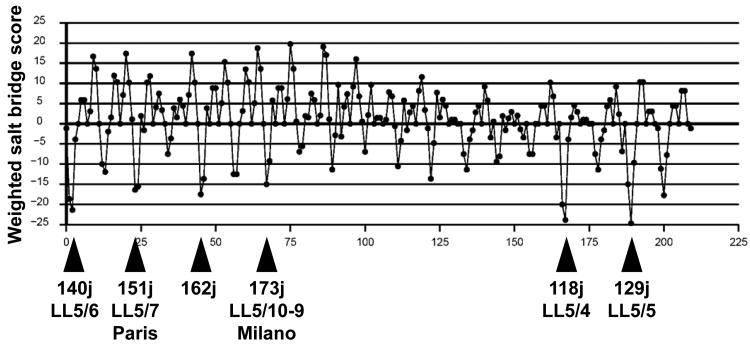
Theoretical considerations of the geometric and physical chemical nature of the apoA-I double belt model resulted in our publication of an atomic resolution antiparallel double belt amphipathic helical model for discoidal HDL [11]. In the model, apoA-I monomers formed a curved amphipathic α helical ring with 11/3 residues per turn — termed an α11/3 helix and since observed in other lipid-associating proteins, such as α-synuclein [12, 13] — so that the hydrophobic surface faced inward toward the lipid disc. In particular, we examined possible interactions between two α11/3 helical rings by docking along, and rotating around, the axis of one ring relative to a second. A modification of the HELNET program [14], ALIGN [15], was used to score the weighted number of salt bridges and charge appositions for all orientations of three possible ring pair interfaces. For clarity, we defined a terminology that denotes the helix-helix stagger and interface in terms of 22 tandem repeats; e.g. LL5/5 refers to a position in which the left interfacial edge of helix 5 in ring-1 is associated with the left interfacial edge of helix 5 in ring-2. Using this terminology, the three most impressive orientations had rank order weighted salt bridge scores of LL5/5 > LL5/4 > LL5/6 (Fig. 1A). Of interest, the LL5/5 registry is identical to the registry seen in the lipid-free x-ray crystal structure [11].
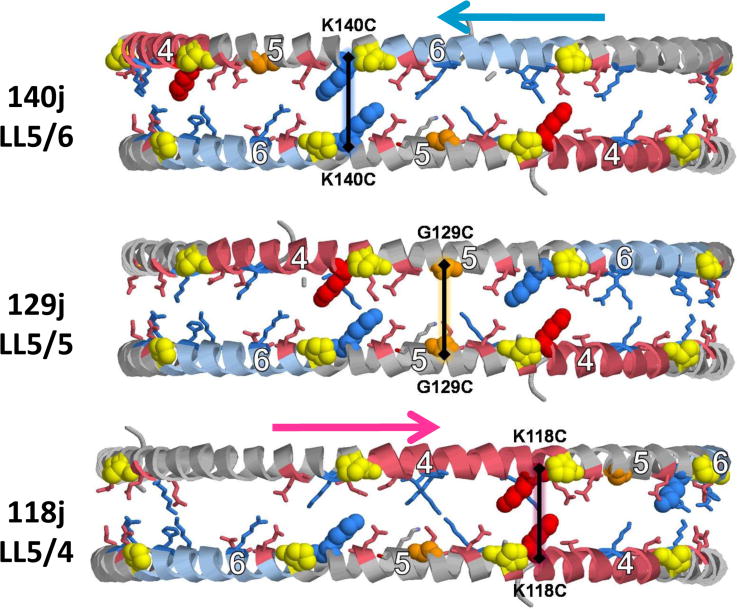
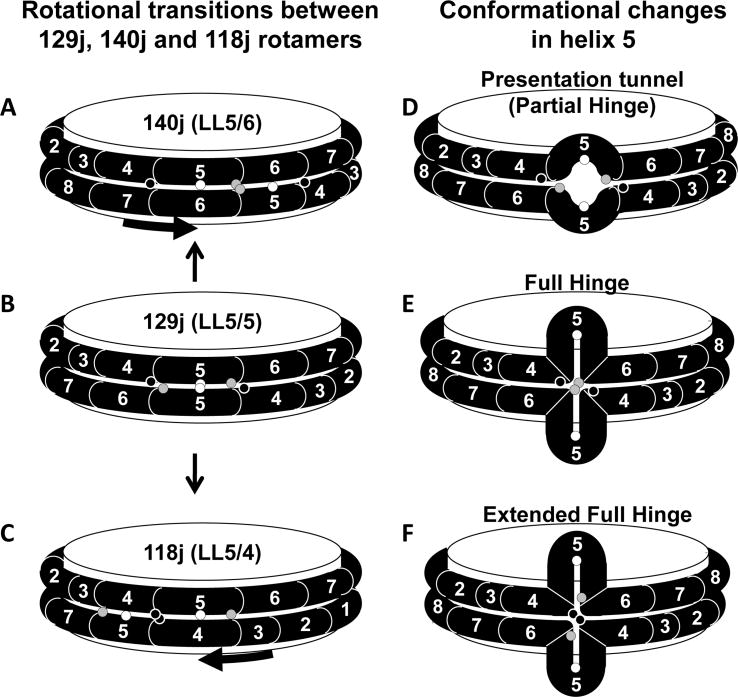
Fig. 1. Geometric rationale for design of cysteine mutants.
A. Weighted salt bridge scores of the LL docking interface determined using ALIGN [11]. B. Schematic of the double belt model for discoidal HDL showing the juxtaposition (within 6 Å) of Cα atoms of pairwise residues 129, 140 and 118 in the LL5/5, LL5/6 and LL5/4 rotamers, respectively. Each residue is in α11/3 wheel position 9. Prolines are yellow spacefilling, G129 is gold spacefilling, K140 is blue spacefilling, K118 is red spacefilling, helix 6 is steel blue, and helix 4 is pink.
In 2000-2001, three laboratories published additional physical chemical studies of apoA-I/HDL that were consistent with the belt model: i) Studies from the Davidson lab measured quenching of tryptophan mutants of apoA-I located at different positions along individual amphipathic helical repeats by nitroxide spin labeled phospholipid (ESR fluorescence quenching) [16, 17]. The belt model was supported because the two Trp residues in each helix exhibited maximal quenching at the same nitroxide group position on the lipid acyl chains; maximal quenching should have occur at two different levels in the bilayer in the picket fence model. ii) Studies from the Sorci-Thomas lab measured fluorescence resonance energy transfer (FRET) between cysteine mutants of apoA-I labeled with thiol-reactive fluorescent probes [18] to demonstrate that residue 132 in two lipid-bound apo A-I monomers were in close proximity, consistent with a double belt conformation for apo A-I on apoA-I/HDL. iii) Initial FRET studies from the Jonas lab excluded the head-to-head configuration of the picket fence model [19]; a second paper excluded all forms of the picket fence model for the conformation of apoA-I on discoidal HDL but considered the intramolecular hairpin-belt, as opposed to the antiparallel intermolecular belt model, to be the more likely of the two possible forms of belt [20].
FRET studies in 2002 by Sorci-Thomas, et al. [21] proposed the concept of a variable rather than a fixed helix-helix registry. Additionally, because spherical particles showed a lowering of acceptor to donor probe quenching when compared with starting apoA-I/HDL, they concluded that as lipid-bound apoA-I adjusts from a discoidal to a spherical surface its intermolecular interactions are significantly reduced. In 2003, the Davidson lab used mass spectrometry technology combined with thiol-cleavable cross-linking chemistry (MSXL) to derive a set of distance constraints on apoA-I/HDL particles [22]. The interhelical distance constraints determined supported the presence of salt-bridge interactions predicted to occur in the double belt model of apoA-I [11], but a helical hairpin model was not ruled out. Also in 2003, the Oda lab used site-directed spin-label electron paramagnetic resonance (EPR) spectroscopy to map apoA-I tertiary and quaternary structure of the apoA-I C terminus in lipid-free and lipid-associated states [23]. Spectra of apoA-I in reconstituted HDL revealed a lipid-induced transition of defined random coils and β strands into α helices. This conformational switch was proposed to serve as a means to overcome the energy barriers of lipid sequestration, a critical step in cholesterol efflux and HDL assembly.
The hinged-domain hypothesis was initially proposed by the Segrest lab as a general mechanism to explain the size heterogeneity of DMPC:apoA-I discoidal complexes [24, 25]. In 2004, the Davidson lab measured ESR fluorescence quenching of tryptophan mutants of apoA-I located at different positions along individual amphipathic helical repeats. The relative exposure of each tryptophan probe with increasing quencher concentrations was determined [26]; only helixes 5, 6, and 7 exhibited significant differences in terms of exposure to lipid between large and small apoA-I/HDL particles compatible with the hinged-domain model for the double belt. In 2006, the Oda lab used EPR spectroscopy and FRET to show that paired apoA-I molecules around the perimeter of apoA-I/HDL align in an extended antiparallel conformation with helix 5 of each apoA-I in juxtaposition (LL5/5 orientation); since the EPR spectra of nitroxide probes positioned at residues 134–145 did not exhibit spin coupling, they proposed a looped belt (or hinged-domain) model for residues 133–146 [27].
In 2005, the Davidson lab used MSXL to suggest a double belt arrangement for apoA-I on apoA-I/HDL in an LL5/5 orientation [28]. In 2008, Davidson lab suggested [29], based upon MSXL of discoidal and spheroidal apoA-I/HDL, that the general structural organization of apoA-I was similar between discs and spheres; Sorci-Thomas, et al. [21] had suggested the opposite. In the same year, the lab of Thomas and Sorci-Thomas used MSXL to suggest that incremental changes in the interaction between the N- and C-terminal ends of apoA-I allow it to unfold and sequester discrete amounts of phospholipid molecules [30, 31]; the N-terminal domain of apoA-I was proposed to loop back on itself maximizing interaction with the C-terminal domain, a model they termed the belt buckle.
The Hazen lab in 2007 reported a molecular model of nascent discoidal HDL, determined using hydrogen/deuterium (H/D) exchange mass spectrometry. The model suggested two apoA-I arranged in an antiparallel double-belt structure, with residues 159-180 (helix 7-8) forming an LCAT-associated solvent-exposed loop (hinged-domain) [32]. In 2009, using small angle neutron scattering (SANS), they proposed a model for apoA-I/HDL particles that is dramatically different from previous discoidal apoA-I/HDL models. In their model, termed a double super helix, apoA-I possesses an open helical shape that twists around a central end-capped hexagonal I phase lipid (rod-shaped micelle) [33]. We discuss pros and cons of the model in Gu, et al. [34].
The rationale for the current study is that, for each of the optimal (minimum) points in the salt bridge scores of the ALIGN plot (Fig. 1A), the defining residue juxtaposition (e.g., 129j (LL5/5)) is also position 9 on the α11/3 wheel [11], i.e., the residue position in the 11-mer repeats that lies directly in the contact edge between antiparallel belt helixes (Fig. S1). Therefore, we reasoned that if the two residues at wheel position 9 in the following juxtapositions, 129j (LL5/5), 140j (LL5/6), 173j (LL5/10-9 Milano) and 118j (LL5/4) (Table 1), were mutated to a Cysteine and kept under reducing conditions until after disc formation, we would have a highly precise method for determining registration in reconstituted PL-rich HDL by measurement of the degree of disulfide-linked apoA-I homodimer formation in discs containing each mutant apoA-I.
Table 1. Relationship between different rotamers, juxtaposed residues, cystine mutations, and ALIGN program scores.
| Rotamer | Wheel Position (wp) | Residues Juxtaposed | Cystine Mutation | Align Score |
|---|---|---|---|---|
| LL5/4 | 9 | 118j | K118C | −23.8 |
| LL5/5 | 9 | 129j | G129C | −24.6 |
| LL5/6 | 9 | 140j | K140C | −21.3 |
| LL5/7 (Paris) | 9 | 151j | R151C | −16.3 |
| 9 | 162j | −17.5 | ||
| LL5/10-9 (Milano) | 9 | 173j | R173C | −15.0 |
Legend: All residues juxtaposed are between two residues at wheel position 9.
2. Materials and methods
2.1. Experimental
2.1.1. Mutant expression and purification
ApoA-I cDNA in plasmid pGEMEX [35] was used as the template for site-directed mutagenesis. Oligonucleotides with desired mutations were designed to introduce mutations in the cDNA by the QuickChange Site-Directed Mutagenesis System (Strategene, La Jolla, CA) according to the manufacture's protocols. Oligonucleotides used in this study were as follows: K118C-apoA-I , the mutant designed to cross-link the LL5/4 rotamer, 5′CTCTACCG-CCAGTGTGTGGAGCCGCTG3′; G129C-apoA-I , the mutant designed to cross- link the LL5/5 rotamer, 5′GAGCTCCAAGA-GTGTGCGCGCCAGAAG3′; K140C-apoA-I , the mutant designed to cross-link the LL5/6 rotamer, 5′GAGCTGCAAGAGTGT-CTGAGCCCACTG3′. After confirming the DNA sequence, the mutant gene product was expressed in E. coli BL21/DE3 cells and purified as described previously [25]. Briefly, the expression of apoA-I and mutants was monitored by Western blotting of the bacterial lysate with anti-apoA-I antibody. The bacterial lysate containing mutant apoA-I was loaded onto a preparative reversed-phase HPLC (C4) column and proteins were eluted and separated by a gradient of acetonitrile with 0.1% trifluoroacetic acid. Preparative HPLC fractions containing apoA-I (identified by immuno-dot-blots) were then subjected to purity and identity analyses, including analytical HPLC (C18), SDS-PAGE, and mass spectrometry. Purified proteins were lyophilized and stored at –20°C or –80°C.
2.1.2. Preparation of wild type and mutant apoA-I solutions
Lyophilized protein was solubilized in 6 M guanidine hydrochloride. The solubilized protein solution was loaded on a desalting column (Econo-Pac 10DG Disposable Chromatography Columns, BioRad, Hercules, CA) and the protein was eluted by PBS buffer (0.02 M phosphate, 0.15 M NaCl, 0.01% NaN3, and 1mM EDTA, pH 7.4) or TBS buffer (0.15 M Tris, 0.15 M NaCl, 0.01% NaN3, and 1mM EDTA, pH 8.0). An extinction coefficient of 1.13 ml/(mg-cm) at 280 nm was used for determining the concentration of apoA-I in 6 M guanidine hydrochloride [35].
2.1.3. Preparation of phospholipids
Multilamellar vesicles (MLV) of 1, 2-dimyristoyl-sn-glycero- 3-phosphocholine (DMPC) were prepared by the procedures described previously [25]. Briefly, dry powder of DMPC (Avanti Polar-Lipids, Alabaster, AL) was dissolved in chloroform and dried under a stream of N2 in a pre-weighted glass tube with periodic mixing to form a lipid film on the wall of the tube. Upon complete removal of chloroform, PBS or TBS buffer was added to give a final DMPC concentration of 10 mg/ml. After hydrated in the buffer for at least 4 hours, DMPC was dispersed by vortexing until a uniform milky suspension was formed. This dispersion was used for preparation of discoidal apoA-I:DMPC complexes.
2.1.4. Preparation and analyses of discoidal apoA-I:DMPC complexes and cross-linking of cysteine-containing apoA-I mutants after disc formation
MLV of DMPC was mixed with apoA-I and mutants in different molar ratios as needed in the presence of a reducing agent, tris(2-carboxyethyl)phosphine (TCEP, final concentration of 5 mM). To ensure that this concentration of TCEP could effectively reduce all cysteine mutants, protein sample without and with TCEP were subjected to SDS-PAGE. The results showed that all cysteine mutants were 100% monomeric in the presence of TCEP (Fig. S2). The goal was to create intermolecular disulfide cross-links specific for the lipid-associated conformation of apoA-I; because each mutation has only one cysteine mutation, intramolecular cross-linking is not possible. The lipid-protein mixture was incubated at room temperature (22-24°C). After an overnight incubation, to completely remove TCEP so as to allow disulfide bond formation, the complex was dialyzed extensively against PBS buffers overnight at 4°C with at least 8 changes of 1 L each. Then, an aliquot of the complex was analyzed by non-denaturing gradient gel electrophoresis (NDGGE) and SDS-PAGE. The rest of the samples were incubated further at 37°C for up to 6 hours before subjected to NDGGE and SDS- PAGE. NDGGE was accomplished on a 4-20% gradient Tris-glycine acrylamide gel and SDS- PAGE on a 12% or 18% acrylamide gel (Invitrogen, Carlsbad, CA). The gels were stained with the colloidal blue staining kit (Invitrogen, Carlsbad, CA). The Stokes diameters (Sd) and percentage of dimerization of cysteine-containing apoA-I mutants were calculated based on density scans of the gels by LabWorks image acquisition and analysis software (UVP Inc., Upland, CA).
2.2. Computational
2.2.1. PL-rich particles
A particle containing 160 DMPC and 2 apoA-I molecules (160:2, R2-2) was created as described by us elsewhere [34]. This initial particle were simulated at 310 K and 1 bar for 5 ns, and then was subjected to the following MDSA protocol (all at 1 bar): heated from 310 K to 500 K in 20 ps, simulated at 500 K for 5 ns, cooled gradually from 500 K to 310 K in 5 ns, and finally simulated at 310 K for 5 ns, giving a total duration of 15 ns. For simulations at a fixed temperature, velocity reassignments occurred every 1 ns to prevent the “flying ice cube” effect [36].
All-atom simulations were performed using NAMD [37] as described in Catte, et al. [38]. Each system was ionized and charge neutralized with NaCl to 0.15 M with the autoionize plug-in of Visual Molecular Dynamics (VMD) [39]. The TIP3P water model was used [40]. The CHARMM 22 [41, 42] and 27 [43, 44] force fields were used for protein and lipid molecules, respectively.
2.2.2. Distances between G129 Ca atoms
The distances between the Cα atoms for pairs of G129 residues were measured over the last 5 ns of the MDSA protocol, and the fractional occurrence for each distance was plotted.
3. Results and Discussion
Fig. 1B diagrammatically illustrates the structural rationale for placement of cysteine residues into wheel position 9 via site-directed mutagenesis. From the diagram, the mutants G129C, K140C or K118C are capable of forming a homodimer if the double belt is in a 129j (LL5/5), 140j (LL5/6) or 118j (LL5/4) rotamer conformation, respectively. This structural feature of the a11/3 double belt conformation explains why the natural human mutations, apoA-I Paris (R151C, 151j (LL5/7) Fig. S3) and Milano (R173C, 173j (LL10-9) Fig. S4), both with cysteine mutations in wheel position 9, are able to form HDL particles containing homodimeric apoA-I [45].
Fig. S5 shows representative NDGGE analysis of particles made with K118C-apoA-I, G129C-apoA-I and K140C-apoA-I at a 180:2 molar ratio. The particles were subjected to SDS gel electrophoresis to distinguish apoA-I monomers and homodimer; the results are shown in Fig. 2A. To test if the percent of dimers is influenced by potential interchain dimerization upon denaturation of particles by SDS, the blocking agent N-ethyl-maleimide (NEM), was used to block all free SH groups before the particles were subjected to SDS-PAGE. The results show no difference in the percentage of dimer formation with or without NEM, indicating no further formation of disulfide bonds after SDS treatment during SDS-PAGE (See Fig. S6). Therefore, NEM was not included in the SDS-PAGE buffer for further experiments.
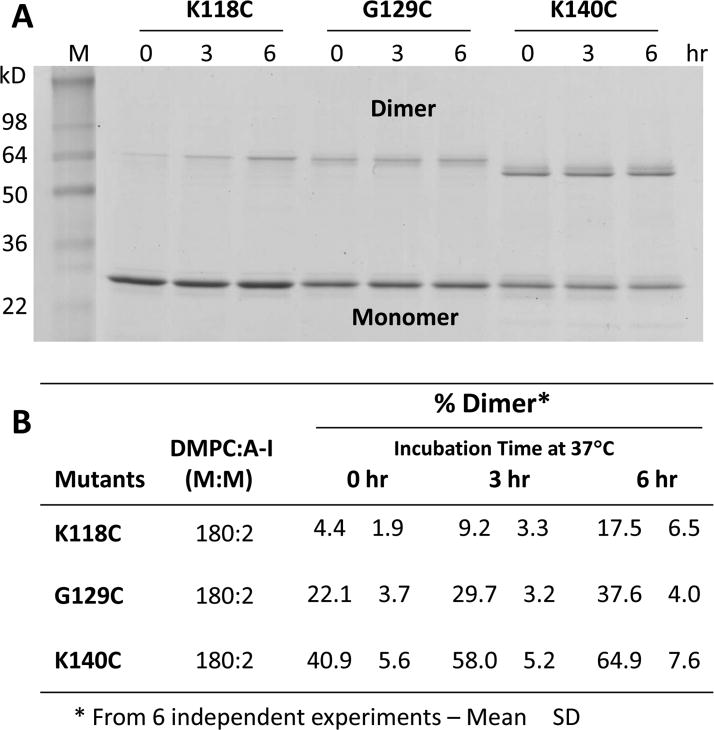
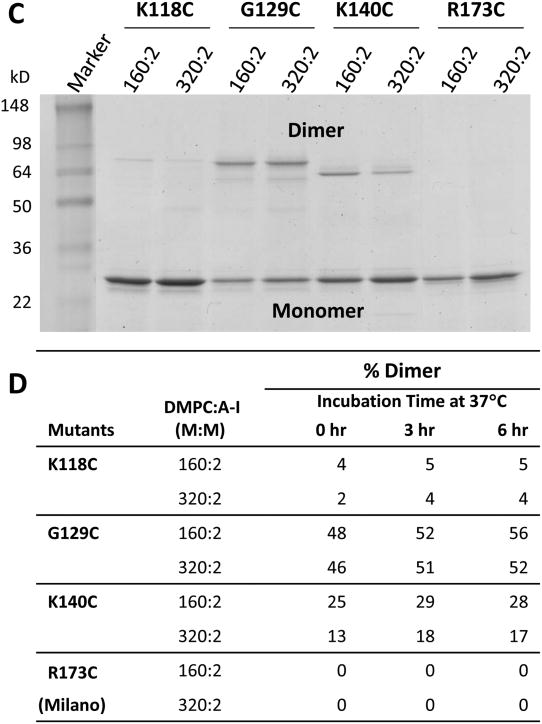
Fig. 2. Formation of disulfide homodimers in preformed DMPC:apoA-I particles made from cysteine mutants of apoA-I.
A. Representative image of SDS-PAGE of 180:2 particles at t = 0, t = 3 and t = 6 hrs after dialysis to induce disulfide homodimers of cysteine mutants K118C-apoA-I, G129C-apoA-I and K140C-apoA-I. B. Percent homodimer formation calculated for each mutant in B from scans as a function of time of incubation at 37°C. C. Representative image of SDS-PAGE of 160:2 and 320:2 particles at 3 hrs after dialysis to induce disulfide homodimers of cysteine mutants K118C-apoA-I, G129C-apoA-I, K140C-apoA-I and R173C-apoA-I. D. Percent homodimer formation calculated for each mutant in D from scans as a function of time of incubation at 37°C.
The bands were scanned and the percent homodimer formation calculated for each mutant as a function of time of incubation at 37°C (Fig. 2B). The mutant K118C-apoA-I forms apoA-I homodimer in low concentrations at t = 0 (4%) and homodimer formation increases 4-fold after 6 hrs of incubation at 37°C (17%). In contrast, the mutants G129C-apoA-I and K140C-apoA-I form homodimer in higher concentrations at t = 0 (22% and 41%, respectively); however, homodimer formation increased less than 2-fold after 6 hrs of incubation at 37°C (38% and 65%, respectively). Therefore, the kinetics of homodimer formation in K118C-apoA-I particles is much slower than in either G129C-apoA-I or K140C-apoA-I particles, while homodimer formation is twice as fast in K140C-apoA-I particles as in G129C-apoA-I particles.
As noted in Fig. 2A, the migration of dimers (apparent molecular weight) differs depending on the site of Cys substitution in the SDS-PAGE. However, all monomer bands migrate as a single band with identical apparent molecular weight. This phenomenon has been observed in other Cys mutants of apoA-I [31, 46], indicating that covalently linked dimer is in a more elongated conformation when the Cys is positioned centrally within the apoA-I primary sequence. Therefore, K118C and G129C dimers appear to be bigger (slower mobility) than K140C dimers on the gel. The migration behavior of proteins in SDS-PAGE was thoroughly studied by Reynolds, et al. [47]. Consistent with what we observe here, these authors suggested that protein in SDS complexes forms double helical rods.
The three apoA-I mutants, K118C, G129C and K140C, were used to make particles under reducing conditions that had differing DMPC:apoA-I molar ratios (160:2 versus 320:2) (Fig. S7). These particles were then subjected to SDS gel electrophoresis to distinguish apoA-I monomers and homodimer and the results are shown in Fig. 2C. The bands were scanned and the percent homodimer formation calculated for all six particles as a function of time of incubation at 37°C (Fig. 2D). The results show that the kinetics of homodimer formation was dependent upon the DMPC to apoA-I ratios (therefore particle size) in K140C-apoA-I (and perhaps K118C-apoA-I) particles; there was half as much homodimer formation in K140C-apoA-I particles containing higher DMPC:apoA-I molar ratios (320:2) as there was in those containing lower molar ratios (160:2). For G129C-apoA-I particles the kinetics of homodimer formation was independent of the DMPC to apoA-I ratios or particle size.
We also made particles from R173C-apoA-I mutants under reducing condition. The resultant particles were subjected to dialysis-induced oxidation to induce interchain homodimer formation and then were subjected to NDGGE and SDS-PAGE. Figs. 2C and D show that R173C was unable to dimerize at either molar ratio.
Our initial expectation based upon our analyses of interhelical salt bridges and the data of other labs was that, under the conditions of our experimental setup, 129j (LL5/5) would start in high abundance, while 140j (LL5/6) and 118j (LL5/4) would be expected to start in low abundance and increase slowly, if at all, with time. However, we found that LL5/5 and LL5/6 both started with high abundance and increased with time.
Based upon our initial rationale for the studies described, it would be tempting to conclude that the observed homodimer formation provides support for a dynamic variability in registry for the antiparallel helical rings in the double belt conformation of apoA-I on discoidal HDL (Figs. 3A-C), a registry that favors 140j (LL5/6) and/or 129j (LL5/5). However, there are several very good reasons to question this possibility: i) The x-ray crystal structure of Borhani, et al. [9] supports the 129j (LL5/5) rotamer; if alternate conformations had been present, crystallization would not have been possible ii) The salt bridge score in Fig. 1A makes it seem unlikely that 129j (LL5/5) could undergo a dynamic rotation to form 140j (LL5/6) or 118j (LL5/4) in the time frame examined by Fig. 2. The free energy barriers between rotamers—the steepness of the slope between minima in Fig. 1A—suggest that a given rotamer would be kinetically trapped. iii) Most importantly, although FRET studies by Sorci-Thomas, et al. [21] proposed the concept of a variable rather than a fixed helix-helix registry, more recent physical chemical studies from three separate laboratories [22,24,26] provide solid evidence for the 129j (LL5/5) rotamer but no evidence at all for either the 140j (LL5/6) or 118j (LL5/4) rotamers.
Fig. 3. Schematic diagrams of rotamer versus hinged domain explanations of the homodimer results.
Mutated residues K118C, G129C and K140C are denoted by black, white and gray circles, respectively. A-C. Positions of cysteine residues in 140j (LL5/6), 129j (LL5/5), and 118j (LL5/4), respectively. D-F. Positions of cysteine residues in 129j (LL5/5) after the pairwise helix 5 domains have formed a partial, full or extended hinged domain, respectively.
An alternate possibility is that formation of different juxtapositions may occur when cysteines are inserted in different positions. Although this possibility cannot be completely ruled out, salt bridges formed by K118-E147 and E125-K140 are what we call solvent-accessible; based upon our MD studies and experimental and theoretical arguments [11, 48]these are much less stable than the solvent-inaccessible salt bridges, like D89-R177. Based upon our MD simulations relating to the possible role of the helix 5 domain as an LCAT presentation tunnel, rather than affecting rotamer distribution, a more likely effect of removal of K140, given the dynamic role of both inter- and intrahelical salt bridges in the stability of this tunnel would be instability of the presentation tunnel and expansion.
Our laboratory in 1984 hypothesized the existence of a hinged domain to explain the discontinuous heterogeneity in size of discoidal HDL particles [24, 25]; we later suggested that the hinge might be located in the pairwise helix 5 domain [32]. Recently, two laboratories have used quenching of tryptophan mutants of apoA-I located within each of its 22 amino acid amphipathic helical repeats by nitroxide spin labeled phospholipid [26] and EPR spectroscopy/FRET scanning mutagenesis [27] to suggest that the pairwise helix 5 domain can form a hinged domain in which the double belt loops off the edge of discoidal HDL (see Fig. 3E).
The kinetics displayed in Fig. 2 can be explained by formation of a hinge in the region of the pairwise helix 5 antiparallel domain. If the pairwise hinge extends the full length of helix 5, the C140 pairs can approach within 6 Å, close enough in particles made with the K140C-apoA-I mutant to form disulfide bonds (Fig. 3E). Further, if the hinge extends a few residues beyond the proline boundaries of the pairwise helix 5 antiparallel domains, then the C118 pairs can also approach closely enough in particles made with the K118C-apoA-I mutant to form disulfide bonds (Fig. 3F).
In the studies reported here, the smallest particle formed and subjected to cross-linking under oxidative conditions was R2-2 (180-160:2). Disulfide bond formation in the G129C particles was not affected by particle size but, for the R140C particles (and perhaps the R118C particles), percent cross-linking was decreased in larger 320:2 compared to smaller R2-2 particles; DMPC at a 320:2 molar ratio forms predominantly larger R2-3 and R2-4 particles [34].
The Davidson lab reported that the hinged domain was present in the smaller R2-0 particles but was not present in larger R2-2 ones [26, 28]. The EPR spectroscopy/FRET studies by the Oda lab [27] were performed only on R2-2 particles; although they speculated that the hinge domain contributes to the folding of apoA-I around smaller R2-0 particles, they did not examine smaller sized particles.
If one assumes that the cysteine mutations have no effects on the energetics of transition from a pairwise antiparallel helix 5 double belt to a hinged domain, the decreased cross-linking in R140C-apoA-I particles observed with increasing particle size can be explained. However, since our studies were performed with R2-2 particles or larger, this possibility is difficult to reconcile with the Davidson lab report that the hinge domain is not obvious in R2-2 particles [26, 28].
If one assumes that the cysteine mutations affect the energetics of transition from a pairwise antiparallel helix 5 double belt to a hinged domain, the results of Fig. 2 are readily explained. Residues R140 and R1 18 are both involved in the putative interhelical salt bridges, R140-E125 and R1 18-E147 [11, 38]; in fact, R140-E125 is the only salt bridge holding the edges of the pairwise antiparallel helix 5 double belt together (Fig. 3D). Removal of this interhelical salt bridge by R140C should destabilize the intermolecular interactions of the pairwise antiparallel helix 5 double belt, allowing an increased rate of transition to the intramolecular interactions of a helix 5 hinged domain. The much lower observed cross-linking of the R118C mutant then would be explained by the persistence of the intervening interhelical salt bridge, R140-E125; thus 4-5% perhaps approximates the actual prevalence of a full hinged domain in R2-2 particles made from wt-apoA-I.
We have suggested, on the basis of MD simulations, that the opposing individual helix 5 portions of the double-belt motif separate to create a gap exposing underlying POPC acyl chains [49]. Specifically, we proposed the working hypothesis that the gap creates an amphipathic presentation tunnel that exposes acyl chain terminal methyls and unesterified cholesterol hydroxyl groups to solvent. After attachment of the enzyme lecithin:cholesterol acyltransferase (LCAT) to HDL, we hypothesized that the pairwise helix 5 domains in apoA-I form amphipathic tunnels for migration of hydrophobic acyl chains and amphipathic unesterified cholesterol hydroxyls from the bilayer to the phospholipase A2-like and esterification active sites of LCAT, respectively [49].
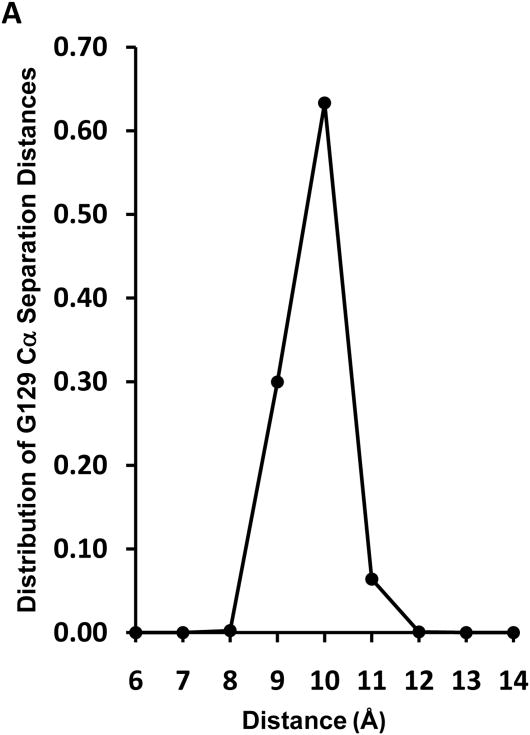
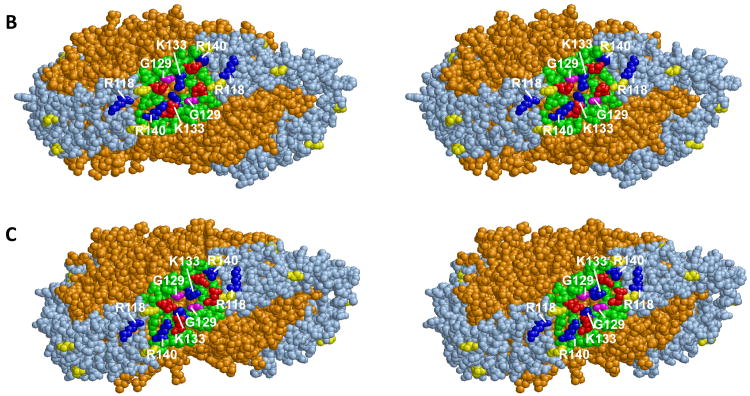
Fig. 4A represents a plot of the distribution of distances between pairwise G129 Cα atoms measured for one R2-2 (160:2) discoidal particle subjected to MDSA simulation. The center of the distribution is 10 Å and the closest pairwise G129 approach in the last 310 K part of the simulation is on the order of 7-8 Å. Two frames whose pairwise G129 Cα atom distances are 11.9 Å and 7.7 Å are shown in Figs. 4B and C, respectively. Given the brief time frame of our MDSA simulation compared to that of the experiments in Fig. 2, it seems reasonable that the left hand edge of the distribution would occasionally approach within the 6 Å distance required for disulfide bond formation. Once within 6 Å, disulfide cross-linking would drive the reaction to the right and over minutes to hours a significant level of homodimer formation would result (see Fig. 4 for details of calculations).
Fig. 4. MD simulation of a particle containing 160 DMPC and 2 apoA-I molecules (160:2).
A. Plot of distribution of distances between Cα atoms of pairwise G129 over the last 5 ns of the MDSA simulation., The fraction of frames is given at each distance d where the measured distance is > d-1 and ≤ d. B. Cross-eyed stereo spacefilling image of one trajectory frame whose pairwise G129 Cα atoms are 11.9 Å apart. C Cross-eyed stereo spacefilling image of one trajectory frame whose pairwise G129 Cα atoms are 7.7 Å apart. Lipids are gold, prolines are yellow, helix 5 domains are green, basic residues K118, K133 and R140 are blue, acidic residues E125 and E136 are red and G129 residues are magenta. Calculations of the probability that G129C can form interchain disulfide bonds between pairwise helix 5 segments. From Fig. 2C, the rate of homodimer formation for G129C particles is approx. 0.05 mole fraction per hour. Assuming a linear rate of disulfide formation, homodimer formation is approx. 1.4 × 10−5 mole fraction per second or 1.4 × 10 −11 mole fraction per usec. In a mole of particles, this is equal to. 8 × 1012 apoA-I/HDL particles per usec. Let's make the assumption that an approach of pairwise G129C residues to within 6 Å results in homodimer formation and further that one MDSA cycle is equivalent to 0.05 μsec of simulation for a single particle (probably an underestimation). On the basis of these assumptions, the plot of distance distributions in Fig. 4A is reasonable: a zero incidence of pairwise G129C residues approaching to within 6 Å and a 5 × 10−5 incidence of pairwise G129C residues approach to within 7 Å, all within tens of nanoseconds. In fact, even one frame in which pairwise G129C residues approach to within 6 Å in the plot would suggest that there should be 100% homodimer formation in one hr or even one minute.
It can be seen from Fig. 4B that the putative presentation tunnel is, in fact, a partial hinged domain as shown in Fig. 3D. We have yet to observe a full hinged domain like that in Fig. 3E in any of our simulations of R2-2 discoidal particles. We have, however, observed helix 5 forming a full hinged domain twice out of eight MD simulations of smaller R2-1 and R2-0 particles (data not shown).
The vertical opening in a full helix 5 hinged domain, like that schematically illustrated in Fig. 3E, would be on the order of 30 Å in length, approximately the thickness of the acyl chain center of the bilayer disc. This observation leads to the speculation that, as part of the mechanism of apoA-I activation of LCAT, interaction of the central helix 5 presentation tunnel with LCAT might drive the formation of a full helix 5 hinged domain, exposing the ester bonds of the underlying phospholipid acyl chains to the phospholipase A2-like active site of the enzyme.
On the basis of the results reported here and that of three other laboratories [22,24,26], 129j (LL5/5) is the major rotamer conformation for discoidal double belt HDL. Although Fig. 4A shows only a tiny fraction of the particle trajectory in which pairwise G129 approach within 7 Å, our MD simulation is representative of only a very small window of time (nanoseconds). It seems reasonable to assume that the Cα atoms of the C129 pair on opposite chains in R2-2 in a time-frame of minutes to hrs approach within 6 Å of one another sufficiently often for disulfide bond formation to occur in 20-40% of the experimental particles; the fact that this does not approach 100% may reflect the polydispersity of the R2-2 particle sizes. Thus, in only a subset of particles does the C129 pair on opposite chains approach close enough to form disulfide bonds. The remainder of the particles appears to form presentation tunnel gaps that are too wide and too stable for cross-linking.
We suggest that the formation of homodimers by R140C-apoA-I in discoidal HDL is the result of formation of a full helix 5 hinged domain, partially as a result of the loss of a pair of presentation tunnel stabilizing interchain salt bridges. Evolution has carefully balanced the helix 5 domain between pairwise helixes, helical hairpins (required for R3 particles) and apparently hinged domains [50]. Decreases in the stability of the pairwise helix 5 domain due to various point mutations employed by the Davidson [26] and Oda [27] labs perhaps explains their reports of the hinged domain. In the relatively rare instance where the hinge extends a few residues beyond a full hinged domain, the C118 pairs also are able to form disulfide bonds.
As a control, C173 does not form homodimers on preformed particles but does when incubated in solution (see Fig. S8). These results suggest that apoA-I:Milano homodimers form in vivo in the lipid-free or lipid-poor state, prior to particle formation [45], or during assembly of nascent HDL, although it is possible that homodimer formation also occurs during intact HDL remodeling. Others have shown that cross-linked C173 dimer can form particles in which the mutant apoA-I adopts a different conformation from that of particles with wild-type apoA-I [51].
We hypothesize that, although R2-2 particles in the 129j (LL5/5) rotamer conformation are primed to create presentation tunnels in the form of gaps between opposing individual helix 5 portions of the double-belt motif like that shown in Figs. 3D and 4B[49], the energy barrier for transition to a full hinged domain (Fig. 3E) is high. Full transition to a stable hinged domain requires additional conditions, such as allosteric binding of the enzyme LCAT to the presentation tunnel domain or a decrease in particle size.
Supplementary Material
Highlights
Interhelical salt bridges for the HDL belt model maximize at wheel position (wp) 9.
Only wp9 lies directly in the contact edge between helical rings.
Wp9 was mutated to Cys and kept under reducing conditions during disc formation.
We show that helix 5, under certain conditions, progresses to a full hinged domain.
We hypothesize that, upon binding of LCAT, helix 5 forms a full hinged domain.
Acknowledgments
We would like to thank the UAB Information Technology's Research Computing Group and the UAB Department of Mechanical Engineering for use of the Cheaha High Performance Computing resource that they jointly maintain. This work was supported in part by the National Institutes of Health grant P01 HL34343 and R01 AG031846.
Footnotes
The abbreviations used are: apo, apolipoprotein; apoA-I/HDL, reconstituted HDL; DMPC, dimyristoylphosphatidylcholine; HDL, high density lipoproteins; LL5/5, double belt rotamer forming a pairwise helix 5 domain; LL5/6, double belt rotamer forming pairwise helix 5/6 domains; LL5/4, double belt rotamer forming pairwise helix 5/4 domains; MD, molecular dynamics; MDSA, molecular dynamics simulated annealing; PATIR-FTIR, Fourier-transform infrared spectroscopy; POPC, palmitoyloleoylphosphatidylcholine; R2 or R3 or R4, reconstituted high density lipoprotein particles containing 2, 3, or 4 molecules of apoA-I; MD, molecular dynamics; VMD, visual molecular dynamics; MDSA, molecular dynamics simulated annealing; NDGGE, non-denaturing gradient gel electrophoresis; SDS, sodium dodecylsulfate.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Linsel-Nitschke P, Tall AR. HDL as a target in the treatment of atherosclerotic cardiovascular disease. Nat Rev Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 2.Tall AR, Lange Y. Interaction of cholesterol, phospholipid and apoprotein in high density lipoprotein recombinants. Biochim Biophys Acta. 1978;513:185–197. doi: 10.1016/0005-2736(78)90172-4. [DOI] [PubMed] [Google Scholar]
- 3.van Antwerpen R, Chen GC, Pullinger CR, Kane JP, LaBelle M, Krauss RM, Luna-Chavez C, Forte TM, Gilkey JC. Cryo-electron microscopy of low density lipoprotein and reconstituted discoidal high density lipoprotein: imaging of the apolipoprotein moiety. J Lipid Res. 1997;38:659–669. [PubMed] [Google Scholar]
- 4.Dory L, Boquet LM, Hamilton RL, Sloop CH, Roheim PS. Heterogeneity of dog interstitial fluid (peripheral lymph) high density lipoproteins: implications for a role in reverse cholesterol transport. J Lipid Res. 1985;26:519–527. [PubMed] [Google Scholar]
- 5.Wlodawer A, Segrest JP, Chung BH, Chiovetti R, Jr, Weinstein JN. High-density lipoprotein recombinants: evidence for a bicycle tire micelle structure obtained by neutron scattering and electron microscopy. FEBS Lett. 1979;104:231–235. doi: 10.1016/0014-5793(79)80821-2. [DOI] [PubMed] [Google Scholar]
- 6.Atkinson D, Small DM, Shipley GG. X-ray and neutron scattering studies of plasma lipoproteins. Ann N Y Acad Sci. 1980;348:284–298. doi: 10.1111/j.1749-6632.1980.tb21308.x. [DOI] [PubMed] [Google Scholar]
- 7.Segrest JP, Jackson RL, Morrisett JD, Gotto AM., Jr A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett. 1974;38:247–258. doi: 10.1016/0014-5793(74)80064-5. [DOI] [PubMed] [Google Scholar]
- 8.Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 9.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koppaka V, Silvestro L, Engler JA, Brouillette CG, Axelsen PH. The structure of human lipoprotein A-I. Evidence for the “belt” model. J Biol Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- 11.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 12.Bussell R, Jr, Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 13.Bussell R, Jr, Ramlall TF, Eliezer D. Helix periodicity, topology, and dynamics of membrane-associated alpha-synuclein. Protein Sci. 2005;14:862–872. doi: 10.1110/ps.041255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones MK, Anantharamaiah GM, Segrest JP. Computer programs to identify and classify amphipathic alpha helical domains. J Lipid Res. 1992;33:287–296. [PubMed] [Google Scholar]
- 15.Segrest JP, Jones MK, Mishra VK, Anantharamaiah GM. Experimental and computational studies of the interactions of amphipathic peptides with lipid surfaces. Current Topics in Membranes. 2002;52:391–429. [Google Scholar]
- 16.Maiorano JN, Davidson WS. The orientation of helix 4 in apolipoprotein A-I-containing reconstituted high density lipoproteins. J Biol Chem. 2000;275:17374–17380. doi: 10.1074/jbc.M000044200. [DOI] [PubMed] [Google Scholar]
- 17.Panagotopulos SE, Horace EM, Maiorano JN, Davidson WS. Apolipoprotein A-I Adopts a Belt-like Orientation in Reconstituted High Density Lipoproteins. J Biol Chem. 2001;276:42965–42970. doi: 10.1074/jbc.M106462200. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Lyles DS, Thomas MJ, Pan W, Sorci-Thomas MG. Structural determination of lipid-bound apo A-I using fluorescence resonance energy transfer. J Biol Chem. 2000;275:37048–37054. doi: 10.1074/jbc.M005336200. [DOI] [PubMed] [Google Scholar]
- 19.Tricerri MA, Behling Agree AK, Sanchez SA, Jonas A. Characterization of apolipoprotein A-I structure using a cysteine-specific fluorescence probe. Biochemistry. 2000;39:14682–14691. doi: 10.1021/bi0014251. [DOI] [PubMed] [Google Scholar]
- 20.Tricerri MA, Behling Agree AK, Sanchez SA, Bronski J, Jonas A. Arrangement of apolipoprotein a-i in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry. 2001;40:5065–5074. doi: 10.1021/bi002815q. [DOI] [PubMed] [Google Scholar]
- 21.Li HH, Lyles DS, Pan W, Alexander E, Thomas MJ, Sorci-Thomas MG. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 22.Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass specrometry study. J Biol Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 23.Oda MN, Forte TM, Ryan RO, Voss JC. The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nature Structural Biology. 2003;10:455–460. doi: 10.1038/nsb931. [DOI] [PubMed] [Google Scholar]
- 24.Brouillette CG, Jones JL, Ng TC, Kercret H, Chung BH, Segrest JP. Structural studies of apolipoprotein A-I/phosphatidylcholine recombinants by high-field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry. 1984;23:359–367. doi: 10.1021/bi00297a027. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Chen J, Mishra VK, Kurtz JA, Cao D, Klon AE, Harvey SC, Anantharamaiah GM, Segrest JP. Double belt structure of discoidal high density lipoproteins: molecular basis for size heterogeneity. J Mol Biol. 2004;343:1293–1311. doi: 10.1016/j.jmb.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Maiorano JN, Jandacek RJ, Horace EM, Davidson WS. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 2004;43:11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- 27.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. Journal of Biological Chemistry. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 28.Silva RA, Hilliard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 29.Silva RA, Huang R, Morris J, Fang J, Gracheva EO, Ren G, Kontush A, Jerome WG, Rye KA, Davidson WS. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc Natl Acad Sci U S A. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 31.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Wagner MA, Zheng L, Parks JS, Shy JM, 3rd, Smith JD, Gogonea V, Hazen SL. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, Gogonea V, Lee X, Wagner MA, Li XM, Huang Y, Undurti A, May RP, Haertlein M, Moulin M, Gutsche I, Zaccai G, Didonato JA, Hazen SL. The Double Super Helix model of high density lipoprotein. J Biol Chem. 2009 doi: 10.1074/jbc.M109.039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu F, Jones MK, Chen J, Patterson JC, Catte A, Jerome WG, Li L, Segrest JP. Structures of Discoidal High Density Lipoproteins: A COMBINED COMPUTATIONAL-EXPERIMENTAL APPROACH. J Biol Chem. 285:4652–4665. doi: 10.1074/jbc.M109.069914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers DP, Brouillette CG, Engler JA, Tendian SW, Roberts L, Mishra VK, Anantharamaiah GM, Lund-Katz S, Phillips MC, Ray MJ. Truncation of the amino terminus of human apolipoprotein A-I substantially alters only the lipid-free conformation. Biochemistry. 1997;36:288–300. doi: 10.1021/bi961876e. [DOI] [PubMed] [Google Scholar]
- 36.Harvey SC, Tan RKZ, Cheatham TE. The flying ice cube: Velocity rescaling in molecular dynamics leads to violation of energy equipartition. Journal of Computational Chemistry. 1998;19:726–740. [Google Scholar]
- 37.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J Comp Phys. 1999;151:283–312. [Google Scholar]
- 38.Catte A, Patterson JC, Jones MK, Jerome WG, Bashtovyy D, Su Z, Gu F, Chen J, Aliste MP, Harvey SC, Li L, Weinstein G, Segrest JP. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen WL, Chandrasekhar J, Madura JD. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 41.MacKerell AD, Jr, Bashford D, Bellot M, Dunbrack RL, Jr, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 42.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. Journal of computational chemistry. 1983;4:187–217. [Google Scholar]
- 43.Feller SE, Pastor RW. Length scales of lipid dynamics and molecular dynamics. Pac Symp Biocomput. 1997:142–150. [PubMed] [Google Scholar]
- 44.Schlenkrich M, Brickmann J, MacKerell A, Jr, Karplus M. In: An empirical potential energy function for phospholipids: criteria for parameter optimization and applications, Biological Membranes: A Molecular Perspective from Computation and Experiment. Merz KM, Roux B, editors. Birkhauser; Boston: 1996. pp. 31–81. [Google Scholar]
- 45.Klon AE, Jones MK, Segrest JP, Harvey SC. Molecular belt models for the apolipoprotein A-I Paris and Milano mutations. Biophys J. 2000;79:1679–1685. doi: 10.1016/S0006-3495(00)76417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oda MN, Bielicki JK, Berger T, Forte TM. Cysteine substitutions in apolipoprotein A-I primary structure modulate paraoxonase activity. Biochemistry. 2001;40:1710–1718. doi: 10.1021/bi001922h. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds JA, Tanford C. The gross conformation of protein-sodium dodecyl sulfate complexes. J Biol Chem. 1970;245:5161–5165. [PubMed] [Google Scholar]
- 48.Blundell T, Barlow D, Borkakoti N, Thornton J. Solvent-induced distortions and the curvature of alpha-helices. Nature. 1983;306:281–283. doi: 10.1038/306281a0. [DOI] [PubMed] [Google Scholar]
- 49.Jones MK, Catte A, Li L, Segrest JP. Dynamics of activation of lecithin:cholesterol acyltransferase by apolipoprotein A-I. Biochemistry. 2009;48:11196–11210. doi: 10.1021/bi901242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bashtovyy D, Jones MK, Anantharamaiah GM, Segrest JP. Sequence conservation of apolipoprotein A-I affords novel insights into HDL structure-function. J Lipid Res. 2010 doi: 10.1194/jlr.R012658. Epub Date 2010/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhat S, Sorci-Thomas MG, Calabresi L, Samuel MP, Thomas MJ. Conformation of dimeric apolipoprotein A-I milano on recombinant lipoprotein particles. Biochemistry. 2010;49:5213–5224. doi: 10.1021/bi1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.