Abstract
The mammary gland is an ideal “model organism” for studying tissue specificity and gene expression in mammals: it is one of the few organs that develop after birth and it undergoes multiple cycles of growth, differentiation and regression during the animal’s lifetime in preparation for the important function of lactation. The basic “functional differentiation” unit in the gland is the mammary acinus made up of a layer of polarized epithelial cells specialized for milk production surrounded by myoepithelial contractile cells, and the two-layered structure is surrounded by basement membrane. Much knowledge about the regulation of mammary gland development has been acquired from studying the physiology of the gland and of lactation in rodents. Culture studies, however, were hampered by the inability to maintain functional differentiation on conventional tissue culture plastic. We now know that the microenvironment, including the extracellular matrix and tissue architecture, plays a crucial role in directing functional differentiation of organs. Thus, in order for culture systems to be effective experimental models, they need to recapitulate the basic unit of differentiated function in the tissue or organ and to maintain its three-dimensional (3D) structure. Mouse mammary culture models evolved from basic monolayers of cells to an array of complex 3D systems that observe the importance of the microenvironment in dictating proper tissue function and structure. In this chapter, we focus on how 3D mouse mammary epithelial cultures have enabled investigators to gain a better understanding of the organization, development and function of the acinus, and to identify key molecular, structural, and mechanical cues important for maintaining mammary function and architecture. The accompanying chapter of Vidi et al. describes 3D models developed for human cells. Here, we describe how mouse primary epithelial cells and cell lines—essentially those we use in our laboratory—are cultured in relevant 3D microenvironments. We focus on the design of functional assays that enable us to understand the intricate signaling events underlying mammary gland biology, and address the advantages and limitations of the different culture settings. Finally we also discuss how advances in bioengineering tools may help towards the ultimate goal of building tissues and organs in culture for basic research and clinical studies.
Keywords: Mouse mammary epithelium, 3D culture, Tissue architecture, Polarity, Tissue-specific signaling
1. Introduction
1.1. Rationale for Using Rodent Culture Models to Understand Human Disease
Biological research has been hampered by the inability to sustain functional differentiation in organs and tissues ex vivo for long enough to allow for adequate experimentation. In addition, tissue samples of human origin are scarce, and the ethical constraints guiding their use necessitate that we find alternative model organisms for study. Rodents and particularly mice have emerged as the species of choice in biological research: they are sufficiently similar to humans anatomically and physiologically, although the differences need to be kept in mind at all times. In addition, they are small and easy to handle and breed, and their short life span and rapid reproductive rate make it possible to study development and disease processes and to generate statistically significant data sets. Standardization of mouse research was possible by the generation of inbred lines of laboratory mice. This was essential initially to circumvent issues of genetic variability among animals, and as such made it possible to replicate experiments more rigorously. Genetic engineering in mice was another milestone for biological research. The versatility of engineered mice allows investigators to probe for the role of specific genes in growth, development, and disease, and thus, these animals have become important model systems for human diseases and an essential component for clinical and pharmaceutical research (1).
Nevertheless, research with rodents is not ideal, not only because it is constrained by ethical guidelines as it should be, but also because of issues of systemic and physiological differences between humans and rodents and variability among different strains of animals, as well as among litter mates. Also, the generation of pure laboratory strains, while useful for genetic analysis, does not encapsulate the heterogeneity of human population. Both concerns are especially relevant when large numbers of samples are required. Furthermore, whole animal experiments make it difficult to dissect molecular events at the cellular level. Thus, the use of cultured cells remains a necessity.
1.2. Structure and Function of the Rodent Mammary Gland
The basic function of the mammary gland is to make milk for the infant. As such the structural unit of mammary function is the acinus: a bilayered tube of two epithelial cell types that surround a hollow lumen where milk is secreted vectorially en route to the nipple via a branched ductal system. Luminal epithelial cells (Leps) are surrounded by contractile myoepithelial cells (Meps) and both cell layers are encapsulated by a basement membrane (BM) and surrounded by stroma including the immune cells (see Fig. 1a). Despite differences between the mouse mammary gland and the human breast, the acinar unit of function is largely similar in the two species, making it a suitable surrogate for the human mammary unit of function. Observations from the mouse mammary gland can be transferred to the human breast, and this exchange of information has been invaluable for the gradual development of mammary culture systems from monotypic 3D cultures, to multicellular cocultures, to in vivo xenograft models.
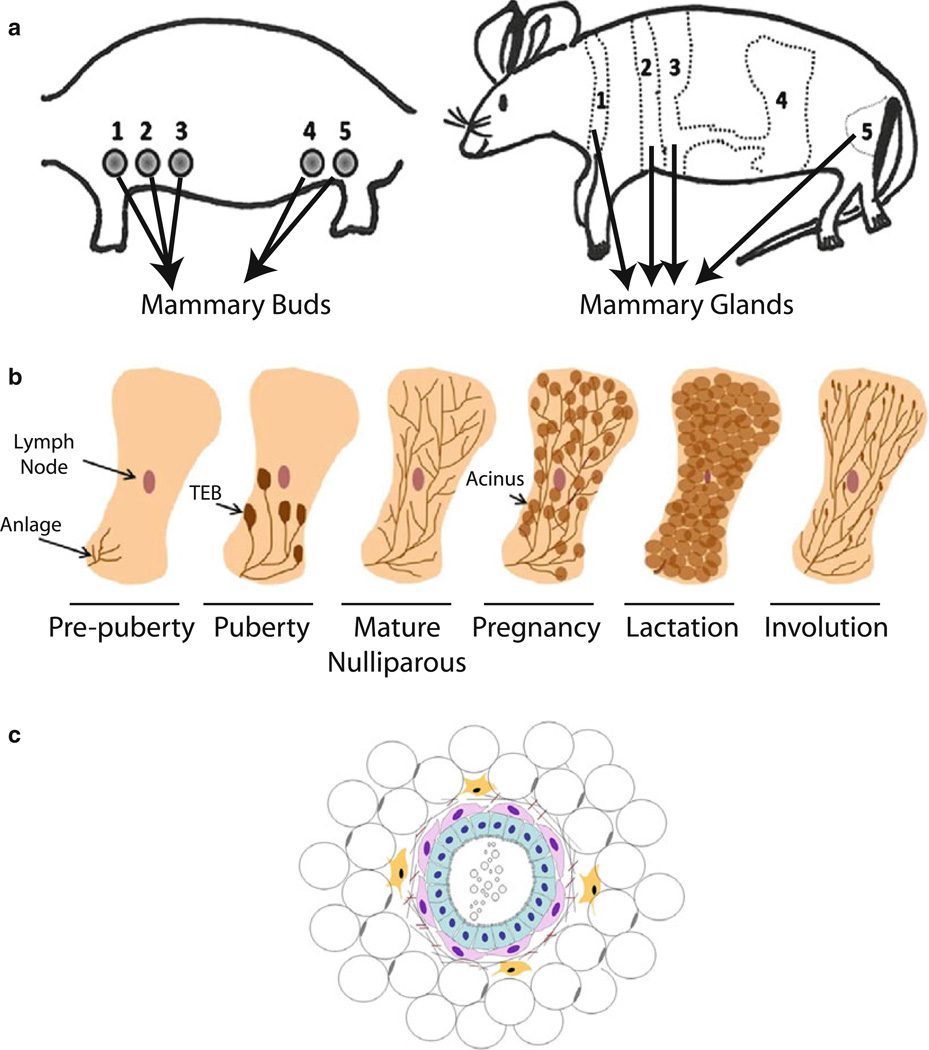
Fig. 1.
Representative schematics of the different stages of mammary gland development and of the mammary acinus. (a) Schematic showing the five mammary buds in the embryo along the milk line which will later develop into mammary glands. (b) Representative drawings of the different stages of mammary gland development, starting from birth when the mammary anlage is present, to puberty where TEB’s extend through the fat pad and the mature nulliparous gland filled with epithelial ducts. Pregnancy and lactation are modeled where the gland fills with alveoli which regress in involution such that the gland remodels to a pre-pregnant like state. (c) Representative drawing of the mammary acinus: Cuboidal luminal epithelial cells (light blue) are surrounded by contractile spindle shaped myoepithelial cells (pink) and facing a lumen on their apical side where milk droplets are deposited during lactation on their way to the ducts that terminate in the nipple. The entire structure is surrounded by a basement membrane with stromal cells including fibroblasts (yellow) and adipose cells (clear round cells).
Each of the 3D models may be used independently and represents a physiologically relevant assay capable of answering a specific question. However, when used as a series and engineered with common components, these models become invaluable tools for identifying and testing disease-related mechanisms, and enable the design or the use of effective drug therapies. Finally, the mouse mammary fat pad may be “humanized” by repopulating it with human breast cells, and may serve then as a more faithful model to the human microenvironment to circumvent the problems pertaining to species differences (2–4).
To use the mammary acinus as an experimental model in culture, it is important to appreciate how the mammary gland develops and attains its function in vivo. In the mouse, there are five pairs of mammary glands, which reside in the fat pads located just below the skin. They extend from the thoracic (three pairs) to the inguinal (two pairs) regions of the animal along the milk line (see Fig. 1b). Each fat pad has an exterior nipple connected to the primary epithelial duct where milk is released after it is pumped by myoepithelial cells from the luminal space of the acini (5, 6). Mammary gland development occurs in two phases: ductal growth and early alveolar development during puberty. In addition, alveolar growth, expansion and differentiation occur with each pregnancy and lactation, followed by a period of epithelial remodeling during post-lactational involution. Before birth, a rudimentary ductal structure that originates from the parenchyma extends in the fat pad (5). This epithelial structure then remains dormant until approximately 3 weeks of age, when it becomes stimulated by the ovarian hormones (7). At this time, a cluster of epithelial cells termed the terminal end buds (TEBs) appears at the end of the mammary ducts and the process of ductal elongation begins (8–10), and continues until approximately 10–12 weeks of age at which time the TEBs reach the limits of the fat pad and regress. With the onset of the estrous cycle at puberty, the gland begins to branch and alveolar buds are formed. During pregnancy, the changes in the hormonal and local environment allow alveolar development to progress and to establish a gland that is filled densely with alveolar bodies (11). Concomitantly, mammary epithelial cells within the gland begin to attain their differential ability to synthesize specific milk constituents, such that by parturition, functional lactogenesis is realized. At parturition, the alveoli begin abundant milk secretion, which continues for about 3 weeks (12–16).
At weaning, the gland begins a process of tissue remodeling (involution) which involves orchestrated apoptosis of mammary epithelial cells and the remodeling back to a state somewhat similar, but not identical, to the pre-pregnant gland architecture (17–20) (see Fig. 1c). The process of involution takes approximately 2 weeks to complete in rodents, after which the gland is ready to initiate another cycle of pregnancy, lactation, and involution.
1.3. Modeling Mammary Gland Functional Differentiation and Morphogenesis in Culture
The mammary acinus exists in the context of a rich stromal and extracellular matrix (ECM) milieu that is constantly changing and is subject to signaling cues from a variety of hormones and growth factors. It has been known for decades that whereas cells in monolayers lose functional differentiation even in the presence of lactogenic hormones, the same cells transplanted into the gland-free fat pads of mice form tubular structures and are able to respond to the correct hormonal cues (21, 22). It is obvious that either the cellular microenvironment surrounding mammary epithelial cells exerts an essential role in driving their functional differentiation, or systemic factors not present in culture are operating in vivo. The work of Michalopoulos and Pitot (1976), who used floating collagen gels for hepatocytes, was followed by studies from Emerman and Pitelka in the 70s and subsequently in our laboratory where both floating collagen-I (Col-I) gels and laminin-rich ECM gels (lrECM) were used to grow mouse primary mammary epithelial cells (23–26). In contrast to cells cultured on attached Col-I gels, mammary epithelial cells grown on floated collagen gels reorganized and formed secretory structures capable of de novo expression of milk proteins (14, 26–28). On floating collagen (but not flat) gels, primary mammary epithelial cells (MECs) clustered with basoapical polarity and junctions, expressed the milk protein β-casein but not whey acidic protein (WAP) and did not form luminal alveolar structures. When cultured in lrECM, primary mouse Leps assumed an in vivo-like structural organization, secreted milk proteins into the lumen and under these conditions, they were capable of expressing WAP (13), not expressed even when Col-I gels were floated (29). Expression of WAP was possible because tight junctions were formed and were necessary to keep TGFα, an inhibitor of WAP, away from its receptor (EGFR). In later studies, laminin-111 (LN1) was shown to be the main component of the ECM that drives milk protein expression (30). Cultures of single mouse cells within the collagen gels required LN1 to express β-casein. It was later shown that LN1 modulates β-casein expression partially by allowing cells to polarize, thus exposing the basolaterally localized prolactin receptor (PrlR) to its ligand (Prl) which signals for β-casein synthesis (30, 31). Interestingly, a study done in collaboration with the Petersen laboratory, using primary human Leps revealed that Leps cultured inside collagen gels form clusters of similar size and quiescence to those formed inside lrECM but were “inside out,” i.e., had reverse polarity. Correct polarity could be established if LN1 was added to collagen gels (32) confirming the role of LN1 in inducing acinar polarity. Whether or not purified mouse Leps also form inside out clusters in collagen has not been reported. In the experiments performed by Streuli et al. (30) with unfractionated epithelial cells (i.e., containing both Leps and Meps) in collagen gels, mouse cells were able to make endogenous LN1 providing the needed ligand for β1 integrin and signaling for milk production.
The structural scaffold that surrounds the cells in their native 3D environment is the basement membrane (BM), a specialized and heterogeneous entity within the ECM (33, 34). In fact, the ECM is not a static standalone entity: its composition is under the control of physiological effectors such as growth factors, cytokines, and hormones and thus is continuously changing during developmental stages, aging, tissue repair, as well as during tumor progression (33, 35). In turn, the ECM structure and its constituents regulate growth, differentiation and survival of cells within tissues. For example in the mammary gland, the specialized BM containing collagen IV and laminin-1 yields better expression of genes encoding milk proteins (36–39), but a Col-I rich ECM would favor tubular growth under the right hormonal stimulation (40–42) For further description of these assays please refer to the upcoming Subheadings 1.3.1 and 1.3.2.
Signals from the ECM are relayed to the cells via surface receptors that translate the biochemical and mechanical stimuli into a cellular and nuclear response (30, 43). ECM receptors are formed largely by the integrin family of proteins although for milk proteins, dystroglycan has been shown to be involved as well (44). As mentioned earlier, the prolactin receptor is also required for milk proteins to be expressed, and in order for these receptors to receive the signal from their ligands, they need to be present in the correct mechanical and structural platform (31, 45).
In addition to ECM and hormone signaling, the role of neighboring cells and intercellular interactions within the tissue are important for functional differentiation (46–48). Currently, mammary culture systems range from monotypic cell culture models grown in the appropriate 3D microenvironment to cocultures of Leps and Meps (32, 49) and cocultures of epithelial and endothelial cells as well as Leps and stromal fibroblasts (50–53), in addition to culturing pieces of the epithelial tissue in different gels referred to as organoid cultures (54, 55). More models still need to be developed to include the entire spectrum of cell–cell interactions found within the mammary gland. Equipped with this growing array of culture systems, it would be easier for investigators to probe for a variety of molecular signaling events that otherwise would be difficult in the whole organism, and artifactual on monolayer cultures (56) (see Subheadings 1.3.1 and 1.3.2).
1.3.1. Differentiation
The mammary acinus in culture provides a reliable functional readout, namely, the expression of milk proteins and vectorial secretion of milk into the lumen (13). The concept was extended to the human cells and used as an assay for distinguishing normal and malignant phenotypes rapidly and reproducibly (57) (for further details please refer to the accompanying chapter by Vidi et al.). Despite our inability to recapitulate the whole organ structure in culture as yet, 3D models of the mammary acinus have provided ample information on the molecular events that contribute to differentiation. These assays have allowed investigators to dissect the hierarchy of the processes that establish tissue function (58). Functional differentiation in the mammary acinus comprises two components: architectural reorganization that allows the formation of a polarized structure, which ultimately enables synthesis and vectorial secretion of milk.
Using the floating Col-I gel methodology developed by Michalopoulos and Pitot (25), Emerman and Pitelka first demonstrated that mammary epithelial cells (MECs) cultured on top of collagen gels which subsequently were allowed to float assumed a cuboidal morphology and expressed some milk proteins (26). In a follow up study, we showed that MECs cultured in floating Col-I gels, were able to become polar and functional by depositing Collagen-IV and LN1 vectorially to the basal surface of the floating gels, which once assembled into a BM would allow the de novo expression of some of the milk proteins the most prominent being β-casein (59). To determine the minimum requirement for expression of β-casein and whether or not single mammary cells could produce this milk protein without cell–cell interactions, we cultured primary mouse Leps inside collagen gels. We then added different BM molecules and showed that LN-1—but not other BM proteins—could induce expression of β-casein (30). Cell–cell interactions however, are essential to allow β-casein expression in the absence of exogenously provided LN1-rich BM (30). This conclusion correlated with the previous findings from floating Col-I gel where cells were able to deposit a BM and make some of the milk proteins (26, 27). When grown on top or inside lrECM, however, primary mouse cells (see Fig. 2a) can express abundant milk proteins including WAP, indicating the importance of the acinar morphology and tight junctions in formation of WAP and presumably other milk-specific molecules not measured in those studies.
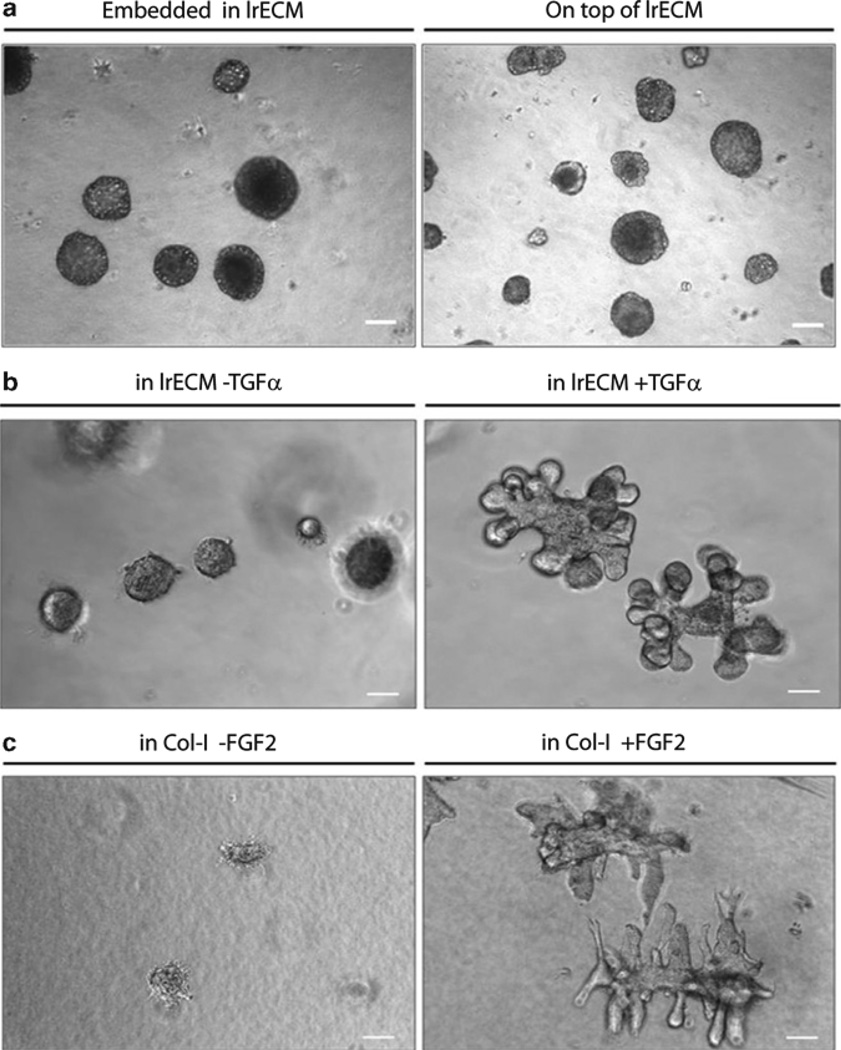
Fig. 2.
Primary mammary organoids in 3D. (a) primary mammary organoids isolated from the mammary glands of 14-week-old nulliparous mice are grown inside/embedded in lrECM (left) and on top of lrECM (right) and imaged by bright-field light microscopy on day 4 of culture, size bar, 50 µm. (b) Primary organoids isolated from the mammary glands of 8–12-week-old mice are embedded in lrECM for 4 days in additive-free medium as control (left), and stimulated with TGFα to induce alveolar growth (right). (c) Primary organoids isolated from the mammary glands of 8–12-week-old mice are embedded in collagen I (1 mg/mL) for 4 days as control (left), and stimulated with FGF2 to induce branch formation (right), size bar, 50 µm.
Signals from the BM and specifically from LN1 were found to be relayed mainly through the integrin family of heterodimeric proteins and specifically through β1- and β4-integrins (60–62). Dystroglycan (DG), another LN1 binding partner, was found to be required along with integrin to induce polarization and β-casein expression associated with LN1 signaling, and functions by helping laminin anchoring at the cell surface (63). Whereas cultures on floating Col-I gels and lrECM gels enabled copious expression of β-casein, WAP, another major milk protein, was poorly expressed even in primary cultures. This is because unlike β-casein which can be expressed even in single cells as long as LN1 and prolactin are present, WAP is not expressed until cells form complete acini with tight junctions, allowing separation of WAP’s inhibitor, TGF-α from its receptor, EGFR (29, 64). This temporally regulated event whereby tight junction sealing separates the distinct apical and basal sides of MEC’s is an important step in the completion of functional differentiation in the tissue. In fact, polarization of cells and tissues is paramount for modulating the reception and transmission of cues within and between cells and is a major contributor to tissue-specific functions. In the mammary acinus, correct polarization of Leps ensures that the basal side of the cells receives the appropriate signaling cues from the surrounding BM, ECM, Meps, and from circulating hormones, and that the apical side of the cell, delivers milk vectorially into the lumen for further transport into the ductal system towards the nipple. Tissue polarity is important for proper presentation of surface molecules and receptors to their ligands. Indeed even in the case of β-casein, we showed this principle again recently: mammary epithelial cells on 2D culture plastic, under differentiation-permissive conditions were unable to sustain activation of transcription factor STAT5 by its upstream regulator, prolactin (Prl). The study revealed that the Prl receptor was localized basally under these conditions and thus it is not available to the ligand, Prl, which is usually placed in the culture medium. However, when cells were grown either on a nonadhesive substratum with LN1 or on top of 3D lrECM gels, Prl was able to bind to the exposed receptor and could activate STAT5 transiently. Nevertheless, continuous exposure to LN1 was further essential to allow reorganization of the chromatin and to sustain activation of STAT5 permitting continued milk protein expression (31).
Using β-casein as the functional readout, it was possible to develop assays to probe the relationship between the extracellular environment and the nucleus and how the cross-talk between the two is essential for tissue-specific functions (31, 65–67). We discovered the first ECM-response element in the promoter of the β-casein gene and later established that the chromatin context and histone acetylation/deacetylation played essential roles in ECM-mediated nuclear changes that in turn lead to the establishment of tissue-specific gene expression supporting the postulated model of “Dynamic Reciprocity” (65, 68–70).
1.3.2. Branching Morphogenesis and Pattern Formation
Branching morphogenesis in the nulliparous mouse results from an intricate interplay between the cells and the extrinsic factors. Earlier studies from our laboratory and others established the importance of ECM remodeling enzymes (metalloproteinases: MMPs) in driving invasion into the fat pad and branching of the mammary epithelium (71–73). The process could be simulated in culture by embedding mammary epithelial cells in collagen-I gels and stimulating them with growth factors that favor branch formation (42, 55, 74).
In the assay described above, it is not easy to investigate the role of positional control in the process of branching. To separate positional control from simple growth and invasion, we developed a micropatterned assay where cells were placed between a collagen “sandwich” and where the results of 50 or more wells could be imaged and binarized, allowing a rapid read out and meaningful statistics to study how positional cues dictate the initiation of branching points (75). Using different geometric contexts to place the cells into, the assay allowed us to discover that geometry influenced the secretion and activation of endogenous TGF-β, which then would inhibit branching depending on the shape of the micropatterns (75).
Morphogenesis in the gland of pregnant mice takes the form of alveologenesis, a process by which lobules of the milk-producing acini are formed. This process can be modeled by replacing the composition of the ECM gel from Col-I to lrECM (see Fig. 2b, c). This is yet another example of how the nature of the ECM molecules allows unique aspects of growth and functional differentiation to be studied. In this latter assay, MECs (primary or established lines that are clustered as described later, or organoids from the mammary gland) are embedded in an lrECM gel and stimulated with growth factors such as TGF-α (76). Under these conditions, mammary epithelial cells undergo a growth and remodeling process that resembles the structure of the “alveoli” in the mammary gland (see Fig. 2c) during pregnancy and lactation, rather than branching ducts (see Fig. 2b). The use of the alveologenesis assay enabled our laboratory to uncover a critical role for MAPK signaling and its downstream molecules in promoting alveolar growth (76).
These studies clearly demonstrate that one can create “designer microenvironments” (77) in 3D to answer questions that are otherwise too difficult to address in the organism. This is made possible by modulating the type and organization of the culture substrata to model different aspects of mammary growth and development. Nevertheless, many challenges remain in making physiologically relevant culture systems and we refer to these in the section below.
1.4. Current Limitations and Future Perspectives
3D mammary epithelial culture systems have greatly evolved since their early inception and are still undergoing refinements aimed ultimately at recreating the entire organ in culture. This will however require concerted efforts from many fields of science. The advent of tools such as bio-engineered materials and nanotechnology has been a driving force in improving available culture systems by allowing investigators to manipulate and control the cellular microenvironment at multiple levels and scales. That is, they have provided means to control the chemistry, geometry, and mechanics of culture settings with increasing precision. These tools will ultimately allow us to replicate the gland in culture, and with rigorous controls it will be possible to answer questions pertaining to particular developmental stages and environmental constraints in vivo.
There are still, however, many challenges ahead and many questions to answer. We still lack a profound knowledge of the complex array of signals generated from the ECM. Signaling from laminin-1 through both integrin and non-integrin (e.g., dystroglycan) receptors is required for BM and acinus formation and differentiation (62, 63). With a few exceptions, currently we have limited knowledge of the functional roles of other BM components. Moreover, it is not yet clear why it is that cells on 2D culture plastic are able to synthesize BM molecules, but why they do not deposit a structurally intact BM that would eventually drive reorganization of cells into their correct conformation in the tissue. However, it is now possible to start to tackle these questions with the use of synthetic inert gels where purified components of the BM are added. Ultimately, different components can be recombined together to study the hierarchy of signaling events from the BM to the epithelium (78).
Unfortunately, simply adding different ECM and BM membrane components is not sufficient to model the complexity of the microenvironment in vivo, especially given the temporal regulation of signaling within a tissue. Our inability to model such complexity confounds our interpretation of data derived from culture studies. One particular example of this complexity is observed with the alveologenesis assay. Even though we are able to model a process that phenotypically resembles alveolar growth in culture, we still do not know if our alveologenesis assay is a true surrogate for alveolar growth and expansion that happens during pregnancy, ultimately leading to lactation. However, it is evident that in the advances we have made in building mammary epithelial culture systems, we have answered a number of questions but also opened a whole array of new questions. It is important to recognize that in order to obtain pertinent knowledge from culture systems, we should ask the relevant questions and admit to the limitations of the models available. In the end the organ itself remains the unit of function, and the integration of signals within the organ is what ultimately provides tissue specificity (79).
2. Materials
The sources listed for the reagents are the current materials used in the Bissell laboratory. We are not advocating their use. Other sources can be substituted, and we do that from time to time depending on the availability, price, and purity.
2.1. Culture Medium for Primary Cells, Organoids, and EpH4 Cells
Dulbecco’s modified Eagle’s medium/Ham’s F-12 1:1 Mix Medium (DMEM/F12): 3.15 g/L Glucose, 0.365 g/L l-Glutamine, 3.754 g/L HEPES, 1.2 g/L NaHCO3 pH 7.3 (DMEM/F12, Life Technologies™).
Fetal Bovine Serum (FBS).
Prolactin (Sigma-Aldrich®): 1 mg/mL (30.3 IU/mL) in 26 mM (2.22 mg/mL) sodium bicarbonate, filter-sterilized (0.22 µm filter), aliquoted, fast-frozen on dry ice, and stored at −80°C. Prolactin is stable for 1 year at −80°C and 1 month at 4°C.
Insulin (Sigma-Aldrich®): dissolved as a 2 mg/mL concentrated stock in 5 mM HCl. The stock solution is filter-sterilized, diluted to 100 µg/mL in filter-sterilized MilliQ (ddH2O) water, aliquoted, fast-frozen on dry ice, and stored at −80°C. Insulin solution is stable for 6 months at −80°C and 1 month at 4°C.
Hydrocortisone (Sigma-Aldrich®): Prepare a 5 mg/mL concentrated stock solution in ethanol. The stock is further diluted to 0.5 mg/mL (1.4 10−3 M) in ethanol and kept at −80°C. Hydrocortisone solution is stable for 1 year at −80°C and 1 month at 4°C.
Insulin-Transferrin-Sodium Selenite media supplement (Sigma-Aldrich®): lyophilized powder, diluted in MilliQ (ddH2O) water and filter-sterilized to a stock of 1,000×. Stock solutions are aliquoted and stored for 6 months at −20°C or 1 month at 4°C.
Penicillin-Streptomycin Solution (Penicillin: 6,370 mg/mL; Streptomycin: 10,000 mg/mL): 100×.
Gentamicin solution (50 mg/mL): 100×.
Poly (2–hydroxyethyl methacrylate), polyHEMA (Sigma-Aldrich®): dissolved in 95% ethanol at 6 mg/mL.
Growth medium for organoids and primary cells (100 mL): 94 mL of DMEM/F12, 100 µL of ITS (1,000× dilution), 5 mL of FBS, 100 µL of gentamycin for a final concentration of 50 µg/mL, 1 mL of Pen/Strep (100× dilution) (see Note 1).
Growth medium for cell lines (100 mL): 95 mL of DMEM/F12, 100 µL insulin for a final concentration of 5 µg/mL, 2 mL of FBS, 100 µL of gentamycin for a final concentration of 50 µg/mL.
Differentiation medium for organoids and primary cells (100 mL): 99 mL of DMEM/F12, 100 µL of ITS (1,000× dilution), 100 µL of gentamycin for a final concentration of 50 µg/mL, 1 mL of Pen/Strep (100× dilution), 100 µL of prolactin for a final concentration of 3 µg/mL, 100 µL of hydrocortisone for a final concentration of 1 µg/mL (see Note 1).
Differentiation medium for cell lines (100 mL): 100 mL DMEM/F12, 100 µL of gentamycin for a final concentration of 50 µg/mL, 100 µL of insulin for a final concentration of 5 µg/mL, 100 µL of prolactin for a final concentration of 3 µg/mL, 100 µL of hydrocortisone for a final concentration of 1 µg/mL.
2.2. Propagation of Primary Organoids, Cells, or EpH4 Cell Lines in 2D
10 cm Cell culture plates.
Solution of trypsin (0.25%) and ethylenediamine tetraacetic acid (EDTA, 1 mM) (Life Technologies™).
Purecol® (Advanced BioMatrix) diluted to a concentration of 1 mL in 44 mL of phosphate buffered saline (PBS).
Cell Freeze solution: 10% Dimethyl Sulfoxide (DMSO) in 90% FBS.
2.3. Monoculture of Primary Cells, Organoids, or EpH4 Cells in 3D
4-Well chamber slides (LabTek, Nunc) are used for immunostaining experiments.
35-mm Tissue culture dishes are used for biochemical assays (protein, RNA, and DNA extracts).
Multiwell plates (6, 12, 24, 48, and 96) are used depending on the type of assay and the amount of cells needed.
DMEM (powder, Sigma-Aldrich®): to prepare 10× solution, dissolve DMEM powder into 0.1 volume of H2O. Filtersterilize and store at −20°C in working aliquots.
Basement membrane matrix from EHS extracts (Matrigel™, BD Biosciences). Matrigel™ is kept at −80°C and thawed on ice 24 h before use. If rigorously kept at 0°C, Matrigel™ is stable for 1 month after thawing and can only be thawed twice. Thus, usually a 10 mL bottle is thawed to prepare 1–2 mL aliquots that will be frozen and kept at −80°C until use. For the optimal culture of human breast cell lines currently available, it was observed that Matrigel™ should have low levels of endotoxins (less than 4 U/mL) and a protein concentration below 13 mg/mL. In addition, certain research teams prefer to use growth-factor depleted lrECM.
Collagen-I/acid-soluble collagen: 7.5 volume of a 0.5% solution mixed gently on ice with 1 volume of 10× DMEM/F12, followed by 1 volume of 0.1 N NaOH. (Cellagen™ AC-5, ICN). Collagen 1 can also be purchased from Cosmo Bio Co., as Native Collagen, Bovine Dermis, 5 mg/mL or 3 mg/mL or from BD Biosciences as acid-extracted rat tail tendon collagen.
2.4. Branching Assay and Alveologenesis Assay
TGFα: 9 nM (Sigma-Aldrich®). The TGFα working solution is prepared by diluting the supplied powder in a balanced salt solution such as PBS under sterile conditions. The working solution is then diluted to desired concentration of 9 nM in desired medium.
FGF2: 9 nM (Sigma-Aldrich®). The FGF2 working solution is prepared by reconstituting the powder in sterile water to 100 µg/mL.
FGF7: 9 nM (Sigma-Aldrich®). The FGF7 working solution is prepared by reconstituting the contents of the vial using sterile PBS. The working solution is then diluted to desired concentration of 9 nM in desired medium.
FGF10: 9 nM (Life Technologies™). The FGF10 working solution is prepared by reconstituting the contents of the vial using sterile PBS. The working solution is then diluted to desired concentration of 9 nM in desired medium.
5. 2% Agarose in 2% DMEM solution: Add 2 g of Agarose to 100 mL of 2% DMEM/F12.
Branching morphogenesis medium for organoids and primary cells (100 mL): 99 mL of DMEM/F12, 100 µL of ITS (1,000× dilution), 100 µL of gentamycin for a final concentration of 50 µg/mL, 1 mL of Pen/Strep (100× dilution), choice of growth factor (see steps 1–4) at designated concentration.
Branching morphogenesis medium for cell lines (100 mL): 100 mL of DMEM/F12, 100 µL insulin for a final concentration of 5 µg/mL, 100 µL of gentamycin for a final concentration of 50 µg/mL, choice of growth factor (see steps 1–4) at designated concentration.
Alveologenesis medium for organoids and primary cells (100 mL): 99 mL of DMEM/F12, 100 µL of ITS (1,000× dilution), 100 µL of gentamycin for a final concentration of 50 µg/mL, 1 mL of Pen/Strep (100× dilution), choice of growth factor (see steps 1–4) at designated concentration.
Alveologenesis medium for cell lines (100 mL): 100 mL of DMEM/F12, 100 µL insulin for a final concentration of 5 µg/mL, 100 µL of gentamycin for a final concentration of 50 µg/mL, choice of growth factor (see steps 1–4) at designated concentration.
2.5. Isolation of Primary Organoids and Fractionation of Mammary Epithelial Cell Types
VWR® Razor Blades (VWR®).
Collagenase Type IV (GIBCO®): 0.2% (w/v), dilute 0.1 g of collagenase in 47.5 mL of DMEM/F12 mixed with 2.5 mL of FBS.
Trypsin 1:250 powder: 0.2% (w/v), dilute 0.1 g of trypsin in 47.5 mL of DMEM/F12 mixed with 2.5 mL of FBS.
DNase-I (Sigma-Aldrich®): Dilute DNase-I powder in DMEM/F12 for a 2 U/mL working solution.
50 mL Conical tube.
Collagenase solution for organoid extraction (50 mL): 0.2% collagenase Type IV (see step 2), 0.2% trypsin (see step 3), 5% FBS, 5 µg/mL gentamycin, 47.5 mL DMEM/F12. Prewarm at 37°C (shake occasionally). Filter-sterilize after the collagenase has gone into solution through a 0.2-µm filter (see Note 2).
Joklik’s Modification of Minimal Essential Medium (Sigma-Aldrich®).
Hanks’ Balanced Salt Solution (HBSS).
L-15 medium (Sigma-Aldrich®).
Alexa Fluor® 647 anti-mouse CD326 (Ep-CAM) Antibody (Biolegend®).
FITC anti-mouse CD104 Antibody (Biolegend®).
Unconjugated anti-mouse Ep-CAM antibody (Abcam®).
Unconjugated anti-mouse CD104/β4 Integrin antibody (R&D systems).
MACS® micro-beads (Miltenyi Biotec).
MACS® Pre-separation filter (Miltenyi Biotec).
MACS® Separation Filter (Miltenyi Biotec).
MACS® Magnet (Miltenyi Biotec).
MACS® Columns (Miltenyi Biotec).
Bovine Serum Albumin (BSA, Sigma-Aldrich®).
Ca2+/Mg2+-free PBS 10×.
MACS Buffer: 0.5% (w/v), BSA in 1× Ca2+/Mg2+-free PBS.
FACS Buffer: 1% BSA in 1× Ca2+/Mg2+-free PBS.
2.6. Biochemical Assays
EDTA (GIBCO®): 0.5 M Stock.
Dispase 100 mL solution (BD Biosciences): recommended concentration is 10 U/cm2 of BD Matrigel™ Matrix (for example: 100 U for a 35 mm culture dish).
Phosphatase inhibitor cocktail 1 (100×) (Sigma-Aldrich®).
Phosphatase inhibitor cocktail 2 (100×) (Sigma-Aldrich®).
100× Protease Inhibitor Cocktail (EMD-Calbiochem Merck®).
PBS–EDTA buffer: 50 mL of cold 1× PBS buffer, 500 µL of 0.5 M EDTA for a final concentration at 5 mM, 500 µL of 100× phosphatase inhibitor cocktail 1 (1×), 500 µL of 100× phsophatase inhibitor cocktail 2 (1×), 500 µL of 100× protease inhibitor cocktail to 1×.
Polyethylene cell lifter.
RLT RNA lysis Buffer (Qiagen).
Ripa Buffer (Sigma-Aldrich®): supplement with 1× protease inhibitor and 1× phosphatase inhibitors (cocktail 1 and 2).
Microcentrifuge tube shaker.
18–21 Gauge syringe.
Sonic Dismembrator Model 100.
2.7. High-Resolution Imaging
Triton™-X 100 (Sigma-Aldrich®): 0.5% (v/v). Add 500 L of Triton™-X 100 to 100 mL of 1× PBS.
16% Paraformaldehyde (Electron microscopy sciences): Dilute 25 mL of 16% paraformaldehyde into 75 mL of PBS to make 4% paraformaldehyde solution.
Superfrost Plus Micro Slides.
Spot RT camera attached to an upright epifluorescence microscope (Carl Zeiss, Inc.).
Zeiss LSM 710 Meta confocal microscope (Carl Zeiss, Inc.).
3. Methods
3.1. Isolation of Primary Organoids and Epithelial Cells
To isolate primary epithelial cells or primary organoids from the mouse mammary gland we use either the third thoracic or the fourth inguinal mammary glands from the mouse. The age of the animal is critical since some assays cannot be performed with older mice (see Subheading 3.5.4). But in general, mammary glands could be removed at different ages, and the choice of age depends on the stage of mammary gland growth desired to answer a specific question (see Notes 3 and 4).
Mince glands with two razor blades on sterile surface (see Subheading 2.5, item 1). For optimal mincing, hold two razor blades and make 10–15 cuts (based on mammary gland size) in two perpendicular directions across the entire gland.
Shake minced tissue in collagenase solution (see Subheading 2.5, item 6) at 100 rpm/37°C for 30 min.
Spin at 405×g for 10 min at room temperature.
The top layer is going to be the fatty layer, but it will often contain uncollected organoids, so collect 10 mL of the top layer in a 15 mL conical tube. Pipette it up and down to break it up and free organoids prior to another round of spinning and add another 5 mL of fresh serum-free medium on top (see Note 5).
The pellet at the bottom will contain epithelial cells, fibroblasts, and debris. Remove all the rest of the medium. Resuspend the pellet at the bottom in 10 mL of serum-free DMEM/F12.
Take the two 15 mL conical tubes, the one that contains the pellet (see step 5) and the one that contains the fatty top layer (see step 4) and spin at 405×g for 10 min at room temperature.
Remove the medium from the two conical tubes and combine the two pellets by resuspending one then the other in the same 4 mL of 0% DMEM/F12. Now there is one 15 mL conical tube of organoids with 4 mL of 0% DMEM/F12. Add 40 µL of DNase, (see Subheading 2.5, item 4) and shake by hand for 2–5 min at room temperature (until clumps are broken up), then add 6 mL of 0% DMEM/F12 to obtain a final volume of 10 mL.
Spin at 405×g for 10 min at room temperature.
Discard supernatant and resuspend the pellet in 10 mL of 0% DMEM/F12.
Spin at 405×g for a brief pulse then cut the machine and hit the brakes as the centrifugation speed reaches 405 × g. This step is done to separate the fibroblasts from the epithelial cells/organoids. Fibroblasts are lighter and will float in the supernatant while the epithelial organoids will pellet down (see Notes 6 and 7).
Resuspend the pellet at the desired concentration into Growth medium (see Subheading 2.1, items 10 and 11, see Note 8).
3.2. Fractionation of Cell Types
To fractionate the epithelial subtypes from each other (i.e., the Leps and Meps from each other) the primary step is the isolation of mammary epithelial organoids as described in Subheading 3.1. Then proceed as follows: Resuspend organoids in JokliK’s Ca-free medium at 37°C for 15 min.
Spin down the cells at 405 × g for 10 min at room temperature and resuspend them in 2 mL of Hank’s Balanced Salt Solution at 37°C for 5 min.
Vigorously flush organoids up and down with Pasteur pipette and incubate for 2 min.
Add 5 mL of serum-free DMEM/F12 supplemented with 50 µL type-I DNase (see Subheading 2.5, item 4) to remove clumps. Incubate for 5 min.
To stop the reaction: add 10% FBS in 5 mL of L-15 medium or in calcium-free DMEM/F12 with DNase (50 µL, 2 U/mL), then spin down the cells at 405 × g for 10 min at room temperature and wash cells 2× in medium to clean.
Remove large multicellular clumps with filtration through preseparation filter (see Subheading 2.5, item 15).
Resuspend cells in 500 µL MACS or 500 µL FACS buffer (see Subheading 2.5, items 21 and 22) depending on the method used (see Subheadings 3.2.1 and 3.2.2).
3.2.1. Fractionation Using MACS Protocol
Pre-wet separation filter (see Subheading 2.5, item 16) with 500 µL MACS buffer (see Subheading 2.5, item 21).
Apply cell suspension to filter.
Wash 3× with MACS buffer.
Count cells.
Label cells by adding 0.25 µg Ab/10,000,000 cells EpCAM (labels Leps) or β4 integrin (labels Meps).
Incubate cells with primary antibody for 30 min in the cold room.
Wash/spin 2× with 10× volume MACS buffer.
Incubate cells with MACS® micro-beads at 1:10 dilution 15 min in the cold room.
Wash/spin 2× with 10× volume MACS buffer.
Resuspend in 500 µL MACS buffer.
Pre-wet MACS columns.
Apply the cell suspension.
Wash columns 3× with 500 µL MACS buffer.
Freeze negative fraction in cell freeze solution (see Subheading 2.2, item 4).
Remove column from magnet (see Subheading 2.5, item 17) to elute positive fraction with 1 mL MACS buffer.
Spin down the positive fraction at 405 × g for 10 min at room temperature and resuspend in 500 µL MACS buffer.
Repeat steps 11–16 with a new column for double purification.
Count the cells in the positive fraction.
3.2.2. Fractionation Using FACS Protocol
Count cells.
Label cells: 0.25 µg Ab/10,000,000 cells EpCAm (luminal epithelial) or β4 integrin (myoepithelial).
Incubate cells with primary antibody for 30 min in the cold room.
Wash/spin 2× with 10× volume FACS buffer (see Subheading 2.5, item 22).
Resuspend in 500 µL FACs buffer.
Run on FACS sorter.
3.3. Maintaining Primary Mouse Mammary Epithelial Cells
Primary mammary epithelial cells are not easy to maintain in culture. To do that, it is preferable first to change the medium the next day and every 2 days after that. Passage the cells at 1:1 or 1:2 split ratio for the first two passages, because the cells are not easy to grow and they have been stressed by the isolation procedure. With time it is possible to increase the split to a 1:5 ratio. They should survive 5–7 passages before reaching senescence.
To passage cells use dispase (see Subheading 2.6, item 2).
Incubate for 20–30 min at 37°C.
Tap culture or pipette up and down to release cells from plate.
3.4. Mouse Mammary Epithelial Cell Lines: EpH4 as a Cell Line Model and as a Surrogate for Primary Cultures
EpH4 cells were originally isolated from the mammary tissue of a mid-pregnant Balb/c mouse yielding a spontaneously immortalized, non-tumorigenic mouse mammary cell line, designated IM-2 (80, 81). They provide a powerful tool for the identification of factors that modulate epithelial architecture. EpH4 cells exhibit a polarized epithelial phenotype (as revealed by their high transepithelial electrical resistance and appropriate localization of apical and basolateral marker proteins), and form branching tubular structures when grown in collagen gels (see Notes 9 and 10).
EpH4 cells are thawed and initially maintained in growth medium (see Subheading 2.1, items 10 and 11).
Cells are plated at a density of 10,000 cells/cm2, and initially allowed to attach for 16–24 h.
Cells are propagated by trypsinization (see Subheading 2.2, item 2) and splitting in 1:4 ratio at around 70% confluency.
3.5. Design of Functional Assays in 3D
The ultimate goal of cell culture models is to allow us to ask biological questions that are either not possible in the organism or that are too costly or inhumane to perform. Useful culture models should recreate at least some aspects of the tissue and organ microenvironment to ask relevant questions, and to provide a functional readout. Mammary epithelial cells (primary and cell lines) provide a variety of functional outputs we can recreate in culture such as recapitulation of formation of acini, milk synthesis, invasion, and branching, etc. We, and others, have taken advantage of this ability and in this next section we try to provide investigators with detailed protocols for culturing these cells under relevant conditions and describe assays that can be used to probe molecular signals.
3.5.1. 3D Laminin-Rich ECM Embedded Assay
In this culture set up, (Barcellos-Hoff et al. (13), mammary epithelial cells are cultured inside lrECM (Matrigel™) such that they are fully surrounded by an lrECM thick coat (see Note 11, Fig. 2a).
Coat tissue culture plates with lrECM and leave them at room temperature for 30 min (see Subheading 2.3, item 5). The desired amount of coating is 100 µL per 0.75 cm2 (48- well plate). In this case, smaller size culture dishes are used because the amount of lrECM needed is larger than in the on top assay.
After obtaining a pellet of EpH4 cells, put cells on ice and add on top of them 200 µL of lrECM and keep on ice while you suspend the cells in lrECM by mixing with a p1000 pipette until the cells are evenly distributed in lrECM. Take the entire volume of cells/lrECM and add it on top of the coated plates and allow cells to attach, then add medium (see Subheading 2.1, items 10 and 11) on top of cells (see Note 12).
Add fresh growth medium (see Subheading 2.1, items 10 and 11) and incubate for 24 h at 37°C, 5% CO2.
The next day, change growth medium into differentiation medium (see Subheading 2.1, items 12 and 13). Make sure to wash the cells from residual growth medium with 1× PBS.
Harvest cells for analysis after 24 h or 48 h from treatment with lactogenic hormones (see Subheading 3.6.).
3.5.2. 3D Laminin-Rich ECM On-Top Assay
In this culture assay, mammary epithelial cells are cultured on top of a thin layer of lrECM and dripped with lrECM (see Note 12, Fig. 2a).
Coat tissue culture plates with lrECM and leave them at room temperature for 30 min. The desired amount of coating is 500 µL/9.62 cm2 (35 mm dish).
Add EpH4 cells on the coated plates at a density of 500 × 103/9.62 cm2 (35 mm dish).
Add fresh growth medium (see Subheading 2.1, items 10 and 11) and drip 2% lrECM (vol/vol) (Matrigel™) on top of cells and incubated for 24 h at 37°C and 5% CO2.
The next day, change growth medium into differentiation medium (with lactogenic hormones) (see Subheading 2.1, items 12 and 13). Make sure to wash the cells from residual growth medium with 1× PBS.
Harvest cells for analysis after 24 h or 48 h from treatment with lactogenic hormones.
3.5.3. Suspension Culture Assay
In this assay cells are allowed to cluster on top of a nonadhesive substratum, polyHEMA.
Dissolve polyHEMA in 95% ethanol at 6 mg/mL to make the working solution.
Coat plates with polyHEMA at 0.25 mg/cm2 as previously described (60) by adding 4 mL of the working solution in a 10 cm plate and drying overnight in an incubator at 55°C.
Plate EpH4 cells on polyHEMA-coated plates at 30,000 cells/cm2.
24–48 h after plating, collect cells by centrifugation (either at 1,154 × g for primary organoids, or at 405 × g for cell lines) and resuspend them in DMEM/F12 either for growth conditions (see Subheading 2.1, items 10 and 11) or for differentiation conditions (see Subheading 2.1, items 12 and 13).
3.5.4. Branching Morphogenesis Assay
Branching assays are divided into two types in concordance with two distinct types of branching that occur in the mammary gland in vivo. The first assay is used to model morphogenesis in the early mammary gland or the phase of tubular growth. Accordingly, the cells in the assay are embedded in a Collagen-I rich matrix to simulate the early status of the epithelium in the mammary gland where cells that are invading the fat pad are constantly remodeling and invading through a layer of collagenous ECM. The second assay (see Subheading 3.5.5) is used to model the alveolar growth in the mammary gland and as such cells are embedded in an lrECM rich matrix similarly to the differentiation assay, but hormones that induce branching are used instead of the lactogenic hormones. The latter assay is referred to from here on as “alveologenesis” (see Subheading 3.5.5).
For the branching morphogenesis assay, EpH4 cells are preclustered before embedding in collagen gels. Pre-clustering can be done on top of polyHEMA (see Subheading 3.5.3) or on top of Agarose coated plates. In both cases a differential centrifugation step is required to remove the single cells and only keep the clustered EpH4 for embedding in Collagen-I (see Note 13 and Fig. 2b).
Day 1: Make 2% Agarose (see Subheading 2.4, item 6) and with a 10 mL pipette drop 1 mL into 4 center wells of a 24-well plate and put medium in surrounding wells.
Let it gel as needed and then add 1.5 mL of 2% medium in the well and incubate at 37°C for ~30 min to 1 h.
Trypsinize cells (90% confluent EpH4).
Spin for 5 min at 115 × g and aspirate the medium and leave 100 µL of medium on top of cell pellet.
Add 5 µL of DNase I (2 U/µL) to the cells. Finger tap for 2 min.
Add 2 mL of 2% medium to the cells.
Aspirate the medium from the four wells. Leave half the amount of medium in the surrounding wells.
Add 500 µL of the 2 mL medium in each of the four wells and incubate in a shaker overnight at 37°C, 100 rpm.
Day 2: differential centrifugation, take 2 mL out of the four wells in the plate and transfer them to 15 mL conical tube, top off to 10 mL with DMEM/F12.
The tube will contain various sizes of clusters and you want to pull out the larger clusters via centrifugation. Centrifuge the sample at 405 × g. When it reaches this speed, stop the centrifugation.
Discard supernatant and resuspend pellet each time in 5–10 mL of DMEM/F12. This step may have to be repeated three times until you get rid of the single cells.
Count and check the number of clusters using a hemocytometer. Calculate how much volume required to get the number of clusters needed (see Note 14). Make Collagen-I (see Subheading 2.3, item 6): 800 µL collagen (take 840 µL because it is viscous to account for loss of volume on the pipette edges) and add: 100 µL 10× DMEM (see Subheading 2.3, item 4) to 100 µL 0.1 N NaOH. Adjust pH to 7.4. Mix well and put 100 µL in each well of 48-well plate and incubate at 37°C to allow Collagen to gel.
Resuspend cells in Collagen-I at concentrations of either 1 mg/mL or 3 mg/mL depending on the experiment (see Note 15). Similarly allow to gel by incubation at 37°C.
Add the branching medium (see Subheading 2.4, items 7 and 8) on top of the cells.
3.5.5. Alveologenesis Assay
lrECM assay (basement membrane mix) is performed in 96-well plates, due to the need for large amounts of Matrigel™. Cells are pre-clustered as per the Collagen-I assay on polyHEMA (see Subheading 3.5.3) or on top of Agarose coated plates (see Subheading 3.5.4, steps 1–12), and then embedded in 50 µL of Matrigel™ in the wells (see Fig. 2c).
Overlay 50 µL of Matrigel™ on the bottom of the well in a 96-well plate.
Decant off the medium from clusters. It is desired that the cells are diluted in Matrigel™ to get 50 clusters per 50 µL of Matrigel™. Mix quickly and swirl but minimize the bubbles when mixing and put 50 µL in each well of the 96-well plate (that was pre-coated with Matrigel™).
Incubate the cells at 37°C for 30 min until the Matrigel™ solidifies.
Add 150–200 µL growth medium (see Subheading 2.1, items 10 and 11) to each 96-well plate.
Change growth medium the next day to alveologenesis medium (see Subheading 2.4, items 9 and 10) and then every other day, and probably after 2–3 days branching will start.
3.6. Biochemical Assays for Isolations of Cells from LrECM (Matrigel™)
Biochemical analysis, including RNA profiling and quantitation and protein analysis, is possible with cells in 3D cultures. However, it is better to isolate the 3D multicellular cell clusters from the surrounding matrix whether it is collagen or Matrigel™. This is usually done by solubilizing the gels using ECM-specific proteases (dispase or PBS–EDTA for Matrigel™). Gel-extracted cells are then subjected to the typical protocols used for RNA extraction and isolation or cell lysis for protein extraction.
3.6.1. Extraction Using Dispase
Remove the medium and add dispase solution (see Subheading 2.6, item 2) (0.75 mL per mL of Matrigel™).
Incubate 30 min at 37°C.
Remove proteases by washing three times in DMEM/F12 medium and once with RNA lysis buffer (see Subheading 2.6, item 8) or protein lysis buffer (see Subheading 2.6, item 9). Each wash is followed by centrifugation (1,154 × g, 5 min at room temperature).
Cell pellets are further processed for RNA or protein analysis, or subcellular fractionation using standard protocols.
3.6.2. Extraction Using PBS–EDTA
Aspirate medium and rinse culture with 2 mL of cold 1× PBS, for a 35 mm dish, and leave the dish on ice.
Add 2–3 mL PBS–EDTA (see Subheading 2.6, item 6) to the culture and use a polyethylene cell lifter to gently scrape the cells and Matrigel™ off the bottom of the dish. Transfer the cell/Matrigel™ mixture with a serological pipet to a 15 mL conical tube.
Repeat 2–3 times and use about 10 mL total PBS–EDTA (depends on amount of Matrigel™ in culture).
Shake the tubes gently in cold room or on ice for 30 min to 1 h to dissolve the Matrigel™ (also dependent on amount of Matrigel™) (see Note 15).
Centrifuge 5 min at 1,154 × g at room temperature.
Transfer the cell pellet to a 1.5 mL Eppendorf tube on ice (see Note 16).
Wash with PBS and spin at full speed for 5 min and get rid of supernatant, and keep the cell pellet (see Note 17). Follow protocols for RNA or protein lysis in Subheading 3.6.2.
3.6.3. Cell Lysis for RNA or Protein Extraction
Add RNA lysis buffer (see Subheading 2.6, item 8) or RIPA buffer (see Subheading 2.6, item 9) (100–150 µL according to pellet size) and keep in mind that a concentrated protein solution is desired.
Sheer cells with an 18–21 gauge syringe (see Subheading 2.6, item 11) or sonicate with sonic dismembrator (see Subheading 2.6, item 12) on ice about 20 s at power setting 3 or 4.
Microcentrifuge for 10 min at full speed (17,000×g) at 4°C.
Transfer supernatant to a new Eppendorf for immediate use or store at −80°C for later use.
3.7. Biochemical Assays for Isolations of Cells from Collagen Gels
To extract RNA or protein from cells and organoids embedded in collagen gels, detach the collagen gel from the culture well and add RNA lysis buffer (see Subheading 2.6, item 8) or Ripa buffer for protein isolation (see Subheading 2.6, item 9).
For collagen gels in 96-well plates:
Add 300 µL of the desired buffer (either RNA or protein).
Shake for 6 h at room temperature in a microcentrifuge tube shaker (see Subheading 2.6, item 10).
Microcentrifuge for 10 min at full speed (17,000×g) at room temperature: collagen will pellet and the RNA or protein will be in the supernatant.
Discard pellet and keep the supernatant with the RNA or the protein either for immediate use or store at −80°C for later use.
3.8. High-Resolution Imaging
Imaging is essential for cell biological studies, and thus, it was important to find the best means to image cells in 3D. Confocal microscopy and live imaging are very important tools for studying cellular behavior in 3D.
Cells grown on tissue culture plastic are fixed with 4% paraformaldehyde (see Subheading 2.7, item 2) and permeabilized with 0.5% Triton™ X-100 (see Subheading 2.7, item 1).
Cells in lrECM gel are either smeared on slides, dried briefly, and fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton™ X-100, or smeared after extraction using PBS–EDTA or dispase solutions (see Subheading 2.6, items 2 and 6). Choice of smearing depends on the strength of the antibody and background considerations with Matrigel™.
Cell clusters on polyHEMA-coated plates are processed for immunofluorescence analysis as follows: cells are pelleted by centrifugation at 500 × g for 2 min and resuspended in PBS. The clusters are plated on slides, dried brie fly, and then fixed with 4% paraformaldehyde.
Stained samples are imaged using a Spot RT camera attached to an upright epifluorescence microscope or viewed and acquired with a digital axiocam camera attached to an upright Zeiss LSM 710 Meta confocal microscope.
Time-lapse movies are collected using a Zeiss LSM 710 Meta confocal microscope. Images of dimensions, 512 × 512 pixels (lateral dimensions) with a maximum axial displacement of 75 µm (axial step size, 2 µm) are acquired using a 0.8 NA 20× air objective at digital zoom 0.6 corresponding to an area of 701 × 701 µm2 at desired rate. Samples were maintained at 37°C and 5% CO2. Immunofluorescence images are exported using Zen 2009 (Zeiss) software.
Acknowledgments
We thank Dr. Joni Mott for critical reading of the manuscript and Dr. Hidetoshi Mori for providing images of cells in Fig. 2b, c. The work from MJB’s laboratory is supported by grants from the US Department of Energy, Office of Biological and Environmental Research, a Distinguished Fellow Award to MJB and Low Dose Radiation Program (contract no. DE-AC02-05CH1123); by National Cancer Institute (awards R37CA064786, U54CA126552, R01CA057621, U54CA112970, U54CA143836, U01CA143233); and by US Department of Defense (W81XWH0810736). RM is supported by US Department of Defense, BCRP fellowship (W81XWH0810481).
Footnotes
Fungizone (Amphotericin B) 100× can be used in primary organoid preparations to reduce the risk of contamination with mold but is not necessary. It is suggested because primary preparations are messy and are easily contaminated, especially if you dissect in the open air. If you do not want to use Fungizone in the medium, the mouse should originally be dissected in the hood and dipped in 70% ethanol including the tail before dissection. In addition, the tools should be sterilized before the preparation. It is necessary to avoid mouse hair and/or any other contamination in the preparation.
The collagenase solution must be made fresh right before the extraction procedure as it will inactivate at room temperature if kept for a long time.
The size and yield of epithelial structures/organoids from the mammary glands is affected by the strain of the mouse used for the preparation.
When performing an organoid/epithelial extraction from the mammary gland it is essential to pre-coat all pipettes and conical (Falcon™ or other) tubes with 5% BSA-PBS to prevent epithelial structures from adhering to the surface of the tubes or pipettes which would decrease the final yield.
After the first round of centrifugation, most organoids will settle in the bottom of the conical tube, but some will remain trapped in the fatty layer floating on top of the supernatant. To extract the organoids from this layer, gently remove about 10 mL of medium from the top layer of the supernatant (with the fatty layer) and transfer to another conical tube, then, add 5 mL of fresh, serum-free medium and flush up and down before spinning.
If you want a fibroblast culture, they are in the supernatant at this step, thus, you can put them into a dish and culture them.
You must clean up the epithelial organoids by repeating the pulse 4–10 times. Each time, monitor the composition of the pellet by examining 10 µL of the resuspended pellet under the microscope. Look for mainly organoids and a few single cells. Single cells are mainly the fibroblasts, so you want to get rid of them. Some epithelial cells are in the single cell form, so you do not have to completely remove all single cells. Ten pulses are generally more than sufficient to get a clean epithelial culture.
If primary cells are to be plated as a monolayer on tissue culture plastic, then it is necessary to plate the cells at a high density (20,000 cells/cm2). Low-density cultures do not do well or maintain proper culture morphology. It is possible to coat the plate before plating with a thin layer of Collagen-I. Our lab uses Purecol® which is diluted as 1 mL in 44 mL of PBS. We add 3 mL of this mixture for every 25-cm2 surface area of culture dish. It must gel over night in an airtight case at 4°C. Before use, it is necessary to aspirate off the excess collagen.
All protocols can be performed with primary organoids with slight modifications that relate to medium composition. Base medium for primary organoids is DMEM/F12 with ITS-PS with 5% FBS (see Subheading 2.1, item 10) while base medium for EpH4 is DMEM/F12 with GI (gentamycin-insulin) with 2% FBS (see Subheading 2.1, item 11).
Primary cells are more difficult to handle than cell lines, mostly due to the inherent variability across different isolations and possible variations in the amount of epithelial cells in each extraction. Thus, it is essential to perform the extraction techniques as rigorously as possible and it is paramount to seed primary cells/organoids at similar numbers when starting the experiment.
Due to variability inherent to biological materials, Matrigel™ lots need to be tested in order to ensure reproducibility between experiments using different lots of Matrigel™. A typical test procedure consists of culturing EpH4 cells using new as well as currently used Matrigel™ lots. After 2 days in culture, cells are extracted from Matrigel™ and their ability to produce milk proteins such as β-casein is assessed by RT-PCR or western analysis. Assessment of morphological characteristics of cell clusters can also be used to infer if the Matrigel™ used is conducive to optimal differentiation or if the batch should be discarded.
LrECM available as Matrigel™ is difficult to handle at room temperature as it gels when removed from 4°C. As it gels, it might create bubbles at the bottom of the culture dish. To avoid that, make sure to keep Matrigel™ on ice inside the cell culture hood.
When using primary organoids for morphogenesis assay, it is crucial that cells be isolated from young animals (less than 14 weeks) to be able to recapitulate the phenotype in culture.
- Cells/mL = the average count per square (count 10 squares) × the dilution factor × 104
- Example: If the average count per square is 45 cells × 100 × 104 = 45,000,000 or 45 × 106 cells/mL.
Using 3 mg/mL versus 1 mg/mL of Collagen-I: Type I collagen is used typically at a concentration of 3 mg/mL. However, it has been observed that in some functional assays, a lower concentration of type-I collagen can be used to induce specific phenotypes. The optimal concentration of type I collagen is to be determined by the researcher when designing the experiment.
If Matrigel™ is not all dissolved, add additional PBS–EDTA, pipet gently, and invert the tube a few times before centrifuging (does not require prolonged shaking this time). If there is still a problem in dissolving the Matrigel™, check your EDTA concentration.
Cell pellet in 15 mL conical tube can also be lysed directly and then lysate can be transferred to Eppendorf.
References
- 1.Babinet C. Transgenic mice: an irreplaceable tool for the study of mammalian development and biology. J Am Soc Nephrol. 2000;11(Suppl 16):S88–S94. [PubMed] [Google Scholar]
- 2.Proia DA, Kuperwasser C. Reconstruction of human mammary tissues in a mouse model. Nat Protoc. 2006;1:206–214. doi: 10.1038/nprot.2006.31. [DOI] [PubMed] [Google Scholar]
- 3.Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 4.Schmeichel KL, Weaver VM, Bissell MJ. Structural cues from the tissue microenvironment are essential determinants of the human mammary epithelial cell phenotype. J Mammary Gland Biol Neoplasia. 1998;3:201–213. doi: 10.1023/a:1018751124382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richert MM, Schwertfeger KL, Ryder JW, Anderson SM. An atlas of mouse mammary gland development. J Mammary Gland Biol Neoplasia. 2000;5:227–241. doi: 10.1023/a:1026499523505. [DOI] [PubMed] [Google Scholar]
- 6.Bolander FF Jr. Differential characteristics of the thoracic and abdominal mammary glands from mice. Exp Cell Res. 1990;189:142–144. doi: 10.1016/0014-4827(90)90266-d. [DOI] [PubMed] [Google Scholar]
- 7.Fendrick JL, Raafat AM, Haslam SZ. Mammary gland growth and development from the postnatal period to postmenopause: ovarian steroid receptor ontogeny and regulation in the mouse. J Mammary Gland Biol Neoplasia. 1998;3:7–22. doi: 10.1023/a:1018766000275. [DOI] [PubMed] [Google Scholar]
- 8.Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ, Soloway P, Itohara S, Werb Z. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. J Cell Biol. 2003;162:1123–1133. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silberstein GB. Postnatal mammary gland morphogenesis. Microsc Res Tech. 2001;52:155–162. doi: 10.1002/1097-0029(20010115)52:2<155::AID-JEMT1001>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 10.Daniel CW, Robinson S, Silberstein GB. The role of TGF-beta in patterning and growth of the mammary ductal tree. J Mammary Gland Biol Neoplasia. 1996;1:331–341. doi: 10.1007/BF02017389. [DOI] [PubMed] [Google Scholar]
- 11.Nandi S. Endocrine control of mammary-gland development and function in the C3H/He Crgl mouse. J Natl Cancer Inst. 1958;21:1039–1063. [PubMed] [Google Scholar]
- 12.Ramanathan P, Martin I, Thomson P, Taylor R, Moran C, Williamson P. Genomewide analysis of secretory activation in mouse models. J Mammary Gland Biol Neoplasia. 2007;12:305–314. doi: 10.1007/s10911-007-9052-6. [DOI] [PubMed] [Google Scholar]
- 13.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee EY, Lee WH, Kaetzel CS, Parry G, Bissell MJ. Interaction of mouse mammary epithelial cells with collagen substrata: regulation of casein gene expression and secretion. Proc Natl Acad Sci U S A. 1985;82:1419–1423. doi: 10.1073/pnas.82.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howlett AR, Bissell MJ. The influence of tissue microenvironment (stroma and extracellular matrix) on the development and function of mammary epithelium. Epithelial Cell Biol. 1993;2:79–89. [PubMed] [Google Scholar]
- 16.Rosen JM, Wyszomierski SL, Hadsell D. Regulation of milk protein gene expression. Annu Rev Nutr. 1999;19:407–436. doi: 10.1146/annurev.nutr.19.1.407. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS, Hennighausen L, Furth PA. Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc Natl Acad Sci U S A. 1997;94:3425–3430. doi: 10.1073/pnas.94.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furth PA, Bar-Peled U, Li M. Apoptosis and mammary gland involution: reviewing the process. Apoptosis. 1997;2:19–24. doi: 10.1023/a:1026454207398. [DOI] [PubMed] [Google Scholar]
- 19.Quarrie LH, Addey CV, Wilde CJ. Apoptosis in lactating and involuting mouse mammary tissue demonstrated by nick-end DNA labelling. Cell Tissue Res. 1995;281:413–419. doi: 10.1007/BF00417859. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Hu J, Heermeier K, Hennighausen L, Furth PA. Apoptosis and remodeling of mammary gland tissue during involution proceeds through p53-independent pathways. Cell Growth Differ. 1996;7:13–20. [PubMed] [Google Scholar]
- 21.Daniel CW, Deome KB. Growth of mouse mammary glands in vivo after monolayer culture. Science. 1965;149:634–636. doi: 10.1126/science.149.3684.634. [DOI] [PubMed] [Google Scholar]
- 22.Daniel CW, Young LJ, Medina D, DeOme KB. The influence of mammogenic hormones on serially transplanted mouse mammary gland. Exp Gerontol. 1971;6:95–101. doi: 10.1016/0531-5565(71)90053-2. [DOI] [PubMed] [Google Scholar]
- 23.Emerman JT, Burwen SJ, Pitelka DR. Substrate properties influencing ultrastructural differentiation of mammary epithelial cells in culture. Tissue Cell. 1979;11:109–119. doi: 10.1016/0040-8166(79)90011-9. [DOI] [PubMed] [Google Scholar]
- 24.Emerman JT, Pitelka DR. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 1977;13:316–328. doi: 10.1007/BF02616178. [DOI] [PubMed] [Google Scholar]
- 25.Michalopoulos G, Pitot HC. Primary culture of parenchymal liver cells on collagen membranes. Morphological and biochemical observations. Exp Cell Res. 1975;94:70–78. doi: 10.1016/0014-4827(75)90532-7. [DOI] [PubMed] [Google Scholar]
- 26.Emerman JT, Enami J, Pitelka DR, Nandi S. Hormonal effects on intracellular and secreted casein in cultures of mouse mammary epithelial cells on floating collagen membranes. Proc Natl Acad Sci U S A. 1977;74:4466–4470. doi: 10.1073/pnas.74.10.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EY, Parry G, Bissell MJ. Modulation of secreted proteins of mouse mammary epithelial cells by the collagenous substrata. J Cell Biol. 1984;98:146–155. doi: 10.1083/jcb.98.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aggeler J, Ward J, Blackie LM, Barcellos-Hoff MH, Streuli CH, Bissell MJ. Cytodifferentiation of mouse mammary epithelial cells cultured on a reconstituted basement membrane reveals striking similarities to development in vivo. J Cell Sci. 1991;99(Pt 2):407–417. doi: 10.1242/jcs.99.2.407. [DOI] [PubMed] [Google Scholar]
- 29.Chen LH, Bissell MJ. A novel regulatory mechanism for whey acidic protein gene expression. Cell Regul. 1989;1:45–54. doi: 10.1091/mbc.1.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streuli CH, Bailey N, Bissell MJ. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu R, Nelson CM, Muschler JL, Veiseh M, Vonderhaar BK, Bissell MJ. Sustained activation of STAT5 is essential for chromatin remodeling and maintenance of mammary-specific function. J Cell Biol. 2009;184:57–66. doi: 10.1083/jcb.200807021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- 34.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 35.Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol. 2004;16:558–564. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci U S A. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wicha MS, Lowrie G, Kohn E, Bagavandoss P, Mahn T. Extracellular matrix promotes mammary epithelial growth and differentiation in vitro. Proc Natl Acad Sci U S A. 1982;79:3213–3217. doi: 10.1073/pnas.79.10.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medina D, Li ML, Oborn CJ, Bissell MJ. Casein gene expression in mouse mammary epithelial cell lines: dependence upon extracellular matrix and cell type. Exp Cell Res. 1987;172:192–203. doi: 10.1016/0014-4827(87)90105-4. [DOI] [PubMed] [Google Scholar]
- 40.Daniel CW, Berger JJ, Strickland P, Garcia R. Similar growth pattern of mouse mammary epithelium cultivated in collagen matrix in vivo and in vitro. Dev Biol. 1984;104:57–64. doi: 10.1016/0012-1606(84)90036-8. [DOI] [PubMed] [Google Scholar]
- 41.Montesano R, Soriano JV, Fialka I, Orci L. Isolation of EpH4 mammary epithelial cell subpopulations which differ in their morphogenetic properties. In Vitro Cell Dev Biol Anim. 1998;34:468–477. doi: 10.1007/s11626-998-0080-3. [DOI] [PubMed] [Google Scholar]
- 42.Niemann C, Brinkmann V, Spitzer E, Hartmann G, Sachs M, Naundorf H, Birchmeier W. Reconstitution of mammary gland development in vitro: requirement of c-met and c-erbB2 signaling for branching and alveolar morphogenesis. J Cell Biol. 1998;143:533–545. doi: 10.1083/jcb.143.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 44.Hynes RO. Integrins: a family of cell surface receptors. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 45.Alcaraz J, Xu R, Mori H, Nelson CM, Mroue R, Spencer VA, Brownfield D, Radisky DC, Bustamante C, Bissell MJ. Laminin and biomimetic extracellular elasticity enhance functional differentiation in mammary epithelia. EMBO J. 2008;27:2829–2838. doi: 10.1038/emboj.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talhouk RS, Elble RC, Bassam R, Daher M, Sfeir A, Mosleh LA, El-Khoury H, Hamoui S, Pauli BU, El-Sabban ME. Developmental expression patterns and regulation of connexins in the mouse mammary gland: expression of connexin30 in lactogenesis. Cell Tissue Res. 2005;319:49–59. doi: 10.1007/s00441-004-0915-5. [DOI] [PubMed] [Google Scholar]
- 47.El-Sabban ME, Sfeir AJ, Daher MH, Kalaany NY, Bassam RA, Talhouk RS. ECM-induced gap junctional communication enhances mammary epithelial cell differentiation. J Cell Sci. 2003;116:3531–3541. doi: 10.1242/jcs.00656. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen DA, Neville MC. Tight junction regulation in the mammary gland. J Mammary Gland Biol Neoplasia. 1998;3:233–246. doi: 10.1023/a:1018707309361. [DOI] [PubMed] [Google Scholar]
- 49.Talhouk RS, Mroue R, Mokalled M, Abi-Mosleh L, Nehme R, Ismail A, Khalil A, Zaatari M, El-Sabban ME. Heterocellular interaction enhances recruitment of alpha and beta-catenins and ZO-2 into functional gap-junction complexes and induces gap junction-dependant differentiation of mammary epithelial cells. Exp Cell Res. 2008;314:3275–3291. doi: 10.1016/j.yexcr.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Sun L, Maffini MV, Soto A, Sonnenschein C, Kaplan DL. A complex 3D human tissue culture system based on mammary stromal cells and silk scaffolds for modeling breast morphogenesis and function. Biomaterials. 2010;31:3920–3929. doi: 10.1016/j.biomaterials.2010.01.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingthorsson S, Sigurdsson V, Fridriksdottir AJ, Jonasson JG, Kjartansson J, Magnusson MK, Gudjonsson T. Endothelial cells stimulate growth of normal and cancerous breast epithelial cells in 3D culture. BMC Res Notes. 2010;3:184. doi: 10.1186/1756-0500-3-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen A, Cuevas I, Kenny PA, Miyake H, Mace K, Ghajar C, Boudreau A, Bissell M, Boudreau N. Endothelial cell migration and vascular endothelial growth factor expression are the result of loss of breast tissue polarity. Cancer Res. 2009;69:6721–6729. doi: 10.1158/0008-5472.CAN-08-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen OW, Nielsen HL, Gudjonsson T, Villadsen R, Rank F, Niebuhr E, Bissell MJ, Ronnov-Jessen L. Epithelial to mesenchymal transition in human breast cancer can provide a nonmalignant stroma. Am J Pathol. 2003;162:391–402. doi: 10.1016/S0002-9440(10)63834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ip MM, Darcy KM. Three-dimensional mammary primary culture model systems. J Mammary Gland Biol Neoplasia. 1996;1:91–110. doi: 10.1007/BF02096305. [DOI] [PubMed] [Google Scholar]
- 55.Hamamoto S, Imagawa W, Yang J, Nandi S. Morphogenesis of mouse mammary epithelial cells growing within collagen gels: ultrastructural and immunocytochemical characterization. Cell Differ. 1988;22:191–201. doi: 10.1016/0045-6039(88)90011-5. [DOI] [PubMed] [Google Scholar]
- 56.Bissell MJ. The differentiated state of normal and malignant cells or how to define a “normal” cell in culture. Int Rev Cytol. 1981;70:27–100. doi: 10.1016/s0074-7696(08)61130-4. [DOI] [PubMed] [Google Scholar]
- 57.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CQ, Bissell MJ. Multi-faceted regulation of cell differentiation by extracellular matrix. FASEB J. 1993;7:737–743. doi: 10.1096/fasebj.7.9.8330681. [DOI] [PubMed] [Google Scholar]
- 59.Streuli CH, Bissell MJ. Expression of extracellular matrix components is regulated by substratum. J Cell Biol. 1990;110:1405–1415. doi: 10.1083/jcb.110.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci U S A. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faraldo MM, Deugnier MA, Lukashev M, Thiery JP, Glukhova MA. Perturbation of beta1-integrin function alters the development of murine mammary gland. EMBO J. 1998;17:2139–2147. doi: 10.1093/emboj/17.8.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muschler J, Lochter A, Roskelley CD, Yurchenco P, Bissell MJ. Division of labor among the alpha6beta4 integrin, beta1 integrins, and an E3 laminin receptor to signal morphogenesis and beta-casein expression in mammary epithelial cells. Mol Biol Cell. 1999;10:2817–2828. doi: 10.1091/mbc.10.9.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weir ML, Oppizzi ML, Henry MD, Onishi A, Campbell KP, Bissell MJ, Muschler JL. Dystroglycan loss disrupts polarity and beta-casein induction in mammary epithelial cells by perturbing laminin anchoring. J Cell Sci. 2006;119:4047–4058. doi: 10.1242/jcs.03103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin CQ, Dempsey PJ, Coffey RJ, Bissell MJ. Extracellular matrix regulates whey acidic protein gene expression by suppression of TGF-alpha in mouse mammary epithelial cells: studies in culture and in transgenic mice. J Cell Biol. 1995;129:1115–1126. doi: 10.1083/jcb.129.4.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Sif S, DeWille J. The mouse C/EBPdelta gene promoter is regulated by STAT3 and Sp1 transcriptional activators, chromatin remodeling and c-Myc repression. J Cell Biochem. 2007;102:1256–1270. doi: 10.1002/jcb.21356. [DOI] [PubMed] [Google Scholar]
- 66.Spencer VA, Xu R, Bissell MJ. Gene expression in the third dimension: the ECM-nucleus connection. J Mammary Gland Biol Neoplasia. 2010;15:65–71. doi: 10.1007/s10911-010-9163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lelievre S, Weaver VM, Bissell MJ. Extracellular matrix signaling from the cellular membrane skeleton to the nuclear skeleton: a model of gene regulation. Recent Prog Horm Res. 1996;51:417–432. [PMC free article] [PubMed] [Google Scholar]
- 68.Xu R, Spencer VA, Bissell MJ. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J Biol Chem. 2007;282:14992–14999. doi: 10.1074/jbc.M610316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidhauser C, Bissell MJ, Myers CA, Casperson GF. Extracellular matrix and hormones transcriptionally regulate bovine beta-casein 5’ sequences in stably transfected mouse mammary cells. Proc Natl Acad Sci U S A. 1990;87:9118–9122. doi: 10.1073/pnas.87.23.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 71.Talhouk RS, Chin JR, Unemori EN, Werb Z, Bissell MJ. Proteinases of the mammary gland: developmental regulation in vivo and vectorial secretion in culture. Development. 1991;112:439–449. doi: 10.1242/dev.112.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Talhouk RS, Bissell MJ, Werb Z. Coordinated expression of extracellular matrix-degrading proteinases and their inhibitors regulates mammary epithelial function during involution. J Cell Biol. 1992;118:1271–1282. doi: 10.1083/jcb.118.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sympson CJ, Talhouk RS, Alexander CM, Chin JR, Clift SM, Bissell MJ, Werb Z. Targeted expression of stromelysin-1 in mammary gland provides evidence for a role of proteinases in branching morphogenesis and the requirement for an intact basement membrane for tissue-speci fi c gene expression. J Cell Biol. 1994;125:681–693. doi: 10.1083/jcb.125.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fata JE, Mori H, Ewald AJ, Zhang H, Yao E, Werb Z, Bissell MJ. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306:193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stoker AW, Streuli CH, Martins-Green M, Bissell MJ. Designer microenvironments for the analysis of cell and tissue function. Curr Opin Cell Biol. 1990;2:864–874. doi: 10.1016/0955-0674(90)90085-s. [DOI] [PubMed] [Google Scholar]
- 78.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bissell MJ, Barcellos-Hoff MH. The influence of extracellular matrix on gene expression: is structure the message? J Cell Sci Suppl. 1987;8:327–343. doi: 10.1242/jcs.1987.supplement_8.18. [DOI] [PubMed] [Google Scholar]
- 80.Reichmann E, Ball R, Groner B, Friis RR. New mammary epithelial and fibroblastic cell clones in coculture form structures competent to differentiate functionally. J Cell Biol. 1989;108:1127–1138. doi: 10.1083/jcb.108.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reichmann E, Schwarz H, Deiner EM, Leitner I, Eilers M, Berger J, Busslinger M, Beug H. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial- fibroblastoid cell conversion. Cell. 1992;71:1103–1116. doi: 10.1016/s0092-8674(05)80060-1. [DOI] [PubMed] [Google Scholar]