Abstract
Estrogen receptor (ER) antagonists are valuable in the treatment of ER-positive human breast cancer. In this study, we designed and synthesized nine new derivatives of 17β-estradiol (E2) with a bulky side chain attached to its C-7α position, and determined their ER antagonistic activity using in vitro bioassays. Four of the derivatives showed a strong inhibition of ERα transactivation activity in a luciferase reporter assay and blocked ERα interactions with coactivators. Similarly, these derivatives also strongly inhibited the growth of the ERα-positive human breast cancer cells. Computational docking analysis was conducted to model the interaction of these antagonists with the human ERα, and showed that they could tightly bind to the ERα in a similar manner as ICI-182,780, a pure ER antagonist. These results provide an example that attachment of a bulky side chain to the C-7α position of E2 can produce ER antagonists with comparable ER affinity as ICI-182,780.
Introduction
Since the 1970s, the incidence of breast cancer remains highest among all cancers for women living in the United States, and at present, it is one of the leading causes of cancer-related mortality for women in this country.1 The use of estrogen receptor (ER) antagonists such as tamoxifen has become a valuable strategy as an adjuvant hormonal therapy for ERα-positive human breast cancer.2 In addition, these antiestrogens are also effective for the prevention of estrogen-inducible breast cancer in high risk populations.3 Tamoxifen, a well-known partial agonist of ERs, has a predominant antiestrogenic activity (i.e., ER antagonist activity) under most conditions, but it also has estrogenic activity (i.e., ER agonist activity) and this is of particular concern relative to tamoxifen resistant breast cancer. Pure ER antagonists, such as ICI-182,780 (fulvestrant)4a and ICI-164,3844b that are devoid of ER agonistic activity, have been developed as alternatives to tamoxifen.4 Studies have shown that the ER-positive human breast cancer cells that become resistant to tamoxifen often are still sensitive to the anticancer effect of fulvestrant, which has been approved for clinical use in the United States. Structurally, most of the pure ER antagonists contain the core structure of 17β-estradiol (E2) with a long side-chain attached to the C-7α position. The 7α-substituted 17β-estradiol derivatives are a class of compounds of considerable pharmaceutical interest because they can serve as pure ER antagonists, and the design and synthesis of these compounds have been a main goal of many investigations.5-11 At the molecular level, it is known that the binding of a pure antagonist to the ERα protein interferes with receptor dimerization and particularly its interaction with co-activators, consequently blocking the transcriptional activity.12 In the present study, we sought to design and synthesize several representative E2 derivatives with a bulky ring-based structure attached to the C-7α position. A number of in vitro bioassays have been used to test the biological functions of these E2 derivatives to determine whether they could function as effective ER antagonists. Out of a total of nine compounds synthesized in this study four showed an effective ER antagonist activity with a rather high binding affinity for the human ERα and ERβ. The results of this study provide an example that attachment of a bulky structure to the C-7α position of E2 can produce ER antagonists with comparable receptor binding affinity as ICI-182,780. Studies are ongoing to further test these ER antagonists for their potential usefulness and efficacy in the treatment and prevention of ER-positive breast cancer using in vivo models.
Design and Chemical Synthesis of Novel Estrogen Receptor Antagonists
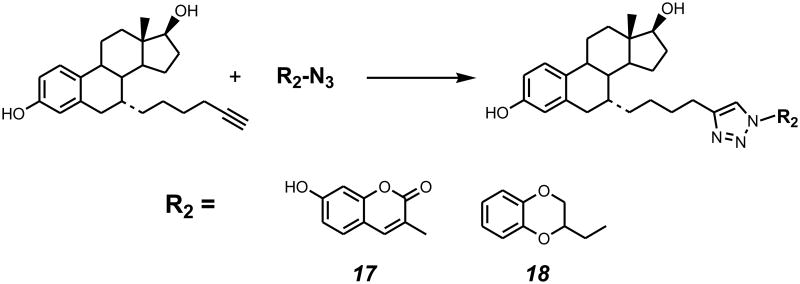
As depicted in Scheme 1, compound 4 was prepared from E2 according to the procedures previously established in our laboratory (described in the Experimental Section). Compound 9 was prepared from compound 8 by reacting with sodium iodide while refluxing in acetone overnight, in 98% yield. Compound 4 was then reacted with compound 9 to give compound 5 in 60% yield using potassium tert-butoxide in THF.6,7,9 Deprotection of THP at the C-3 and C-17β positions from compound 5 using HCl (6 N) in THF gave compound 6 in 91% yield. Then compound 6 was reduced to compound 7 using triethylsilane in the presence of boron trifluoride etherate in methylene chloride in 75% yield. From this rather versatile intermediate (compound 7), the palladium-catalyzed coupling reaction (typically, compound 7, aryl iodide, PdCl2(PPh3)2, CuI, and TEA in DMF) gave compounds 10-16 in yield ranging from 54 to 85% (Schemes 2, 3). Using the “click reaction” (typically, compound 7, azide, sodium ascorbate, and CuSO4 in water and ethyl alcohol, v/v = 1:1), compounds 17 and 18 were synthesized in 81% and 74% yield, respectively (Schemes 2, 4).
Scheme 1. The flow chart for the synthesis of 3,17β-bis(hydroxy)-7α-(6-hexynyl)-estra-1,3,5(10)-trien (compound 7) using E2(compound 1) as the starting material.
The reagents and the reaction conditions used are summarized below: a. DHP, pyridinium p-toluenesulfonate, DCM, reflux. b. (1) LDA, t-BuOK, THF, −78°C. (2) B(OMe)3, 0°C. (3) H2O2, H2O, 25°C. c. Swern oxidation. d. t-BuOK, THF, 0°C, compound 9, −78°C. e. 6 N HCl, THF. f. (Et)3SiH, BF3•Et2O, DCM. g. NaI, acetone, reflux, 12 h.
Scheme 2.
Scheme 3.
Typical conditions: compound 7, aryl iodide, PdCl2(PPh3)2, CuI, TEA in DMF.
Scheme 4.
Typical conditions: compound 7, azide, sodium ascorbate, CuSO4 in water and ethyl alcohol (v:v = 1:1).
Biological Activity Study
After adequate amount of the new E2 derivatives had been synthesized and purified, we performed a series of in vitro experiments to test their biological activity, which included the ERα/ERβ binding assays, cell proliferation assay for both ER-positive and ER-negative human breast cancer cell lines, and the reporter assays for ERα trans-activation and for receptor interaction with coactivators. The data are summarized below.
The new E2 derivatives retain high binding affinity for human ERα and ERβ
First, we determined the relative binding affinity (RBA) of each newly-synthesized E2 derivative for human ERα and ERβ in vitro by using the radioligand-receptor competition assay. In this in vitro assay, the recombinant human ERα and ERβ proteins were used as the receptor proteins, and a final concentration of 10 nM [3H]E2 was used the radioligand. Each of the competing ligands (i.e., the new E2 derivatives) was tested at a wide range of final concentrations (0, 0.24, 0.98, 3.9, 15.6, 62.5, 250, and 1000 nM). The RBA value for each competing estrogen derivative was then calculated according to the RBA of E2 (see the Experimental Section for details).
Although each of the E2 derivatives has a rather bulky structure attached to the C-7α position, the binding affinity of these derivatives is very high (see Figure 2 and Table 1). It was predicted beforehand that these compounds most likely would still retain the ability to interact with the ligand-binding domains of human ERα and ERβ in similar ways as would E2, by forming hydrogen bonds between the C-3 and C-17β hydroxyl groups of the ligand molecules and the amino acid residues in the binding domains of the receptors. Experimental data showed that while each of the nine newly-synthesized E2 derivatives did not show significant preference for binding to human ERα vs ERβ, significant differences were noted in the binding affinities of different derivatives for the human receptors. While J1 and J9 showed the highest binding affinity for human ERs (their RBAs > 20% of E2), the RBAs of J2, J5, J7 and J8 were 3-5% of E2, and the RBAs of J3, J4 and J6 were only approximately 1% of E2.
Figure 2. Comparison of the relative binding affinities (RBAs) of nine new E2 derivatives (J1 to J9) synthesized in this study.
Eight concentrations (0.06, 0.24, 0.98, 3.9, 15.6, 62.5, 250 and 1000 nM) of each competing ligand were tested to determine the concentration-dependent inhibition of the binding of 10 nM [3H]E2 to the recombinant human ERα and ERβ. Each point was the mean of duplicate measurements (average variations were <5%). The incubation mixture in the absence of a competing chemical was set as control. The IC50 values were calculated according to the inhibition curves, and their respective RBAs were then calculated according to the method described in the Experimental Section.
Table 1. The IC50 and RBA values of the new E2 derivatives for ERα and ERβ.
The IC50 values for each competing ligand was calculated according to the sigmoidal inhibition curves as shown in Figure 2, and the relative binding affinity (RBA) values were calculated against E2 by using the following equation: RBA = The IC50 for E2/The IC50 for the test compound.
| Compounds | ERα | ERβ | ||
|---|---|---|---|---|
| IC50 (nM) | RBA (%) | IC50 (nM) | RBA (%) | |
| E2 | 6.0 | 100.0 | 8.7 | 100.0 |
| J1 (compound 14) | 28.3 | 21.2 | 19.1 | 45.7 |
| J2 (compound 16) | 175.7 | 3.4 | 120.7 | 7.2 |
| J3 (compound 11) | 481.8 | 1.2 | 331.1 | 2.6 |
| J4 (compound 10) | 567.4 | 1.1 | 436.4 | 2.0 |
| J5 (compound 12) | 96.5 | 6.2 | 185.0 | 4.7 |
| J6 (compound 15) | 637.8 | 0.9 | 565.9 | 1.5 |
| J7 (compound 17) | 116.0 | 5.2 | 111.9 | 7.8 |
| J8 (compound 13) | 126.4 | 4.7 | 150.6 | 5.8 |
| J9 (compound 18) | 24.4 | 24.6 | 41.2 | 21.1 |
Several new E2 derivatives can inhibit estrogen-dependent growth of ER-positive human breast cancer cells
It is known that the growth of ERα-positive MCF-7 cells can be stimulated by ER agonists and inhibited by ER antagonists.13,14 Of the nine derivatives made in this study, four of them (i.e., J3, J4, J7 and J9) showed a concentration-dependent inhibition of the growth of MCF-7 cells, effective at as low as 1 or 10 nM concentrations (Figure 3A). The IC50 values of J3, J4, and J9 for growth inhibition were approximately 50 nM, and the IC50 value for J7 is approximately 100 nM. Although these compounds are not as potent as ICI-182,780 (IC50 of approximately 2 nM), their inhibitory potency is higher than that of tamoxifen (IC50 of approximately 200 nM) (Figure 3A). Using J3 and J4 as representative compounds, we also tested their activity in the ER-negative MDA-MB-231 cells. As predicted, no appreciable effect (inhibition or stimulation) on the growth of these ER-negative cells was observed when J3 or J4 was present (Figure 3B). However, the other five compounds (J1, J2, J5, J6, and J8) showed either no effect or a stimulatory effect on MCF-7 cell growth (Figure 3C).
Figure 3. Effect of the new E2 derivatives on the proliferation of human breast cancer cell lines.
The ERα-positive MCF-7 and the ERα-negative MDA-MB-231 human breast cancer cell lines were cultured and seeded in 96-well plates. Different final concentrations of the ER ligands (at 0, 0.1, 0.32, 1.6, 8, 40, 200, and 1000 nM) were added to the cell culture medium for 6 days with one medium change at the end of the third day. The cell density was determined by using the crystal violet staining method as described in the methods section. Each point was the mean ± S.E.M. (N = 6). Note that for the y-axis, 100% refers to the cell density after 6 days of culture in the presence of vehicle only, and 0% means the initial seeding density of the cells on day 0. In these experiments, since no additional estrogen was added to the cell culture medium, all the estrogens came with the 10% FBS contained in the cell culture medium. A. Four of the E2 derivatives (J3, J4, J7 and J9) showed a concentration-dependent inhibition of the growth of ER-positive MCF-7 cells, and their effect was compared with ICI-182,780 and tamoxifen. B. Two representative ER antagonists (J3 and J4) were further tested in the ER-negative MDA-MB-231 cells, and they did not show an appreciable effect on the growth of these cells. C. Five of the other E2 analogs (J1, J2, J5, J6 and J8) did not show an appreciable effect on the growth of the ER-positive MCF-7 cells.
Upon comparing the structures of J3, J4, J7 and J9 with the other five chemicals, we noticed that the two closely-placed rings and the presence of nitrogen and oxygen atoms in the side chain seem important for the antagonist activity. Therefore, the precursors in Scheme 4 appear to be more promising for the design and synthesis of pure ER antagonists than the precursors in Scheme 3.
Regulation of ERα transcriptional activity by the new E2 derivatives
While E2 is known to be an agonist for ERα in all cell environments, other compounds, particularly partial agonists/antagonists vary in their ability to stimulate ERα transcriptional activity depending upon cell types. In this study, the ability of the new E2 derivatives to affect the transcriptional activity of ERα was tested in three different cell lines. In the MCF-7 human breast cancer cells, we observed that 100 nM of J1, J2 or J5 exerted a similar activation of the ERα-based transcription of a target gene as did 1 nM E2, but J6 and J8 at the same concentration showed a markedly weaker effect (Figure 4A). In comparison, J3 and J4 were very poor agonists, and J7 and J9 had no agonist activity. While the profile of the transcriptional activity of these E2 derivatives in HeLa cells (Figure 4C) was very similar to that obtained for MCF-7 cells, several of the derivatives produced more agonist activity in HepG2 cells than in HeLa or MCF-7 cells. For instance, J3 and J9 are poor agonists in the latter cell types, but exert approximately 50% of the agonist activity of E2 in HepG2 cells (Figure 4B), suggesting that these compounds may regulate some of the ERα functions in a cell-type dependent manner. The ER activity in HepG2 cells is positively influenced by the activity of the N-terminal portion of the receptor which encompasses the activation function 1 (AF-1) domain12 and the ability of J3 and J9 to stimulate luciferase activity in these cells suggests that these compounds do not block this ligand-independent activity. The compounds that exerted the least agonist activity (J3, J4, J7, and J9) were tested for their ability to block E2-induced ERα function in HeLa cells (Figure 4D). Compounds J7 and J9 were the strongest antagonists of the E2-stimulated luciferase expression, while J3 and J4 were less effective inhibitors, consistent with their partial agonist activity observed in Figure 4C.
Figure 4. Assessment of the ERα agonist and antagonist activities of the new E2 derivatives.
A. MCF-7 cells were transfected with the ERE-Elb-Luc reporter plasmid and treated with the vehicle (0.1% ethanol), 1 nM E2, 100 nM ICI-182,780, or 100 nM of each of the new E2 derivatives. HepG2 (B) and HeLa (C) cells were transfected with the expression vector pCR3.1-ERα and the reporter plasmid ERE-Elb-Luc, and they were also treated with the ligands as described for panel A. D. HeLa cells were transfected as described above and were treated in the absence (white bars) or presence (stripped bars) of 1 nM E2 with the ligands as indicated. For each of these experiments, the luciferase values are normalized to those obtained for E2 treatment. Bars represent the average ± S.E.M. of at least 3 experiments.
The transcriptional activity of ERα is dependent upon coactivators such as SRC-1 which bind to ERα in an agonist-dependent manner and help to remodel chromatin via their intrinsic or associated histone acetyltransferase activities.15 To determine if the transcriptional activity induced by each of the E2 derivatives was reflected in their ability to regulate the recruitment of coactivators to ERα, the mammalian two-hybrid assays were performed. As shown in Figure 5A, the new derivatives with the greatest agonist activity (i.e., J1, J2, J5, J6, and J8) effectively promoted the interaction between the ER and the SRC-1 coactivator, while those with antagonist activity (i.e., J3, J4, J7, and J9) did not induce significant SRC-1 binding. As expected, the pure ER antagonist ICI-182,780 also did not promote detectable ERα-SRC-1 interaction. Thus, the relative ability of these new E2 derivatives to stimulate ERα transcriptional activity correlates well with their ability to promote ERα interaction with the SRC-1 coactivator. Finally, a prior report had demonstrated that pure antagonists such as ICI-182,780 can strongly promote ERα interaction with the CBP coactivator at a domain distinct from the agonist binding site.16 As these new compounds are C7α side-chain derivatives of E2 as is ICI-182,780, the ability of these derivatives to promote CBP binding to ERα was determined. In comparison to ICI-182,780, none of the derivatives effectively promoted binding of the CH3 domain of CBP to the ERα LBD (Figure 5B). This effectively distinguishes the molecules with prominent antagonist activity (J3, J4, J7, and J9) from the pure ER antagonist ICI-182,780, and there by reinforces the concept that the chemical nature of the C7α side chain is an important determinant of whether the CH3 domain of CBP can be recruited to ERα.17
Figure 5. The use of mammalian two hybrid assay to examine the interactions between the ERα LBD and SRC-1 or the CBP CH3 domain.
The luciferase activity of HeLa cells transfected with the expression vectors for GAL-SRC-1 (A) or GAL-CBP-CH3 (B) in the presence of the VP16-ERα-LBD expression vector and the pG5-Luc reporter plasmid. Cells were treated with vehicle (0.1% ethanol), 1 nM E2, 100 nM ICI-182,780, or 100 nM of each of the E2 derivatives. The values are normalized to those obtained for VP16-ERα-LBD and the respective GAL-coactivator in the presence of E2. Bars represent the average ± S.E.M. of at least 3 experiments.
Computational Molecular Modeling Study
The three-dimensional structures of the ERα LBD in association with both an agonist and an antagonist have been resolved using X-ray crystallography.18 The most notable difference between these two conformations is that in the antagonist-binding conformation, the helix-12 (H12), which is critical for formation of the docking site for the LXXLL-containing coactivators, is forced to adapt an alternative position.18 With this conformation, the ERα lacks transcriptional activity. Using the ERα LBD in complex with raloxifene (an ER antagonist) as a template, we analyzed the binding interaction of the ERα LBD with the new ER antagonists developed in this study by using computational docking tools.
As shown in Figure 6 (A and B), both ICI-182,780 and J9 can bind inside the binding pocket of the human ERα in a very similar manner, and their steroidal rings, especially the C-3 and C-17β hydroxyl groups, overlapped with each other extensively. Their hydroxyl groups form hydrogen bonds with the amino acid residues E353, R394 and H524 of ERα in a similar fashion as those formed with E2. In addition to the formation of hydrogen bonds between the C-3 or C-17β hydroxyl groups of J9 and ERα, it appears that its side chain also forms an additional hydrogen bond with ERα-N532, although it would be difficult to estimate its relative contribution to the overall binding interaction. Notably, the high degree of overall similarity between the binding modes of ICI-182,780 and J9 inside the ERα binding pocket as predicted by computational docking analysis is consistent with their relative binding affinities for the human ERα (the RBA of ICI-182,780 is 45% of E2, and the RBA of J9 is 24.6%). Using the same method, we also docked J3, J4 and J7 into the ERα LBD, and similar binding interactions between the ligands and the receptor were observed (data not shown).
Figure 6. Molecular docking of representative E2 derivatives with the ligand binding domain of human ERα.
A. The structures of the ERα ligand binding domain in complex with ICI-182,780 and J9. B. Formation of hydrogen bonds between several amino acid residues in the ERα ligand binding domain and ICI-182,780 or J9. The protein structure was shown in colored ribbons according to the secondary structures (red for α-helix, blue for β-sheets, green for turns, and white for coils). Amino acids and ligands are shown in stick and balls. Oxygen, nitrogen, carbon and fluorine atoms are colored red, blue, gray, and cyan, respectively. Hydrogens are omitted from all structures.
It is of note that based on the docking models developed in this study, we noticed that the pure antagonists (ICI-182,780 and the newly-synthesized J3, J4 J7 and J9) adopt a unique binding mode compared to E2 by flipping their core steroidal structure 180° along the C-3 hydroxyl and C-17β hydroxyl axis. In this way the long C-7α side chain was positioned in the C-11β channel and then exited the ligand binding pocket in a similar manner as raloxifene. However, the characteristic hydrogen bonds of C-3 and C-17β hydroxyl groups with the receptor binding pocket basically remain unchanged. This predicted binding mode is rather intriguing and it is consistent with the reported crystal structure of human ERβ in complex with ICI-164,384 (an analog of ICI-182,780).19
In conclusion, we have designed and synthesized in this study nine E2-based new derivatives with a bulky side chain attached to its C-7α position. While all these derivatives have considerable binding affinity for human ERα and ERβ, only four of them have pure ER antagonistic activity, based on in vitro analyses using the ERα transactivation activity assay and the proliferation assay in the ER-positive human breast cancer cells. Molecular computational modeling analysis showed that these new ER antagonists can bind inside the ERα binding pocket in a similar way as other known ERα antagonists such as ICI-182,780. The results of this study showed that these new ER antagonists are good candidates for further structural modifications as well as testing of their usefulness and efficacy in treating and preventing ERα-positive breast cancers.
Experimental Section
Chemicals and reagents
Most of the commercially-available reagents were obtained from Sigma-Aldrich (St. Louis, MO) or ACROS (through Fisher Scientific, Atlanta, GA). 17β-Estradiol (E2) was purchased from Steraloids (Newport, RI). Tetrahydrofuran (THF) was distilled over sodium benzophenone ketyl and dichloromethane distilled over calcium hydride in our laboratory prior to use. Most of the chemicals and solvents used in this study were of ACS grade and used directly without additional steps of purification.
In all of the in vitro bioactivity assays, the new E2 derivatives synthesized in this study were first purified and then dissolved in 200-proof ethanol to a stock concentration of 1 mM. They were then further diluted to desired concentrations with the binding buffer (for ER binding assays) or with the cell culture medium (for treating cultured cells).
Spectrometric analyses
Mass spectra were determined using a VG70S analytical mass spectrometer. An aliquot of the ethanol solution of the test compound was used for the direct-probe mass analysis. The nuclear magnetic resonance (NMR) spectra of all test compounds were determined in deuterochloroform or dimethyl sulfoxide-d6 solution using a Varian Mercury/VX 300 spectrometer with tetramethylsilane (TMS) as an internal standard (δ = 0) unless noted otherwise. The purity of all synthesized final products was determined to be ≥95% using a Waters analytical HPLC unit (model Alliance 2695) coupled with a photodiode array detector (model 2996) and a Restek Pinnacle II reverse phase analytical column (C18, 5 μ, 150 × 4.6 mm). The HPLC elution included a linear gradient of 5-95% methanol in H2O + 0.1 % formic acid over a 30 min period with a flow rate of 1.5 mL/min.
Synthesis of 3,17β-bis(tetrahydropyranyloxy)-7α-(6-hexynyl)-estra-1,3,5(10)-trien-6-one (compound 5)
The synthesis of compound 4 was described in our recent study.21,22 For the synthesis of compound 5, a 1.0 M solution of t-BuOK in THF (5.35 mL, 5.35 mmol) was added to a cooled solution (at 0°C) of compound 4 (1.2 g, 2.67 mmol) in THF (20 mL). The reaction mixture was stirred at 0°C for 40 min and then cooled to −78°C. 6-Iodo-1-hexyne (compound 9, 1.48 g 7.1 mmol) was added drop-wise to the solution. The reaction mixture was allowed to warm to 0°C and stirred for 2 hours, and then to room temperatures and stirred overnight. The reaction was quenched with water and extracted with ethyl acetate. The organic phase was dried over Na2SO4 and solvent was evaporated in vacuo. Flash chromatography (hexane:ethyl acetate = 4:1) afforded compound 5 in 60% yield (0.86 g). Data from spectrometric analyses of the afforded compound are summarized below. 1H NMR (300 MHz, CDCl3): 7.61 (1H, m), 7.23 (1H, m), 7.13 (1H, dd, J = 2.7, 8.7 Hz), 5.38 (1H, m), 4.57 (1H, m), 3.82 (2H, m), 3.69 (1H, t, J = 8.1 Hz), 3.53 (1H, m), 3.41 (1H, m), 2.61 (1H, m), 2.31 (2H, m), 2.07∼1.72 (10H, m), 1.61∼1.18 (20H, m), 0.73 (3H, s). MS (TOF MS ES+, C34H46O5, M 534): 535 (M + 1) (base peak). HRMS [535 (M + 1)]: 535.3423 (calculated) and 535.3422 (observed).
Synthesis of 3,17β-bis(hydroxy)-7α-(6-hexynyl)-estra-1,3,5(10)-trien-6-one (compound 6)
A solution containing compound 5 (0.86 g 1.6 mmol), HCl (15 mL, 6 M) and THF (15 mL) was stirred at room temperature overnight. The reaction mixture was poured into water and then extracted with ethyl acetate (4 × 50 mL). The combined organics were dried over Na2SO4, concentrated, and then chromatographed (hexane:ethyl acetate = 2:1) to afford compound 6 (0.54 g, 91% yield). Data from spectrometric analyses of the afforded compound are summarized below. 1H NMR (300 MHz, DMSO-d6): 9.56 (1H, s), 7.26 (2H, m), 6.96 (1H, dd, J = 2.7, 8.1 Hz), 3.54 (1H, t, J = 8.1 Hz), 2.59 (1H, m), 2.48 (2H, m), 2.31 (2H, m), 2.08 (2H, m), 1.85 (4H, m), 1.47∼1.13 (11H, m), 0.73 (3H, s). MS (TOF MS ES+, C24H30O3, M 366): 367 (M + 1) (base peak). HRMS [367 (M + 1)]: 367.2277 (calculated) and 367.2273 (observed).
Synthesis of 3,17β-bis(hydroxy)-7α-(6-hexynyl)-estra-1,3,5(10)-trien (compound 7)
Compound 6 (0.26 g, 0.70 mmol) was dissolved in methylene chloride (20 mL). To this solution was added triethylsilane (3.5 mL), followed by boron trifluoride etherate (12.5 mL) dropwise. The reaction was stirred for 2 days at room temperatures, cooled to 0°C, and quenched with 10% potassium carbonate (20 mL). The heterogeneous solution was filtered through a short silica plug and the organic layer separated. The aqueous layer was extracted twice with methylene chloride (20 mL). The organic fractions were combined, dried over sodium sulfate, and evaporated in vacuo to a yellow solid. Purification by flash chromatography (hexane:ethyl acetate = 2:5) provided compound 7 (0.185 g 75%). Data from spectrometric analyses of the afforded compound are summarized below. 1H NMR (300 MHz, DMSO-d6): 8.96 (1H, s), 7.02 (1H, d, J = 8.4 Hz), 6.48 (1H, dd, J = 2.7, 8.4 Hz), 6.40 (1H, d, J = 2.7 Hz), 4.46 (1H, d, J = 5.1 Hz), 3.51 (1H, m), 2.75 (1H, dd, J = 5.1, 16.5 Hz), 2.70 (1H, t, J = 2.7 Hz), 2.57 (1H, d, J = 16.5 Hz), 2.48 (2H, m), 2.24∼2.07 (4H, m), 1.85 (1H, m), 1.76 (1H, m), 1.62 (2H, m), 1.47∼1.13 (11H, m), 0.64 (3H, s). MS (C24H32O2, M 352): 157 (base peak), 269, 352. HRMS [352 (M)]: 352.2402 (calculated) and 352.2398 (observed).
Synthesis of 6-iodo-1-hexyne (compound 9)
Under nitrogen, 5 mL (41.3 mmol) of 6-chloro-1-hexyne (compound 8) was added into a mixture of 39 g (262 mmol) of sodium iodide and 100 mL acetone. The mixture was reflux over night. After completion, the reaction mixture was allowed to cool to room temperatures. The organic solvent was removed in vacuo. Then, to the residue 150 mL hexane was added. After the solid was removed by filtration, the filtrate was concentrated. Flash chromatography (hexane as the eluent) afforded compound 9 (8.4 g 98%). Data from spectrometric analyses of the afforded compound are summarized below. 1H NMR (300 MHz, CDCl3): 3.21 (2H, t, J = 6.9 Hz), 2.33 (2H, t-d, J = 2.7, 6.9 Hz), 1.97 (2H, m), 1.64 (2H, m).
Experimental procedures for the palladium-catalyzed coupling reaction
Under nitrogen protection, a mixture of compound 7 (20 mg 0.057 mmol) and R-I (0.063 mmol) was dissolved in dry DMF (1 mL). PdCl2(PPh3)2 (2 mg, 2.85 μmol) and CuI (1.08 mg, 5.7 umol) were added to the mixture solution. The resulting mixture was cooled down to −20°C and TEA (48 μL, 0.342 mmol) was added. After 15 min, the cold bath was removed, the reaction solution was warmed to room temperatures for another 24 hours. Water (10 mL) was added to the reaction mixture. The aqueous layer was extracted with ether acetate (3 × 20 mL). The combined organic layer was dried over Na2SO4, concentrated in vacuo. Flash chromatograph (ether acetate:hexene = 1:2) gave the desired products. Data from spectrometric analyses of the afforded compounds are summarized below.
4-{4-[6-((7α,17β)-3,17-Dihydroxyestra-1,3,5(10)-trien-7-yl)-hex-1-ynyl]-phenyl}-piperazine-1-carboxylic acid tert-butyl ester (compound 10)
Yield 85%. 1H NMR (300 MHz, CDCl3): 7.21 (1H, d, J = 8.4 Hz), 7.07 (1H, d, J = 8.4 Hz), 6.75 (2H, d, J = 8.4 Hz), 6.56 (1H, dd, J = 2.7, 8.4 Hz), 6.45 (1H, d, J = 2.7 Hz), 4.87 (1H, s), 3.66 (1H, brs), 3.45 (4H, t, J = 5.1 Hz), 3.08 (4H, t, J = 5.1 Hz), 2.81 (1H, dd, J = 4.8, 16.5 Hz), 2.65 (1H, d, J = 16.5 Hz), 2.29 (2H, m), 2.03 (1H, m), 1.85 (1H, m), 1.68 (1H, m), 1.55∼1.16 (23H, m), 0.71 (3H, s). MS (TOF MS ES+, C39H52N2O4, M+1 613): 613 (base peak). HRMS [(M+1, 613)]: 613.4005 (calculated) and 613.4017 (observed).
3,17β-Bis(hydroxy)-7α-[6-(4-Isoxazol-5-yl-phenyl)-hex-5-ynyl]-estra-1,3,5(10)-trien (compound 11)
Yield 82%. 1H NMR (300 MHz, CDCl3): 8.27 (1H, d, J = 2.1 Hz), 7.70 (2H, d, J = 8.4 Hz), 7.42 (2H, d, J = 8.4 Hz), 7.13 (1H, d, J = 8.4 Hz), 6.61 (1H, dd, J = 2.7, 8.4 Hz), 6.52 (1H, d, J = 2.7 Hz), 6.49 (1H, d, J = 2.1 Hz), 4.78 (1H, brs), 3.72 (1H, t, J = 7.8 Hz), 2.87 (1H, dd, J = 5.4, 16.8 Hz), 2.72 (1H, d, J = 16.8 Hz), 2.39 (2H, t, J = 6.9 Hz), 2.29 (2H, m), 2.09 (1H, m), 1.90 (1H, m), 1.59∼1.21 (15H, m), 0.76 (3H, s). MS (TOF MS ES+, C32H37NO3, M 495): 496 (M + 1) (base peak). HRMS [496 (M + 1)]: 496.2851 (calculated) and 496.2849 (observed).
3-{4-[6-((7α,17β)-3,17-Dihydroxyestra-1,3,5(10)-trien-7-yl)-hex-1-ynyl]-phenyl}-3 -oxo-propionic acid ethyl ester (compound 12)
Yield 64%. 1H NMR (300 MHz, CDCl3): 7.80 (2H, d, J = 8.4 Hz), 7.36 (2H, d, J = 8.4 Hz), 7.08 (1H, d, J = 8.4 Hz), 6.56 (1H, dd, J = 2.7, 8.4 Hz), 6.45 (1H, d, J = 2.7 Hz), 5.58 (1H, s), 4.77 (1H, s), 4.14 (2H, q, J = 7.2 Hz), 3.90 (2H, s), 3.67 (1H, brs), 2.82 (1H, dd, J= 4.8, 16.2 Hz), 2.66 (1H, d, J = 16.2 Hz), 2.35 (2H, m), 2.24 (2H, m), 2.03 (1H, m), 1.83 (1H, m), 1.69 (1H, m), 1.56∼1.16 (16H, m), 0.71 (3H, s). MS (C35H42O5, M 542): 542, 496, 466, 189 (base peak). HRMS [542 (M)]: 542.3032 (calculated) and 542.3010 (observed).
2-{4-[6-((7α,17β)-3,17-Dihydroxyestra-1,3,5(10)-trien-7-yl)-hex-1-ynyl]-benzyl}-malonic acid diethyl ester (compound 13)
Yield 76%. 1H NMR (300 MHz, CDCl3): 7.25 (2H, d, J = 8.1 Hz), 7.12 (3H, m), 6.62 (1H, dd, J = 2.7, 8.4 Hz), 6.45 (1H, d, J = 2.7 Hz), 5.51 (1H, brs), 4.15 (4H, m), 3.72 (1H, t, J = 7.2 Hz), 3.62 (1H, t, J = 8.1 Hz), 3.17 (2H, d, J = 7.2 Hz), 2.85 (1H, dd, J= 5.1, 16.5 Hz), 2.70 (1H, d, J = 16.5 Hz), 2.35 (2H, t, J = 6.9 Hz), 2.29 (2H, m), 2.09 (1H, m), 1.89 (1H, m), 1.62∼1.11 (21H, m), 0.76 (3H, s). MS (C35H42O5, M 600): 600 (base peak). HRMS [600 (M)]: 600.3451 (calculated) and 600.3471 (observed).
3,17β-Bis(hydroxy)-7α-[6-(4-pyrrol-1-yl-phenyl)-hex-5-ynyl]-estra-1,3,5(10)-trien (compound 14)
Yield 75%. 1H NMR (300 MHz, CDCl3): 7.36∼7.19 (4H, m), 7.10 (1H, d, J = 8.7 Hz), 7.02 (2H, t, J = 2.4 Hz), 6.56 (1H, m), 6.48 (1H, m), 6.28 (2H, t, J = 2.4 Hz), 4.59 (1H, brs), 3.67 (1H, brs), 2.83 (1H, dd, J = 3.9, 16.2 Hz), 2.67 (1H, d, J = 16.2 Hz), 2.33 (2H, m), 2.24 (2H, m), 2.06 (1H, m), 1.81 (1H, m), 1.69 (1H, m), 1.57∼1.09 (14H, m), 0.71 (3H, s). MS (C34H39NO2, M 493): 493 (base peak), 272, 157. HRMS [493 (M)]: 493.2981 (calculated) and 493.2977 (observed).
3,17β-Bis(hydroxy)-7α-[6-(4-piperidin-1-ylmethyl-phenyl)-hex-5-ynyl]-estra-1,3,5 (10)-trien (compound 15)
Yield 54%. 1H NMR (400 MHz, CDCl3): 7.36 (2H, d, J = 8.0 Hz), 7.25 (2H, d, J = 8.0 Hz), 7.14 (1H, d, J = 8.4 Hz), 6.63 (1H, dd, J = 2.8, 8.4 Hz), 6.55 (1H, d, J = 2.8 Hz), 3.66 (1H, t, J = 6.8 Hz), 2.77 (1H, dd, J= 5.2, 16.8 Hz), 2.60 (1H, d, J = 16.8 Hz), 2.31∼2.21 (4H, m), 2.16 (2H, t, J = 6.8 Hz), 2.07 (2H, m), 1.84 (2H, m), 1.64∼1.11 (24H, m), 0.71 (3H, s). MS (TOF MS ES+, C36H47NO2, M+1 526): 526 (base peak). HRMS [(M+1, 526)]: 526.3685 (calculated) and 526.3691 (observed).
3,17β-Bis(hydroxy)-7α-(6-naphthalen-2-yl-hex-5-ynyl)-estra-1,3,5(10)-trien (compound 16)
Yield 82%. 1H NMR (300 MHz, CDCl3): 8.24 (1H, m), 7.75 (2H, m), 7.53∼7.32 (4H, m), 7.07 (1H, d, J = 8.4 Hz), 6.56 (1H, dd, J = 2.7, 8.4 Hz), 6.42 (1H, d, J = 2.7 Hz), 3.62 (1H, t, J = 8.7 Hz), 2.81 (1H, dd, J = 5.4, 16.8 Hz), 2.68 (1H, d, J = 16.8 Hz), 2.48 (2H, t, J = 5.7 Hz), 2.24 (2H, m), 1.99 (1H, m), 1.79 (1H, m), 1.66∼1.15 (16H, m), 0.70 (3H, s). MS (C34H38O2, M 478): 478 (base peak), 157. HRMS [478 (M)]: 478.2872 (calculated) and 478.2866 (observed).
Experimental procedures for the “click reaction” (Scheme 4)
In a mixture of compound 7 (20 mg, 0.057 mmol) and sodium azide (0.063 mmol) in water and ethyl alcohol (v/v = 1:1, 2 mL), sodium ascorbate (17 μL, 0.017 mmol of freshly prepared 1 M solution in water was added, followed by the addition of copper (II) sulfate pentahydrate 7.5% in water (14 μL, 0.0042 mmol). The mixture was stirred vigorously at room temperature for 24 hours. Ethanol was removed and the residue was diluted with water, cooled in ice, and then the precipitate was collected by filtration. After washing the precipitate with cold water, it was dried under vacuum to afford the desired product. Data from spectrometric analyses of the afforded compounds are summarized below.
3-{4-[4-((7α,17β)-3,17-Dihydroxyestra-1,3,5(10)-trien-7-yl)-butyl]-1,2,3-triazol-1 -yl}-7-hydroxy-1-benzopyran-2-one (compound 17)
Yield 81%. 1H NMR (300 MHz, DMSO-d6): 8.51 (1H, s), 8.24 (1H, s), 7.69 (1H, d, J = 8.7 Hz), 7.03 (1H, d, J = 8.4 Hz), 6.85 (1H, dd, J = 2.4, 8.7 Hz), 6.78 (1H, d, J = 2.4 Hz), 6.48 (1H, dd, J = 2.4, 8.4 Hz), 6.40 (1H, d, J = 2.4 Hz), 4.46 (1H, brs), 3.41 (1H, t, J = 6.9 Hz), 2.67 (2H, m), 2.25 (2H, m), 1.88∼1.15 (20H, m), 0.64 (3H, s). MS (C33H78N3O5, M 555): 555 (base peak). HRMS (TOF MS ES+) [556 (M+1)]: 556.2811(calculated) and 556.2811(observed).
3,17β-Bis(hydroxy)-7α-{4-[1-(2,3-dihydro-1,4-benzodioxin-2-ylmethyl)-1H-1,2,3-triazol-4-yl]-butyl}-estra-1,3,5(10)-trien (compound 18)
Yield 74%. 1H NMR (300 MHz, CDCl3): 7.99 (1H, s), 7.29 (1H, s), 7.09 (1H, d, J = 8.4 Hz), 6.86 (4H, m), 6.64 (1H, m), 6.53 (1H, m), 4.57 (1H, m), 4.29 (1H, m), 3.83 (1H, m), 3.75 (1H, t, J = 8.4 Hz), 2.79 (1H, m), 2.67 (3H, m), 2.26 (2H, m), 2.10 (2H, m), 1.90 (1H, m), 1.71∼1.10 (15H, m), 0.75 (3H, s). MS (C33H41N3O4, M 543): 543 (base peak, direct exposure probe). HRMS [543 (M), direct exposure probe]: 543.3097(calculated) and 543.3088(observed).
Synthesis of 2-azidomethyl-2,3-dihydro-1,4-benzodioxine (compound 22)

To a solution of 2-hydroxymethyl-1,4-benzodioxane (compound 20; 20 mmol) in toluene (150 mL) were added, with vigorous stirring, imidazole (22 mol), triphenylphosphine (21 mmol), and iodine (22 mmol). After 2 hours at ambient temperature, the solution was decanted from the gummy precipitate. The decantate was decolorized by shaking it with aqueous Na2S2O3 and water, dried (Na2SO4) and evaporated to dryness to afford crude product, 2-iodomethyl-1,4-benzodioxane (compound 21). This crude product was used in next step without further purification.
To a solution of 2-iodomethyl-1,4-benzodioxane (compound 21; 10 mmol) in 40 mL DMF was added sodium azide (40 mmol). The mixture was heated to 40°C overnight then the resulting solid was removed by filtration, and the filtrate was poured into a mixture of hexane and ethyl acetate (400 mL; 3:1). The solid that formed was filtered and the filtrate was concentrated in vacuo. The crude product was purified by silica gel column (eluting with a mixture of hexane and ethyl acetate (15 - 40%) to afford 2-azidomethyl-2,3-dihydro-1,4-benzodioxine (compound 22) in 62% yield (over two steps). Data from spectrometric analyses of compound 22 are summarized below. CDCl3, 400MHz, 6.91 (4H, m), 4.36 (1H, dd, J = 2.0, 11.2), 4.27 (1H, m), 4.14 (1H, dd, J = 6.0, 11.2 Hz), 3.32 (2H, m).
Synthesis of 3-azido-7-hydroxycoumarin (compound 26)

3-Azido-7-hydroxycoumarin was prepared according to the published procedures.23 Typical procedures are as follows: A mixture of 2,4-dihydroxy benzaldehyde (2.76 g 20 mmol), N-acetylglycine (2.34 g 20 mmol), anhydrous sodium acetate (60 mmol) in acetic anhydride (100 mL) was refluxed under stirring for 4 hours. The reaction mixture was poured onto ice to give a yellow precipitate. After filtration, the yellow solid was washed by ice water before it was refluxed in a solution of concentrated HCl and ethanol (2:1, 30 mL) for 1 hour, then ice water (40 mL) was added to dilute the solution. The solution was then cooled in an ice bath and NaNO2 (40 mmol) was added. The mixture was stirred for 10 minutes and NaN3 (60 mmol) was added in portions. After stirring for another 15 minutes, the resulting precipitate was filtered off, washed with water, and dried under reduced pressure to afford a brown solid; 3-azido-7-hydroxycoumarin (compound 26) 2.2 g (54% overall yield).
ERα and ERβ binding assays
For the in vitro ERα and ERβ radioligand binding assays, [2,4,6,7,16,17-3H]E2 (specific activity of 110 Ci/mmol) was used as the radioligand. It was obtained from PerkinElmer (Waltham, MA) and was purified in our laboratory using a high-pressure liquid chromatography (HPLC)-based method20 before it was used in the binding assays. The recombinant human ERα and ERβ proteins were obtained from PanVera (Madison, WI). According to the supplier, the recombinant human ERα and ERβ were produced in a baculovirus-mediated expression system, and they were soluble and functionally active, with post-translational modifications similar to those found in mammalian cells.
The following buffer solutions were used in the ERα and ERβ binding assay, and they were prepared beforehand and stored at 4°C. The binding buffer consisted of 10% glycerol, 2 mM dithiothreitol, 1 mg/mL BSA and 10 mM Tris-HCl at pH 7.5. The ERα washing buffer contained 40 mM Tris-HCl and 100 mM KCl (pH 7.4), but the ERβ washing buffer contained only 40 mM Tris-HCl (adjusted to pH 7.4). The 50% hydroxylapatite slurry was prepared first by vigorously mixing 10 g hydroxylapatite with 60 mL of the Tris-HCl solution (50 mM, pH 7.4). Hydroxylapatite was then allowed to settle for 20 min at room temperatures, and the supernatant was decanted. The above procedures were repeated 10 times, and afterwards hydroxylapatite was kept in the 50 mM Tris-HCl solution overnight at 4°C. Hydroxylapatite slurry (EMD Biosciences, Inc. San Diego, CA) was then adjusted to an approximate final concentration of 50% (v/v) using the same Tris-HCl solution and stored at 4°C.
On the day of performing the ER binding assay, [3H]E2 solution was freshly diluted in the binding buffer, and an aliquot (45 μL) of the [3H]E2 solution was added to a 1.5 mL microcentrifuge tube, intended for a final [3H]E2 concentration at 10 nM. Each of the competing ligands (in 50 μL volume of the binding buffer) was then added to the mixture for the intended final concentrations at 0, 0.24, 0.98, 3.9, 15.6, 62.5, 250, and 1000 nM. Immediately before addition of the ERα or ERβ protein, it was diluted in the binding buffer and mixed gently with repetitive pipetings. An aliquot (5 μL) of the diluted ERα or ERβ solution was precisely added to the mixture containing 45 μL of the [3H]E2 and 50 μL of the competing ligand and then mixed with repetitive pipetings. Nonspecific binding (NSB) by the [3H]E2 was determined in separate tubes by inclusion of a 400-fold concentration of the nonradioactive E2 (at a final concentration of 4 μM). The binding mixture was incubated at 4°C for overnight.
At the end of the incubation, 100 μL of the hydroxylapatite slurry was added to each tube and the tubes were incubated on ice for 15 min with 3 times of brief vortexing. An aliquot (1 mL) of the appropriate washing buffer was added, mixed, and centrifuged at 10,000 g for 5 min, and the supernatants were discarded. This wash step was repeated twice. Hydroxylapatite pellets were then resuspended in 200 μL ethanol (followed by another rinse with 200 L ethanol), and then the content was transferred to scintillation vials (containing 3 mL scintillation fluid) for measurement of 3H-radioactivity with a liquid scintillation counter (Packard Tri-CARB 2900 TR Downers Grove, IL).
The specific binding (pmol/mL) of the human ERα or ERβ protein at each concentration point was calculated using the following equation:
The IC50 value for each competing estrogen was calculated according to the four parameter sigmoidal inhibition curve in regression wizard of SigmaPlot software (all the curve regressions had a Rsqr higher than 0.95) by setting y value in the four parameter sigmoidal equation to 50. Relative binding affinity (RBA) was calculated using the following equation:
In vitro cell proliferation assay to determine the antiestrogenic activity in human breast cancer cells
The ER-positive MCF-7 and ER-negative MDA-MB-231 human breast cancer cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The culture conditions for these two cell lines were described in detail in our earlier study.24 Note that in these experiments, no additional estrogen was added to the cell culture medium, and all the estrogens came from the 10% FBS contained in the cell culture medium. The human breast cancer cells were first propagated in the 75 cm2 flasks to 80% confluence under 37°C air with 5% CO2 and 95% humidity to 80% confluence. They were then detached from the flask by treatment with 3 mL of the trypsin-EDTA solution for 5 min. Cell suspensions were centrifuged and the cell sediments were resuspended in phenol red-free culture medium containing 10% FBS at the desired density of 105 cells/mL. A 0.1 mL aliquot of the cell suspension was then added to each well of the 96-well microplates usually at a final density of 104 cells per well. After the cells were allowed to attach and grow for 24 h, the cell culture medium was changed and different drug treatments were introduced at that time. In most experiments, the drug treatment lasted for 6 days with one medium change on the fourth day following the initial drug treatment.
The cell density in each well was determined by using the crystal violet staining method24. Briefly the culture medium in the microplates was first removed by aspiration, and then the cells in each well were fixed with 1% glutaraldehyde for 15 min. After removing the fixation solution, each well was rinsed with PBS buffer and allowed to dry at room temperature. The cells in each well were then stained with 50 μL of 0.1% crystal violet (dissolved in 20% methanol and 80% deionized water) for 15 min at room temperature, and the plates were rinsed carefully with tap water to remove residual crystal violet. The stained dye was then dissolved in 100 μL of 0.5% Triton X-100 for two hours. After addition of 50 μL of 200-proof ethanol, the absorbance values of each well were measured at 560 and 405 nm with a UVmax microplate reader (Molecular Device, Palo Alto, CA), and the difference in the absorbance values at these two wavelengths were used to represent the cell density.
ERα trans-activation assays
The HeLa (human cervical cancer), HepG2 (human hepatoma), and MCF-7 cell lines employed for these experiments were maintained in DMEM supplemented with 10% FBS. For ERα trans-activation assays, cells were plated in a 6-well plate and grown overnight in phenol-red free DMEM containing 10% FBS, followed by transfection with the indicated plasmids using Lipofectamine-2000 (Invitrogen). Thereafter, cells were treated with vehicle or the indicated concentrations of the ERct ligands, and 20-24 h later cells were harvested for luciferase activity assay using a Luciferase Assay Kit (Promega) and a Luminoskan Ascent Thermo Labsystems apparatus (Thermo Electron, Milford, MA). Relative luciferase units were normalized to total protein content of the cell lysate measured by Bio-Rad protein assay (Bio-Rad, Hercules, CA).
ERα-coactivator interaction assays
Interactions between the ERα ligand binding domain (LBD) and either the SRC-1 or CBP coactivators were assessed by mammalian two-hybrid assay essentially as described previously16, 25. HeLa cells were transfected with 100 ng of the expression vectors for the GAL4 DNA binding domain fused to either SRC-1 (pBIND-SRC-1e) or the CBP CH3 domain (pBIND-CBP). Along with pBIND expression vectors, 1000 ng of the expression vector for VP16-ERα-LBD (pACT-hERα-LBD) and 1000 ng of the pG5-Luc reporter plasmid were used. Control experiments employed equivalent amounts of the pACT and pBIND empty vectors. Cells were treated with either 0.1% ethanolic vehicle or the indicated concentrations of ligands for 20-24 h prior to cell harvest and assay as described above.
Computational modeling
Docking study was performed with the InsightII modeling software (Version 2005, Accelrys Inc. San Diego, CA) installed in a Dell Precision 690 workstation with the Red Hat Enterprise Linux WS4.0 operating system (Red Hat Inc. Raleigh, NC). Energy minimization and molecular dynamics simulation were performed with the Discovery Studio modeling program (Version 1.7, Accelrys Inc. San Diego, CA) installed on the same computer. The CHARMm force field in DiscoveryStudio was used for molecular dynamics simulation.
Because the crystal structure of human ERα in complex with a full antagonist is presently not available, the crystallographic structure of ERα in complex with raloxifen (PDB code: 1ERR26), which is a partial agonist/antagonist, was used as a template for the docking study of ICI-182,780 and the newly-synthesized ER antagonists. The structures of ICI-182,780 and ER antagonists were first built using the Builder module and then minimized with the Discover module of InsightII. The docking method was described in our earlier study.14 In brief, the flexible docking was carried out using the Simulated Annealing Docking method in the Affinity module of InsightII. The binding pocket was defined to include all residues within the 7-Å reach of the original ligand raloxifen. A combination of Monte Carlo and simulated annealing methods was used to explore all possible conformations of the ligands. One hundred conformations were obtained and the one with the lowest potential binding energy was chosen for further minimization using the Standard Dynamics Cascade protocol in the Discovery Studio. The backbone of the protein and its key residues in the binding pocket (namely, E353, R394 and H524) were fixed during the docking procedures.
For energy minimizations, the steepest descent method was employed first to 10 kcal/(molÅ) root mean square (RMS) energy gradient and followed by the Polak and Ribiere conjugate gradient method until the final convergence criterion reached 0.01 kcal/(molÅ) RMS energy gradient. Then the whole system was heated from 100 to 300 K in 2 ps and equilibrated in 300 K for 100 ps. One hundred conformations were collected in 20 ps production phase at 300 K. The conformation with the lowest potential energy was further minimized and used for Binding Energy Calculation protocol. The backbone of the protein and its key residues in the binding pocket (namely, E353, R394 and H524) were constrained during the simulation process.
Figure 1. Structures of the newly-synthesized E2 derivatives.
This figure shows the structures of all nine E2 derivatives that were synthesized in this study. The short names are given for convenience. J1 or compound 14: 3,17β-bis(hydroxy)-7α-[6-(4-pyrrol-1-yl-phenyl)-hex-5-ynyl]-estra-1,3,5(10)-trien; J2 or compound 16: 3,17β-bis(hydroxy)-7α-(6-naphthalen-2-yl-hex-5-ynyl)-estra-1,3,5(10)-trien; J3 or compound 11: 3,17β-bis(hydroxy)-7α-[6-(4-Isoxazol-5-yl-phenyl)-hex-5-ynyl]-estra-1,3,5(10)-trien; J4 or compound 10: 4-{4-[6-((7α,17β)-3,17-dihydroxyestra-1,3,5(10)-trien-7-yl)-hex-1-ynyl]-phenyl}-piperazine-1-carboxylic acid tert-butyl ester; J5 or compound 12: 3-{4-[6-((7α,17β)-3,17-dihydroxyestra-1,3,5(10)-trien-7-yl)-hex-1-ynyl]-phenyl}-3-oxo-propionic acid ethyl ester; J6 or compound 15: 3,17β-bis(hydroxy)-7α-[6-(4-piperidin-1-ylmethyl-phenyl)-hex-5-ynyl]-estra-1,3,5(10)-trien; J7 or compound 17: 3-{4-[4-((7α,17β)-3,17-dihydroxyestra-1,3,5(10)-trien-7-yl)-butyl]-1,2,3-triazol-1 -yl}-7-hydroxy-1-benzopyran-2-one; J8 or compound 13: 2-{4-[6-((7α,17β)-3,17-dihydroxyestra-1,3,5(10)-trien-7-yl)-hex-1-ynyl]-benzyl}-malonic acid diethyl ester; J9 or compound 18: 3,17β-bis(hydroxy)-7α-{4-[1-(2,3-Dihydro-1,4-benzodioxin-2-ylmethyl)-1H-1,2,3-triazol-4-yl]-butyl}-estra-1,3,5(10)-trien.
Table 2. The computed binding energy values (ΔEbinding) for the binding of the E2 derivatives with human ERα.
The ligand-ERα interaction energy values (ΔEbinding) were calculated using the following equation: ΔEbinding = Ecomplex - (Ereceptor + Eligand), where Ecomplex is the potential energy for ERα in complex with a ligand, Ereceptor is the potential energy of ERα alone, and Eligand is the potential energy for the ligand alone.
| Ligand | Ecomplex | Eligand | Ereceptor | ΔEbinding |
|---|---|---|---|---|
| ICI-182,780 | −8924.06 | 103.58 | −8834.09 | −193.57 |
| J3 (compound 11) | −9575.35 | 42.78 | −9464.53 | −153.60 |
| J4 (compound 10) | −9630.81 | 26.26 | −9468.65 | −188.42 |
| J7 (compound 17) | −8020.15 | 84.23 | −7936.76 | −167.60 |
| J9 (compound 18) | −8924.53 | 88.85 | −8828.70 | −164.69 |
Acknowledgments
This study was supported by grants from the National Institutes of Health (Grant No. CA097109 and ES015242 to BTZ and DK53002 to CLS).
Abbreviations Used
- ER
estrogen receptor
- ERα and ERβ
the α and β subtype of ER, respectively
- E2
17β-estradiol
- RBA
relative binding affinity
- LBD
ligand binding domain
Footnotes
The authors declare no competing financial interest.
Contributor Information
Xiang-Rong Jiang, Department of Pharmacology, Toxicology and Therapeutics, University of Kansas Medical Center, Kansas City, KS 66160, USA.
Pan Wang, Department of Pharmacology, Toxicology and Therapeutics, University of Kansas Medical Center, Kansas City, KS 66160, USA.
Carolyn L. Smith, Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA
Bao Ting Zhu, Department of Pharmacology, Toxicology and Therapeutics, University of Kansas Medical Center, Kansas City, KS 66160, USA; Department of Biology, South University of Science and Technology of China, Shenzhen 518055, China.
References
- 1.Cancer Facts & Figures 2009 American Cancer Society. http://www.cancer.org/docroot/STT/content/STT_1x_Cancer_Facts_Figures_2009.asp?from=fast.
- 2.Jordan VC. Antiestrogens and selective estrogen receptor modulators as mutifunctional medicines. 1. Receptor interactions. J Med Chem. 2003;46:883–908. doi: 10.1021/jm020449y. [DOI] [PubMed] [Google Scholar]
- 3.Fisher B, Costantino JP, Wickerham DL, Cecchini RS, Cronin WM, Robidoux A, Bevers TB, Kavanah MT, Atkins JN, Margolese RG, Runowicz CD, James JM, Ford LG, Wolmark N. Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J Natl Cancer Inst. 2005;97:1652–1662. doi: 10.1093/jnci/dji372. [DOI] [PubMed] [Google Scholar]
- 4.(a) Howell A, Osborne CK, Morris C, Wakeling AE. ICI 182,780 (Faslodex™): Development of a novel, “pure” antiestrogen. Cancer. 2000;89:817–825. doi: 10.1002/1097-0142(20000815)89:4<817::aid-cncr14>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]; (b) Wakeling AE, Bowler J. Steroidal pure antioestrogens. J Endocrinol. 1987;112:R7–R10. doi: 10.1677/joe.0.112r007. [DOI] [PubMed] [Google Scholar]
- 5.Lin HR, Safo MK, Abraham DJ. Identification of a series of tetrahydroisoquinoline derivatives as potential therapeutic agents for breast cancer. Bioorg Med Chem Lett. 2007;17:2581–2589. doi: 10.1016/j.bmcl.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Hussey SL, He E, Peterson BR. Synthesis of chimeric 7α-substituted estradiol derivatives linked to cholesterol and cholesterylamine. Org Lett. 2002;4:415–418. doi: 10.1021/ol0171261. [DOI] [PubMed] [Google Scholar]
- 7.Marc BS, Frank RW, John AK. Synthesis and Binding Affinities of Novel Re-Containing 7α-Substituted Estradiol Complexes. Models for Breast Cancer Imaging Agents J Org Chem. 1999;64:8108–8121. doi: 10.1021/jo990641g. [DOI] [PubMed] [Google Scholar]
- 8.Termblay MR, Simard J, Poirier D. Parallel solid-phase synthesis of a model library of 7α-alkylamide estradiol derivatives as potential estrogen receptor antagonists. Bioorg Med Chem Lett. 1999;9:2827–2832. doi: 10.1016/s0960-894x(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 9.Tedesco R, Katzenellenbogen JA, Napolitano E. An expeditious route to 7α-substituted estradiol derivatives. Tetrahedron Lett. 1997;38:7997–8000. [Google Scholar]
- 10.French AN, Wilson SR, Welch MJ, Katzenellenbogen JA. A synthesis of 7α-subatituted estradiols: synthesis and biological evaluation of a 7α-pentyl-substituted BODIPY fluorescent conjugate and a fluorine-18-labeled 7α-pentylestradiol analog. Steroids. 1993;58:157–169. doi: 10.1016/0039-128x(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 11.Bowler J, Lilley TJ, Pittam JD, Wakeling AE. Novel steroidal pure antiestrogens. Steroids. 1989;54:71–99. doi: 10.1016/0039-128x(89)90076-7. [DOI] [PubMed] [Google Scholar]
- 12.Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- 13.Lippman ME, Bolan G. Oestrogen-responsive human breast cancer in long-term tissue culture. Nature. 1975;256:592–593. doi: 10.1038/256592a0. [DOI] [PubMed] [Google Scholar]
- 14.Lippman ME, Bolan G, Huff K. The effects of estrogens and antiestrogens on hormone-responsive human breast cancer in long-term tissue culture. Cancer Res. 1976;36:4595–4601. [PubMed] [Google Scholar]
- 15.Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/P300in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 16.Jaber BM, Gao T, Huang L, Karmakar S, Smith CL. The pure estrogen receptor antagonist ICI 182,780 promotes a novel interaction of estrogen receptor-α with the 3′,5′-cyclin adenosine monophosphate response element-binding protein-binding protein/p300 coactivators. Mol Endocrinol. 2006;20:2695–2710. doi: 10.1210/me.2005-0218. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman KL, Foster EA, Smith CL. The terminal substituents of 7α,6-hexanyl derivatives of estradiol determine their selective estrogen receptor modulator versus agonist activities. Steroids. 2012;77:496–503. doi: 10.1016/j.steroids.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu BT, Shim JY, Nagai M, Bai HW. Molecular modelling study of the mechanism of high-potency inhibition of human catechol-O-methyltransferase by (-)-epigallocatechin-3-O-gallate. Xenobiotica. 2008;38:130–146. doi: 10.1080/00498250701744641. [DOI] [PubMed] [Google Scholar]
- 19.Pike AC, Brzozowski AM, Walton J, Hubbard RE, Thorsell AG, Li YL. Structural insights into the mode of action of a pure antiestrogen. Structure. 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 20.Lee AJ, Cai MX, Thomas PE, Conney AH, Zhu BT. Characterization of the oxidative metabolites of 17β-estradiol and estrone formed by fifteen selectively-expressed human cytochrome P450 isoforms. Endocrinology. 2003;144:3382–3398. doi: 10.1210/en.2003-0192. [DOI] [PubMed] [Google Scholar]
- 21.Jiang XR, Wang P, Fu X, Zhu BT. Chemical synthesis and biochemical characterization of a biotinylated derivative of 17β-estradiol with a long side chain covalently attached to its C-7alpha position. Steroids. 2008;73:1252–1261. doi: 10.1016/j.steroids.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang XR, Sowell JW, Zhu BT. Synthesis of 7α-substituted derivatives of 17β-estradiol. Steroids. 2006;71:334–342. doi: 10.1016/j.steroids.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Krishnamoorthy S, Fang X, Brandom MC, Su L, Hannah NB, Qian W. A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 24.Wang P, Wen Y, Han GZ, Sidhu PK, Zhu BT. Characterization of the oestrogenic activity of non-aromatic steroids: are there male-specific endogenous oestrogen receptor modulators? Br J Pharmacol. 2009;158:1796–1807. doi: 10.1111/j.1476-5381.2009.00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaber B, Mukopadhyay R, Smith CL. Estrogen receptor-α interaction with the CREB bind protein coactivator is regulated by the cellular environment. J Mol Endocrinol. 2004;32:307–323. doi: 10.1677/jme.0.0320307. [DOI] [PubMed] [Google Scholar]
- 26.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1977;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]