Abstract
The Mycobacterium tuberculosis genome contains an unusually high number of toxin–antitoxin modules, some of which have been suggested to play a role in the establishment and maintenance of latent tuberculosis. Nine of these toxin–antitoxin loci belong to the mazEF family, encoding the intracellular toxin MazF and its antitoxin inhibitor MazE. Nearly every MazF ortholog recognizes a unique three- or five-base RNA sequence and cleaves mRNA. As a result, these toxins selectively target a subset of the transcriptome for degradation and are known as “mRNA interferases.” Here we demonstrate that a MazF family member from M. tuberculosis, MazF-mt6, has an additional role—inhibiting translation through targeted cleavage of 23S rRNA in the evolutionarily conserved helix/loop 70. We first determined that MazF-mt6 cleaves mRNA at 5′UU↓CCU3′ sequences. We then discovered that MazF-mt6 also cleaves M. tuberculosis 23S rRNA at a single UUCCU in the ribosomal A site that contacts tRNA and ribosome recycling factor. To gain further mechanistic insight, we demonstrated that MazF-mt6–mediated cleavage of rRNA can inhibit protein synthesis in the absence of mRNA cleavage. Finally, consistent with the position of 23S rRNA cleavage, MazF-mt6 destabilized 50S–30S ribosomal subunit association. Collectively, these results show that MazF toxins do not universally act as mRNA interferases, because MazF-mt6 inhibits protein synthesis by cleaving 23S rRNA in the ribosome active center.
Toxin–antitoxin (TA) systems have the potential to control Mycobacterium tuberculosis growth rate and persistence. One of the best characterized TA modules is mazEF in Escherichia coli (1–7), an autoregulated operon that encodes the intracellular toxin MazF and its antitoxin inhibitor MazE. Under unstressed conditions, the MazE protein forms a stable complex with MazF to neutralize its toxicity (1). During times of stress, however, proteases degrade MazE and allow the relatively stable MazF toxin to disrupt protein synthesis (1, 2), which can induce a state of reversible dormancy (3). Expression of MazF triggers this quasi-dormant state, during which cells stop dividing but are able to transcribe mRNA and synthesize proteins (4). The striking similarities between this state of quasi-dormancy and the slow-growing or nonreplicating state of M. tuberculosis during latent tuberculosis (TB) have led to the suggestion that TA modules are involved with persistence and dormancy in M. tuberculosis (8).
The genome of M. tuberculosis has >80 putative TA pairs (9), a remarkably large number relative to most other prokaryotes. Although TA modules are ubiquitous in bacteria and archaea, few prokaryotes have more than 15 loci (10). Although the physiological role of the large repertoire of TA loci in M. tuberculosis is largely unknown, some clues have emerged. First, there is an inverse correlation between the number of chromosomal TA loci and growth rate (10). Second, more than 60 TA toxins are conserved between five members of the M. tuberculosis complex but not in 13 other closely related mycobacteria (9), raising the possibility that the large number of TA systems in M. tuberculosis is specifically important for TB pathogenesis. Third, TA systems have intriguing connections to stress responses and persistence, both in E. coli (1, 2, 11, 12) and in M. tuberculosis (9, 13–23). Various TA loci in M. tuberculosis are induced during heat shock (23), hypoxia (9, 21), DNA damage (20), nutrient starvation (13), macrophage infection (9, 14, 18), and antibiotic treatment (19, 22), whereas other TA loci are down-regulated during macrophage infection (16) or salicylate treatment (15). Finally, 10 TA loci are induced in M. tuberculosis persister cells (17), whereas three RelE toxins increase M. tuberculosis persister recovery when expressed and two RelE toxins decrease persister recovery in response to specific drugs when deleted (22).
Nine of the TA systems in M. tuberculosis belong to the mazEF family. This is a large repertoire, because the majority of bacteria have only one or two mazEF operons (10). All of the MazF orthologs characterized from bacteria and archaea arrest growth by cleaving single-stranded RNA at specific three-, five-, or seven-base recognition sequences (6, 7, 24–29). Nearly every characterized MazF toxin, including each of the three in M. tuberculosis (28, 29), recognizes a unique RNA sequence. Toxins in the MazF family have also been labeled “mRNA interferases,” because they target single-stranded RNA but apparently do not cleave tRNAs or rRNAs (7, 25, 26, 30, 31). However, Moll, Engelberg-Kulka, and coworkers recently demonstrated that E. coli MazF cleaves 16S rRNA in vivo to form a subpopulation of “stress ribosomes” that selectively translate leaderless mRNAs (5). Although the cleavage specificity of three M. tuberculosis MazF toxins is known and potential physiological roles for MazF orthologs in other bacteria have been proposed, the mode of action and physiological roles of MazF toxins in M. tuberculosis are not fully understood.
As with other M. tuberculosis TA loci, mazEF-mt6 (“mt” refers to M. tuberculosis; locus Rv1103c-Rv1102c) seems to play a role in stress responses and persistence. The antitoxin gene mazE-mt6 is up-regulated during nutrient starvation (13), whereas expression of MazF-mt6 increases the level of persisters (32) and expression or deletion of mazF-mt6 alters persister recovery in a drug-specific manner in M. tuberculosis (22). To better understand the function of the mazEF family in M. tuberculosis, we studied the biochemical properties of the mazEF-mt6 TA module. We determined the precise MazF-mt6 recognition sequence, 5′UU↓CCU3′, and discovered that its mode of action includes cleavage of 23S rRNA at a highly conserved UUCCU site in M. tuberculosis. This activity serves as a potent disruptor of ribosome function and is unique among TA toxins characterized to date.
Results
MazF-mt6 Cleaves mRNA at the Five-Base Recognition Sequence UUCCU.
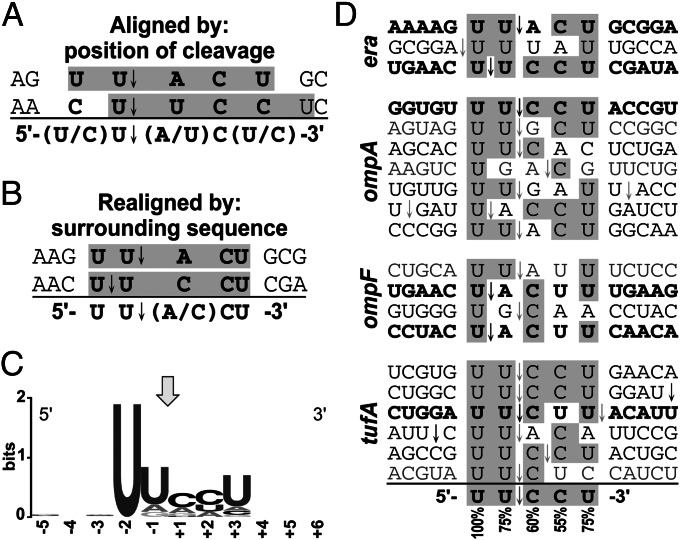
After limited analysis, Zhu et al. (29) reported the mRNA recognition sequence for the MazF-mt6 toxin as 5′(U/C)U↓(A/U)C(U/C)3′ or “U-rich” (Fig. 1A). Upon realignment of these published cleavage sites, we identified a putative five-base mRNA consensus of 5′UU↓(A/C)CU3′ (Fig. 1B). We then performed a more thorough primer extension analysis to identify 17 additional MazF-mt6 cleavage sites in vivo (Fig. S1 A–L), enabling us to pinpoint the predominant MazF-mt6 recognition sequence as 5′UU↓CCU3′ (Fig. 1 C and D).
Fig. 1.
MazF-mt6 cleaves mRNA at the five-base recognition sequence UUCCU. (A) Zhu et al. (29) determined the 5′(U/C)U↓(A/U)C(U/C)3′ recognition sequence by aligning the two in vivo sites at the point of cleavage. Gray boxes indicate the five-base sequences that we used to determine the realignment in Fig. 1B. Bold letters (A and B) or gray shading (D) indicate the consensus sequence (below the line) and nt in each cleavage site identical to the consensus. (B) A realignment of the cleavage sites reveals a more specific putative recognition sequence. (C) WebLogo (48) representation of the consensus sequence of mRNA sites cleaved by MazF-mt6 in E. coli. Numbering reflects the nt position relative to the point of cleavage, indicated by the arrow. (D) The mRNA recognition sequence for MazF-mt6 was deduced as 5′UU↓CCU3′ upon alignment of the 20 total sites. Percentages of the most abundant nucleotide indicated below each position. The three era cleavage sites were published previously (29). Bold text and black arrows indicate major mRNA cleavage sites, regular black text and gray arrows indicate minor cleavage sites, and gray text and arrows are rare sites.
Because this MazF family member has a five-base recognition sequence, its expression should not lead to wholesale mRNA cleavage as for the ACA-cleaving E. coli MazF toxin. Transcripts lacking a particular recognition sequence are stable in the presence of endoribonuclease toxins (24, 25, 27), and there is also a direct correlation between the number of potential cleavage sites within a given transcript and its susceptibility to degradation (24, 27). Therefore, we investigated the impact of MazF-mt6 expression on the M. tuberculosis transcriptome.
We performed this statistical analysis using the MazF-mt6 recognition sequence illustrated by the sequence logo shown in Fig. 1C. This sequence, UUMHU (where M = C or A and H = C, U, or A), includes UUCCU but also accounts for the degeneracy in the consensus at positions three and four (20% identity cutoff). First, upon analysis of all ORFs and noncoding RNAs in the M. tuberculosis genome, we determined that 31% lack the UUMHU cleavage motif and were predicted to be resistant to MazF-mt6 cleavage. Second, we analyzed RNAs predicted to be preferentially degraded by MazF-mt6. Among the top 20 RNAs that contain more UUMHU motifs than expected, nine are from the proline-glutamate (PE)/proline-proline-glutamate (PPE) family (Table S1). This large family of proteins contains conserved PE or PPE motifs at the N terminus and is only present in mycobacteria (33). Finally, of the top 20 RNAs with the highest actual number of MazF-mt6 cleavage motifs (Table S2), the first six encoded PE/PPE proteins.
MazF-mt6 Also Cleaves 23S rRNA at a Single UUCCU in the Ribosomal A site.
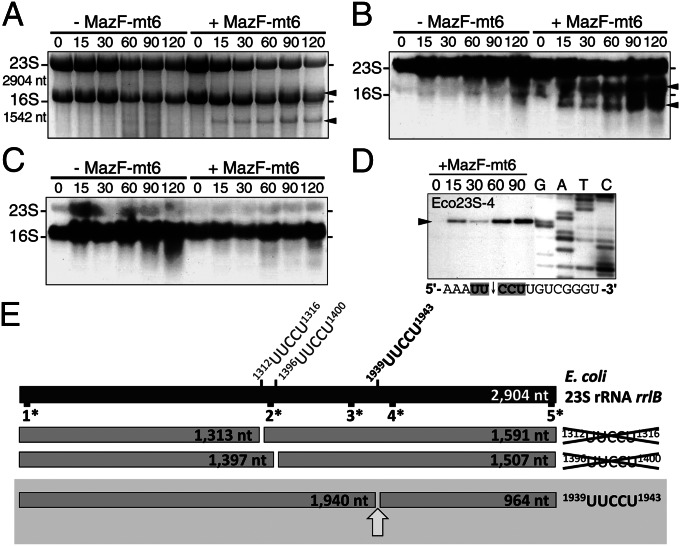
Expression of MazF-mt6 leads to growth arrest in E. coli (29, 32), Mycobacterium smegmatis (9, 32) (Fig. S2A), and M. tuberculosis (22) (Fig. S2B). Because all MazF toxins exhibit substantial mRNA cleavage activity, the resulting growth defects have generally been attributed to this property alone. However, upon checking the quality of total RNA from E. coli cells expressing MazF-mt6 by gel electrophoresis, we noted the appearance of two cleavage products, suggesting rRNA was also subject to MazF-mt6 cleavage (Fig. 2A). Although it seemed that 23S rRNA was the only rRNA target for cleavage because it decreased after MazF-mt6 induction (Fig. 2A), we used Northern analysis with radiolabeled DNA complementary to 23S or 16S rRNA to confirm the source (Fig. 2 B and C). We did not detect any cleavage products from 16S rRNA, nor did we see a reduction in 16S rRNA with time (Fig. 2C), consistent with the absence of UUCCU in the primary sequence of E. coli 16S rRNA. However, we observed a gradual accumulation of cleavage products from 23S rRNA (Fig. 2B).
Fig. 2.
MazF-mt6 also cleaves 23S rRNA at a single UUCCU in the ribosomal A site. Ethidium bromide staining (A) or Northern analysis (B and C) of total RNA from E. coli cells expressing MazF-mt6 (Right) for the times shown (in min), relative to the uninduced control (Left). The radiolabeled DNA fragments were complementary to E. coli 23S rRNA (B) or 16S rRNA (C). Black arrows highlight the position of cleavage products. (D) Primer extension analysis of E. coli 23S rRNA. Numbered lanes indicate time of MazF-mt6 induction (in min), whereas G, A, T, and C lanes denote DNA sequencing ladders. Oligonucleotide used for sequencing and primer extension reactions is in the top left corner, and the RNA sequence surrounding the cleavage site is listed below. (E) Scale schematic of E. coli 23S rRNA, indicating the positions of the three UUCCUs and potential cleavage products. Top bar, 23S rRNA. 1*-5* represent the positions of the radioactive oligonucleotides used for individual Northern blots (Fig. S3) to estimate the location of the MazF-mt6 cleavage site. The three lower rows of bars represent the possible cleavage products for each UUCCU. Our Northern data coincided with the cleavage products represented in the bottom row of bars boxed in gray, with the cleavage site shown by an arrow.
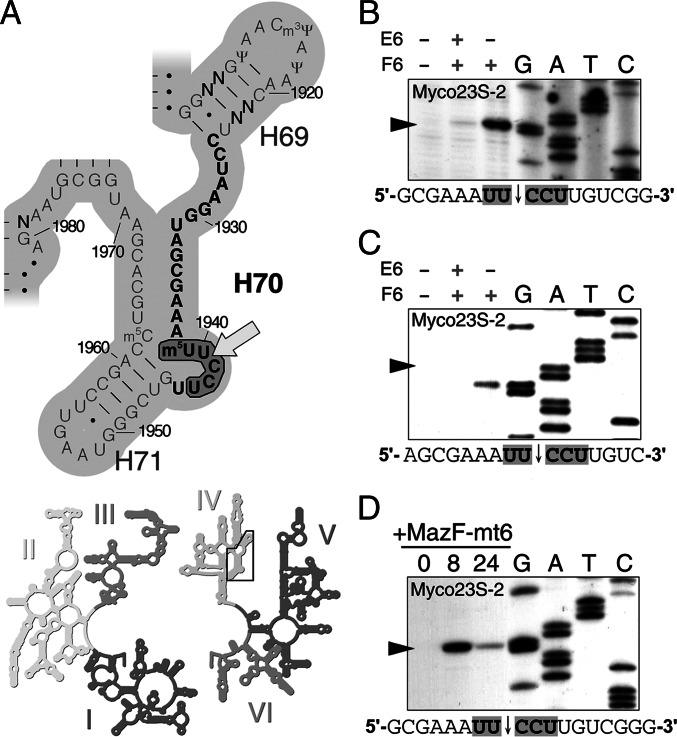
Examination of the primary sequence revealed that there are three potential MazF-mt6 cleavage sites in 23S rRNA (Fig. 2E, above the top bar). We designed five oligonucleotides between each UUCCU site in 23S rRNA to detect potential cleavage fragments and performed Northern analysis (Fig. S3) to compare the relative sizes of cleavage products. Of the three potential outcomes for a single cleavage event (Fig. 2E, three lower rows of bars), the observed pattern of cleavage implicated 1939UUCCU1943 as the lone target of MazF-mt6. Upon analysis of the secondary structure of E. coli 23S rRNA using the Comparative RNA Web Site (www.rna.icmb.utexas.edu/), 1939UUCCU1943 is the only MazF-mt6 recognition sequence that maps to a single-stranded region, in agreement with the requirements of MazF family members that (i) the substrate be single-stranded RNA and (ii) contain the correct recognition sequence for cleavage (6, 7). We then used primer extension analysis to map the precise position of 23S rRNA cleavage (Fig. 2D), which confirmed that the cleavage site coincides with our predicted site, 1939UUCCU1943, in helix 70 (H70, also referred to as helix/loop 70 or the “1942 loop” because this region only has a single Watson–Crick base pair) of domain IV (Fig. 3A). Interestingly, residues 1939–1943 are surface-accessible and have several vital roles in translation, including the following: (i) interaction with tRNA in the A site (34–37), (ii) stabilization of ribosome recycling factor (RRF) binding and function (38, 39), and (iii) 50S-30S subunit association (40–42).
Fig. 3.
MazF-mt6 cuts M. smegmatis and M. tuberculosis 23S rRNA at the same conserved UUCCU. (A) Secondary structure of a conserved region of domain IV (Inset) in 23S rRNA (Lower). Helix/loop 70 (H70), whose nucleotides are in bold, is 100% identical between E. coli, M. smegmatis, and M. tuberculosis. Nonconserved residues are labeled N, E. coli position number adjacent to tick marks, MazF-mt6 recognition sequence encircled by a bold black line, and the lone MazF-mt6 cleavage site indicated by an arrow. Structure adapted from E. coli 23S rRNA courtesy of the University of California, Santa Cruz (http://rna.ucsc.edu/rnacenter/ribosome_images.html). (B–D) Primer extension analysis of M. smegmatis (B) or M. tuberculosis (C) 23S rRNA incubated with MazF-mt6 in vitro or of M. smegmatis 23S rRNA upon MazF-mt6 expression in vivo (D). Labeling is similar to that in Fig. 2D. M. smegmatis (B) or M. tuberculosis (C) total RNA was incubated with or without MazF-mt6 (F6) or MazE-mt6 (E6) as indicated above each box on the left. (D) Time of MazF-mt6 induction (in h) in M. smegmatis shown above left.
MazF-mt6 Cuts M. smegmatis and M. tuberculosis 23S rRNA at the Same Conserved UUCCU.
We compared the sequence and structure of E. coli 23S rRNA with that in M. smegmatis and M. tuberculosis. Overall, they are highly conserved, and the sequence containing the 1939UUCCU1943 cleavage site, helix/loop 70, is 100% identical (Fig. 3A, labeled H70 and with nt in bold). Incubation of increasing concentrations of recombinant MazF-mt6 with M. smegmatis total RNA (Fig. S4A) generated a cleavage pattern similar to that seen in RNA from E. coli cells expressing MazF-mt6 (Fig. 2A). The minor size differences of the cleavage products are in agreement with the ∼200-nt longer M. smegmatis 23S rRNA. Comparison of these results with the two possible M. smegmatis 23S rRNA cleavage patterns (Fig. S4D, two lower rows of light blue bars) implicated 2166UUCCU2170 in helix/loop 70 as the single MazF-mt6 cleavage site. Indeed, primer extension analysis confirmed this (Fig. 3B).
We then tested the effect of MazF-mt6 on M. tuberculosis RNA. Of the three UUCCU sites that are potential MazF-mt6 targets in M. tuberculosis 23S rRNA (Fig. S4E), primer extension analysis (Fig. 3C) confirmed that position 2177UUCCU2181 in helix/loop 70—analogous to the cleavage positions in E. coli and in M. smegmatis—was the single MazF-mt6 cleavage site (Fig. S4E, yellow arrow).
We then examined the effect of ectopic MazF-mt6 expression on rRNA in the mycobacterial model organism M. smegmatis wild-type strain mc2155. We did not detect cleavage products even though MazF-mt6 induction resulted in a decrease of 23S rRNA after 24 h. To reconcile the discrepancies between the cleavage patterns resulting from MazF-mt6 induction in E. coli vs. in M. smegmatis, we checked for differences in RNases involved in RNA turnover. Notably, although E. coli seems to lack a 5′-to-3′ exonuclease, M. smegmatis has at least one, called RNase J (rnj, locus MSMEG_2685), which is also present in M. tuberculosis (locus Rv2752c) (43). Consequently, we hypothesized that MazF-mt6 cleavage fragments that are generally stable in E. coli might be degraded rapidly in mycobacteria. To test this, we obtained an mc2155 derivative with a deletion of the rnj gene and repeated the MazF-mt6 induction in this strain background. We were then able to resolve cleavage products with a migration pattern similar to those previously seen in E. coli, M. smegmatis, and M. tuberculosis. Using Northern (Fig. S4 B and C) and primer extension analyses (Fig. 3D), we unequivocally determined that MazF-mt6 cleaved the same single 2166UUCCU2170 site in M. smegmatis 23S rRNA (Fig. S4D, yellow arrow) as that obtained in vitro with M. smegmatis total RNA and analogous to the 1939UUCCU1943 position in E. coli and the 2177UUCCU2181 position in M. tuberculosis.
MazF-mt6 Disables Translation by Targeting 23S rRNA in 50S Ribosomal Subunits.
As previously mentioned, the 23S rRNA cleavage site for MazF-mt6 maps to the ribosomal A site and is functionally associated with many essential features of protein synthesis. Therefore, any perturbation at this site is expected to have dire consequences for the translation machinery. To assay for overall ribosome function, we modified a standard cell-free transcription/translation system composed of purified ribosomes and all necessary translation factors.
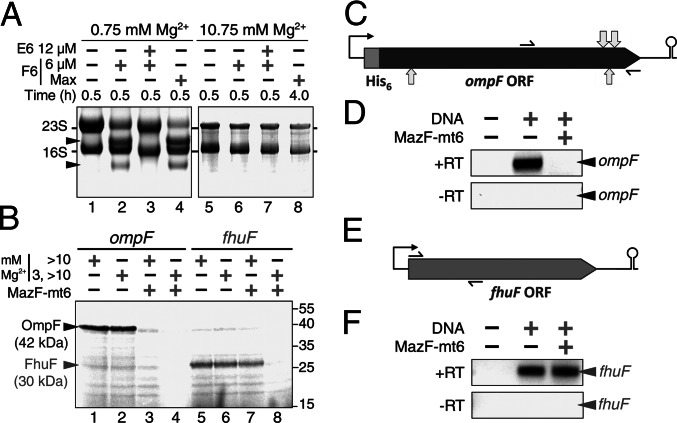
We first enlisted this system to determine whether MazF-mt6 can cleave 23S rRNA in a fully assembled ribosome. The ribosome fraction is separate from other essential translation factors, which enabled us to manipulate ribosome association and dissociation. The 1939UUCCU1943 site in E. coli 23S rRNA overlaps nucleotides 1940–1944, which are protected from Pb(II)-induced cleavage in the intact ribosome but not the 50S subunit (42). Therefore, having first demonstrated that MazF-mt6 is active at high and low Mg2+ concentrations (Fig. S5), we then tested the 23S rRNA cleavage activity of MazF-mt6 on dissociated ribosomes (0.75 mM Mg2+) vs. intact ribosomes (10.75 mM Mg2+). MazF-mt6 only cleaved 23S rRNA when the 50S and 30S ribosomal subunits were dissociated (Fig. 4A, lanes 2 and 4), and the cleavage was toxin-specific because it was inhibited by preincubation of the toxin with its cognate antitoxin (lane 3). No cleavage occurred when MazF-mt6 was incubated with intact ribosomes (Fig. 4A, lanes 6 and 8), even upon a concomitant increase in incubation time and in the molar ratio of MazF-mt6 to ribosomes (lane 8) to compensate for the lower MazF-mt6 cleavage efficiency at 10.75 mM Mg2+ (Fig. S5).
Fig. 4.
MazF-mt6 disables translation by targeting 23S rRNA in 50S ribosomal subunits. (A) 70S ribosomes from a cell-free transcription/translation system were either completely dissociated into 50S and 30S subunits (lanes 1–4) or kept intact (lanes 5–8). Reactions were incubated with or without the indicated concentration of MazF-mt6 (F6) or MazE-mt6 (E6). Max indicates 12 μM (lane 4) or 24 μM (lane 8) MazF-mt6 incubated for the times indicated. (B) MazF-mt6 not only inhibits in vitro translation of OmpF (lanes 3 and 4), whose transcript is cleaved by MazF-mt6 in vivo but also inhibits translation of the FhuF protein (lane 8), whose transcript does not contain any MazF-mt6 recognition sequences. Molecular mass in kDa noted by tick marks on the right. (C and E) Scale schematics of transcripts encoded by the pET-28a-ompF (C) and pET-21c-fhuF (E) plasmids used in the transcription/translation system. Thin black arrows correspond to primers used for RT-PCR. (C) Light gray arrows above the ompF ORF show the two major sites cleaved by MazF-mt6 in vivo, whereas arrows below the ORF show the two minor cleavage sites (Fig. S1 G and H). (D and F) RT-PCR of RNA extracted from the transcription/translation system. The mRNA encoding the His6-OmpF protein was cleaved by MazF-mt6 (D), whereas the mRNA encoding FhuF was not (F). Data shown are representative of three independent experiments.
Having determined the conditions required for MazF-mt6 activity on the ribosome, we then tested whether MazF-mt6 cleavage of 23S rRNA affects translation. Using the transcription/translation system, we dissociated the ribosome fraction at 3 mM Mg2+, added MazF-mt6, and adjusted to 13 mM Mg2+ to facilitate ribosome reassociation for translation. Because of the transcriptional and translational requirements of 13 mM Mg2+ and a limited discretionary reaction volume, Mg2+ could only be diluted to 3.0 mM, dissociating ∼70% of 70S monosomes (44) before the transcription step. However, a dynamic equilibrium exists between associated 70S ribosomes and dissociated subunits (45), enabling us to compensate with both a fourfold excess of MazF-mt6 and a relatively long incubation time (60 min). We then added one of two custom templates to compare the effect of MazF-mt6 on protein synthesis with or without concomitant mRNA cleavage. One is a template for ompF (outer membrane porin F) mRNA, which is cleaved by MazF-mt6 at four sites in vivo (Fig. S1 G and H, positions illustrated by light gray arrows in Fig. 4C). The other is a template for fhuF, which has no MazF-mt6 cleavage recognition sequences (Fig. 4E). Therefore, MazF-mt6-mediated inhibition of translation from the ompF template should be the result of both mRNA and 23S rRNA cleavage, whereas translation inhibition from the fhuF transcript should result solely from cleavage of 23S rRNA.
We first measured the effect of MazF-mt6 on translation using the ompF template. In the absence of toxin, the 42-kDa OmpF protein was produced using the recommended Mg2+ concentration (Fig. 4B, lane 1) and with exposure of the ribosome to two Mg2+ concentrations to enable ribosome dissociation and reassociation (lane 2). However, no OmpF product was detected upon addition of MazF-mt6 when ribosomes were initially dissociated (Fig. 4B, lane 4), and translation was significantly inhibited with intact ribosomes (lane 3). Using RT-PCR, we measured the level of ompF transcript under the same reaction conditions as in Fig. 4B, lanes 2 and 4, except [35S]-Met was not added. No ompF mRNA was detected in the sample exposed to MazF-mt6 (Fig. 4D), indicating MazF-mt6 cleaves this mRNA as expected. Therefore, the cleavage activity of MazF-mt6 on both targets—23S rRNA in the ribosome and mRNA (Fig. 4B, lane 4)—results in translation inhibition that is more potent than inhibition resulting from MazF-mt6-mediated cleavage of only the ompF transcript (lane 3).
Next, we repeated the same series of experiments using the fhuF transcript. This template produced the 30-kDa FhuF protein at the standard Mg2+ concentration (Fig. 4B, lane 5) or upon Mg2+-mediated ribosome dissociation and reassociation (lane 6). As with the ompF template, preincubation of MazF-mt6 with dissociated ribosomes resulted in total inhibition of translation (lane 8). However, unlike the results with ompF, inhibition of FhuF protein synthesis occurred even when the fhuF transcript was not cleaved (Fig. 4F) and did not occur when the ribosomes were kept intact (Fig. 4B, lane 7). Therefore, MazF-mt6–mediated cleavage of 23S rRNA results in potent inhibition of protein synthesis.
MazF-mt6 Expression Inhibits Association of Ribosomal Subunits.
The 23S rRNA cleavage site for MazF-mt6 is in helix/loop 70, identical between E. coli, M. smegmatis, and M. tuberculosis. Domain IV, comprising helices 61 through 71 (Fig. 3A), is a flat surface that forms most of the interface between the 50S and 30S subunits (40). Consistent with their position at the subunit interface, helices 67 through 71 constitute a large region of rRNA devoid of contact with ribosomal proteins. These structural features are in agreement with two functional studies that identified a link between the 1939UUCCU1943 cleavage site and stable subunit association. First, mutation U1940A results in 50S subunits defective for 50S–30S ribosomal subunit association (41). Second, U1940, C1491, C1492, and U1943 are protected from Pb(II)-induced cleavage during 50S–30S subunit association (42).
Because helix/loop 70 containing the MazF-mt6 cleavage site is important for subunit association, we determined the effect of MazF-mt6 expression on the abundance of dissociated 50S and 30S subunits relative to the 70S monosome. We expressed MazF-mt6 in E. coli for up to 120 min, prepared S30 extracts, and performed polysome profile analysis (Fig. 5). We observed a steady increase in the percentage of free 50S and 30S subunits (shown in bold for each panel in Fig. 5) after MazF-mt6 induction, from ∼25% in the 0-h induced time point and uninduced controls (Fig. 5 A–C) to 42% at 60 min after induction (Fig. 5E) and 56% at 120 min after induction (Fig. 5G). Therefore, another functional consequence of MazF-mt6 activity was the progressive loss of assembled ribosomes in vivo.
Fig. 5.
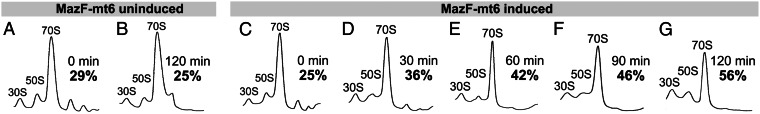
MazF-mt6 expression prevents association of ribosomal subunits. (A–G) Ribosome profiles of BW25113Δ6 cells harboring pBAD33-mazF-mt6, either without induction of MazF-mt6 at 0 (A) and 120 min (B) or expressing MazF-mt6 at 0 (C), 30 (D), 60 (E), 90 (F), and 120 min (G) after induction. The time in regular black text and the percent of ribosomes dissociated in bold text are indicated on the right side of each profile. The profiles are representative of three independent experiments.
Discussion
Given the large body of data demonstrating that MazF toxin family members cleave mRNA, Inouye and colleagues coined the term “mRNA interferases” to describe them (6, 25–29), and this term is now almost universally accepted. However, we demonstrated that the MazF-mt6 toxin from M. tuberculosis also directly targets 23S rRNA for cleavage at an evolutionarily conserved single UUCCU site in M. tuberculosis. An in vitro translation assay revealed that MazF-mt6 required ribosomal dissociation for activity on rRNA, which enabled us to dissect the functional consequences of combined mRNA and 23S rRNA cleavage by MazF-mt6 vs. 23S rRNA cleavage alone. We observed complete translation inhibition in both cases, indicating that MazF-mt6–mediated cleavage of only 23S rRNA was capable of disabling protein synthesis. This result is in agreement with the critical function of the target region, helix/loop 70 of domain IV, which facilitates tRNA binding in the ribosomal A site.
The vital role of the region in E. coli 23S rRNA cleaved by MazF-mt6, 1939UUCCU1943, is well established and consistent with the growth and translation phenotypes we observed. Indeed, a U1940A mutation displays defects in translation associated with the function of helix/loop 70, yielding 50S ribosomal subunits defective in assembly (41). Moreover, the functional significance of the 1939UUCCU1943 site in helix/loop 70 was illuminated in several structural studies. First, the X-ray crystal structure of a 50S subunit revealed that helices 67–71 form the front rim of the peptidyl transferase active site cleft (40). Second, the crystal structure of the 70S ribosome bound to mRNA in the presence of tRNAs revealed that helix/loop 70 positions the CCA tail of the tRNA acceptor and that the C1942 and U1943 residues of the MazF-mt6 consensus sequence contact the acceptor stem of tRNA (37). Third, crystallographic studies enlisting tRNA mimics showed that C1942 and U1943 contact the tRNA acceptor stem mimic and that C1941 contacts the tRNA CCA tail mimic (34). Fourth, this region also directly interacts with RRF, because C1941 and C1942 form hydrogen bonds to distinct amino acids of RRF (38). Higher resolution X-ray crystal structures of the 50S subunit with a domain of RRF also uncovered an interaction between U1941 and a highly conserved amino acid in RRF that is important for 50S binding and function (39). Finally, the X-ray structure of a 23S rRNA fragment suggested that the helix/loop 70 has the potential to base pair with tmRNA (46).
Interestingly, the eukaryotic counterpart of 23S rRNA is a target for several deadly bacterial toxins. Ricin, saporin, Shiga toxin, and pokeweed antiviral toxin all separate A4324 from the phosphodiester backbone (i.e., depurinate) in 28S rRNA. In every case, this cleavage occurs in intact ribosomes, in contrast to MazF-mt6, which only cleaves rRNA in the free 50S subunit. There are also several antibiotics—sparsomycin, clindamycin, chloramphenicol, linezolid, the pleuromutilins, and the macrolides (e.g., erythromycin)—that bind to 23S rRNA in intact E. coli 50S subunits. Chloramphenicol specifically binds to A2451 and A2452 residues in the 23S rRNA of the 50S subunit. Therefore, there is ample precedent for potent translation inhibition through targeted disruption of the rRNA of the large ribosomal subunit.
The physiological activities of MazF-mt6 and other family members in M. tuberculosis were thought to exclusively stem from their mRNA cleavage activity (28, 29). Consequently, the characterized M. tuberculosis MazF toxins with five-base recognition sequences were proposed to function in selective editing of the transcriptome to alter protein expression (28). Our data revealed that this is not the complete extent of MazF-mt6 activity, because cleavage at a functionally essential and evolutionarily conserved region of 23S rRNA led to a block in translation. Among the MazF orthologs that have been studied in E. coli, M. tuberculosis, and other bacteria, only MazF-mt6 exhibits this unique activity. In E. coli, 23S and 16S rRNA are stable upon MazF induction at all intervals reported up to 30 min, despite the presence of numerous target ACA sequences in those rRNAs (7, 30). Additionally, preincubation of MazF with components of an E. coli cell-free transcription/translation system prior to transcription does not inhibit protein synthesis, suggesting MazF either does not cleave rRNA and tRNA or that their cleavage does not impair translation (7). Finally, during MazF expression, the translation machinery supports robust and sustained protein production from an expression plasmid containing a gene devoid of ACA sequences (4). As for MazF orthologs in other bacteria or on plasmids, Staphylococcus aureus MazF-sa expression does not alter the abundance of 23S or 16S rRNA when expressed endogenously (31), nor does expression of plasmid-borne toxin PemK (26) or archaeal Haloquadra walsbyi toxin MazF-hw (25) in E. coli.
In addition to its comprehensive mRNA cleavage activity at ACA sequences, E. coli MazF can cleave 16S rRNA in vivo at a single site 43 bases from the 3′ end at 1500ACA1502 (5). Interestingly, this modification is not lethal but seems to modify the specificity of a subpopulation of ribosomes to selectively translate leaderless mRNAs. Although 16S rRNA contains other ACA sequences, they are not cleaved in vivo because they are in double-stranded regions or inaccessible to MazF. Our data, as well as that of Vesper et al., demonstrate that MazF toxins do not possess inherent specificity for exclusive mRNA cleavage. Instead, the primary determinants for MazF substrate specificity are RNA sequence, single-strandedness, and accessibility of the RNA.
The evolution of this MazF family member into a toxin that selectively targets both mRNA and ribosomes is intriguing. Because 23S rRNA cleavage alone can arrest translation, the significance of the dual activities and the consequences of their interplay are unclear. Our statistical analyses identified several PE/PPE proteins with a higher than expected number of MazF-mt6 cleavage sites. Unfortunately, the precise roles of the PE/PPE transcripts predicted to be preferential targets by MazF-mt6 are unknown. PE/PPE proteins are either secreted or localized near the cell membrane and are involved in pathogenesis and virulence (33). Approximately half of the PE/PPE family members—those in the PE-PGRS (polymorphic GC-rich-repetitive sequence) and PPE-MPTR (major polymorphic tandem repeat) subfamilies—have characteristic (GGAGGN)n or GN-rich amino acid repeats, respectively. As a consequence, our statistical predictions would be biased if the consensus sequence consistently appeared within a repetitive region. However, for 11 of the top 12 MazF-mt6–susceptible PE/PPE genes, with the exception of PPE34, the distribution of MazF-mt6 cleavage motifs was random. In fact, we found that the MazF-mt6 UUMHU recognition sequence cannot appear in the most abundant PE-PGRS or PPE-MPTR repetitive regions, because adjacent glycine, alanine, and asparagine codons do not contain consecutive U nucleotides. Therefore, the abundance of PE/PPE family members susceptible to MazF-mt6 cleavage was not merely due to motif overrepresentation within repetitive regions.
MazF-mt6 seems to exclusively cleave 23S rRNAs in free 50S ribosomal subunits but not those assembled into 70S ribosomes. Therefore, simultaneous targeting of a subset of mRNAs for cleavage may facilitate the rapid release of ribosomal subunits that are then inactivated. This “one-two punch” by the MazF-mt6 enzyme would enable a faster physiological response—by reducing the level of ribosomes below the threshold required for active growth—than by mRNA cleavage alone. In fact, a recent study forged a connection between cleavage of 23S rRNA at helix/loop 70 and a slowdown in cell growth. Deutscher and colleagues (47) demonstrated that an unknown RNase cleaves 23S rRNA in E. coli cells during glucose starvation and upon quality control during steady-state growth. Interestingly, the cleavage occurs within the same UUCCU sequence cleaved by MazF-mt6. Their study provides clues as to how cleavage of 23S rRNA by MazF-mt6 may affect M. tuberculosis physiology and is consistent with the hypothesis that TA toxins in M. tuberculosis contribute to the establishment or maintenance of the nonreplicating persistent state characteristic of latent TB.
Materials and Methods
All bacterial strains and plasmids are listed in Table S3, and oligonucleotides are listed in Table S4. The MazF-mt6 cleavage specificity and its cleavage of 23S rRNA were determined by Northern and primer extension analyses. The effect of MazF-mt6–mediated cleavage of 23S rRNA in ribosomal subunits on ribosome association was determined by polysome profile analysis, and the effect on translation was determined using transcripts with or without MazF-mt6 recognition sequences in a cell-free transcription/translation system. A full description of methods is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ling Zhu for providing pET-21c-mazF-mt6 and pBAD33-mazF-mt6; Jonathan W. Cruz for providing pET-28a-ompF; Jonathan W. Cruz and Bryce E. Nickels for critical reading of the manuscript; Chang-Chih Wu and Estela Jacinto for use of their density gradient fractionation system; and Harald Putzer for providing the M. smegmatis deletion strain rnj102. This work was supported in part by National Institutes of Health Grant R21 AI072399 from the National Institute of Allergy and Infectious Diseases (NIAID) (to R.N.H. and N.A.W.) and National Institutes of Health Training Grant 5T32AI007403-18, Virus-Host Interactions in Eukaryotic Cells, from the NIAID (to J.M.S. and J.D.S., awarded to G. Brewer).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222031110/-/DCSupplemental.
References
- 1.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: A model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93(12):6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332(4):809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 3.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45(2):501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Mol Cell. 2005;18(2):253–261. doi: 10.1016/j.molcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Vesper O, et al. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147(1):147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280(5):3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, et al. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12(4):913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 8.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: Success through dormancy. FEMS Microbiol Rev. 2012;36(3):514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: Implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5(12):e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33(3):966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7(6):359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43(3):717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 14.Cappelli G, et al. Profiling of Mycobacterium tuberculosis gene expression during human macrophage infection: Upregulation of the alternative sigma factor G, a group of transcriptional regulators, and proteins with unknown function. Res Microbiol. 2006;157(5):445–455. doi: 10.1016/j.resmic.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Denkin S, Byrne S, Jie C, Zhang Y. Gene expression profiling analysis of Mycobacterium tuberculosis genes in response to salicylate. Arch Microbiol. 2005;184(3):152–157. doi: 10.1007/s00203-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 16.Fontán P, Aris V, Ghanny S, Soteropoulos P, Smith I. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect Immun. 2008;76(2):717–725. doi: 10.1128/IAI.00974-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio. 2011;2(3):e00100–e00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korch SB, Contreras H, Clark-Curtiss JE. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J Bacteriol. 2009;191(5):1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Provvedi R, Boldrin F, Falciani F, Palù G, Manganelli R. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology. 2009;155(Pt 4):1093–1102. doi: 10.1099/mic.0.024802-0. [DOI] [PubMed] [Google Scholar]
- 20.Rand L, et al. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol Microbiol. 2003;50(3):1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 21.Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PloS One. 2008;3(1):e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh R, Barry CE, 3rd, Boshoff HI. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J Bacteriol. 2010;192(5):1279–1291. doi: 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart GR, et al. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148(Pt 10):3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 24.Rothenbacher FP, et al. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J Bacteriol. 2012;194(13):3464–3474. doi: 10.1128/JB.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi Y, Nariya H, Park JH, Inouye M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat Commun. 2012;3:607. doi: 10.1038/ncomms1621. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Zhang Y, Zhu L, Suzuki M, Inouye M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem. 2004;279(20):20678–20684. doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, et al. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J Bacteriol. 2009;191(10):3248–3255. doi: 10.1128/JB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu L, et al. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol Microbiol. 2008;69(3):559–569. doi: 10.1111/j.1365-2958.2008.06284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu L, et al. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem. 2006;281(27):18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 30.Baik S, Inoue K, Ouyang M, Inouye M. Significant bias against the ACA triplet in the tmRNA sequence of Escherichia coli K-12. J Bacteriol. 2009;191(19):6157–6166. doi: 10.1128/JB.00699-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Z, Tamber S, Memmi G, Donegan NP, Cheung AL. Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J Bacteriol. 2009;191(7):2051–2059. doi: 10.1128/JB.00907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han JS, et al. Characterization of a chromosomal toxin-antitoxin, Rv1102c-Rv1103c system in Mycobacterium tuberculosis. Biochem Biophys Res Commun. 2010;400(3):293–298. doi: 10.1016/j.bbrc.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Gey van Pittius NC, et al. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11(1):91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 35.Knutsson Jenvert RM, Holmberg Schiavone L. Characterization of the tRNA and ribosome-dependent pppGpp-synthesis by recombinant stringent factor from Escherichia coli. FEBS J. 2005;272(3):685–695. doi: 10.1111/j.1742-4658.2004.04502.x. [DOI] [PubMed] [Google Scholar]
- 36.Noller HF. RNA structure: Reading the ribosome. Science. 2005;309(5740):1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 37.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 38.Agrawal RK, et al. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci USA. 2004;101(24):8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson DN, et al. X-ray crystallography study on ribosome recycling: The mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24(2):251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 41.Leviev I, Levieva S, Garrett RA. Role for the highly conserved region of domain IV of 23S-like rRNA in subunit-subunit interactions at the peptidyl transferase centre. Nucleic Acids Res. 1995;23(9):1512–1517. doi: 10.1093/nar/23.9.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merryman C, Moazed D, Daubresse G, Noller HF. Nucleotides in 23S rRNA protected by the association of 30S and 50S ribosomal subunits. J Mol Biol. 1999;285(1):107–113. doi: 10.1006/jmbi.1998.2243. [DOI] [PubMed] [Google Scholar]
- 43.Taverniti V, Forti F, Ghisotti D, Putzer H. Mycobacterium smegmatis RNase J is a 5′-3′ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol Microbiol. 2011;82(5):1260–1276. doi: 10.1111/j.1365-2958.2011.07888.x. [DOI] [PubMed] [Google Scholar]
- 44.van Duin J, van Dieijen G, van Knippenberg PH, Bosch L. Different species of 70S ribosomes of Escherichia coli and their dissociation into subunits. Eur J Biochem. 1970;17(3):433–440. doi: 10.1111/j.1432-1033.1970.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 45.Ball LA, Johnson PM, Walker IO. The dissociation of Escherichia coli ribosomes. Eur J Biochem. 1973;37(1):12–20. doi: 10.1111/j.1432-1033.1973.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120(5):599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 47.Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: Comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17(2):338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.