Abstract
Neuropeptides are signaling molecules that commonly act via G protein-coupled receptors (GPCRs) and are generated in neurons by proneuropeptide (pNP) cleavage. Present in both cnidarians and bilaterians, neuropeptides represent an ancient and widespread mode of neuronal communication. Due to the inherent difficulties of analyzing highly diverse and repetitive pNPs, the relationships among different families are often elusive. Using similarity-based clustering and sensitive similarity searches, I obtained a global view of metazoan pNP diversity and evolution. Clustering revealed a large and diffuse network of sequences connected by significant sequence similarity encompassing one-quarter of all families. pNPs belonging to this cluster were also identified in the early-branching neuronless animal Trichoplax adhaerens. Clustering of neuropeptide GPCRs identified several orthology groups and allowed the reconstruction of the phyletic distribution of receptor families. GPCR phyletic distribution closely paralleled that of pNPs, indicating extensive conservation and long-term coevolution of receptor–ligand pairs. Receptor orthology and intermediate sequences also revealed the homology of pNPs so far considered unrelated, including allatotropin and orexin. These findings, together with the identification of deuterostome achatin and luqin and protostome opioid pNPs, extended the neuropeptide complement of the urbilaterian. Several pNPs were also identified from the hemichordate Saccoglossus kowalevskii and the cephalochordate Branchiostoma floridae, elucidating pNP evolution in deuterostomes. Receptor–ligand conservation also allowed ligand predictions for many uncharacterized GPCRs from nonmodel species. The reconstruction of the neuropeptide-signaling repertoire at deep nodes of the animal phylogeny allowed the formulation of a testable scenario of the evolution of animal neuroendocrine systems.
Keywords: Platynereis, proenkephalin, Petromyzon
Neuropeptides are diverse neuron-secreted peptides with neuromodulatory, neurotransmitter, or hormonal functions. Most neuropeptides signal via G protein-coupled receptors (GPCRs) (1), with a few exceptions (2–8). As modulators of neuronal activity, neuropeptides contribute to the generation of different outputs from the same neuronal circuit in a context-dependent manner (9), or orchestrate complex motor programs (10). Many neuropeptides act as hormones and are released into the haemolymph by neurohemal organs, such as the vertebrate pituitary gland, or the insect corpora cardiaca (11). These peptide hormones regulate various aspects of physiology, including growth, metabolism, and reproduction. Active neuropeptides are generated from an inactive proneuropeptide (pNP) that contains a single or multiple copies of active peptides. Active peptides are commonly short, with only a few families adopting well-defined, but unrelated, folds (e.g., prolactin and glycoprotein hormones). pNPs have a signal peptide (SP) and enter the secretory apparatus where dedicated proteases cleave them at mono- or dibasic cleavage sites (12) and where the maturing peptides are often further modified (13). pNPs are ubiquitous in eumetazoans, and genomics and mass spectrometry revealed the full neuropeptide repertoire of several species (14–20). Given their wide occurrence in metazoans, and importance in neuronal regulation, a global pNP phylogeny would further our knowledge of nervous system evolution. pNP phylogenetics is challenging because pNP evolution has patterns and constraints different from the evolution of folded proteins (21). pNPs are often repetitive; the number and length of repeats change during evolution, or the sequences diverge into distinct peptides within a precursor (21–23). Conserved sequence stretches in pNPs often constitute only a few residues corresponding to biologically active short peptides (24–27). Neuropeptides showing such limited conservation were nevertheless shown to be ligands of orthologous GPCRs in different phyla, confirming that the pNPs are orthologous (28–30).
The high diversity and repetitive sequence of pNPs hamper multiple alignment-based molecular phylogeny analyses. Although a pNP catalog exists (31), it is not clear how the diverse families are related. Furthermore, the paucity of pNP information from placozoans, ambulacrarians, and cephalochordates obscures the evolutionary origins of distinct pNP families. I have performed a comprehensive analysis of pNP and neuropeptide GPCR evolution using sequence similarity-based clustering. Clustering and mining of poorly studied genomes clarified the interrelatedness and origin of pNP families. GPCR clustering revealed several bilaterian orthologous groups containing receptors for orthologous peptides. These results provide a global view of the evolution of metazoan neuropeptide signaling and uncover the stable evolutionary association of GPCR–ligand pairs.
Results
Cluster Representation of pNP and GPCR Diversity.
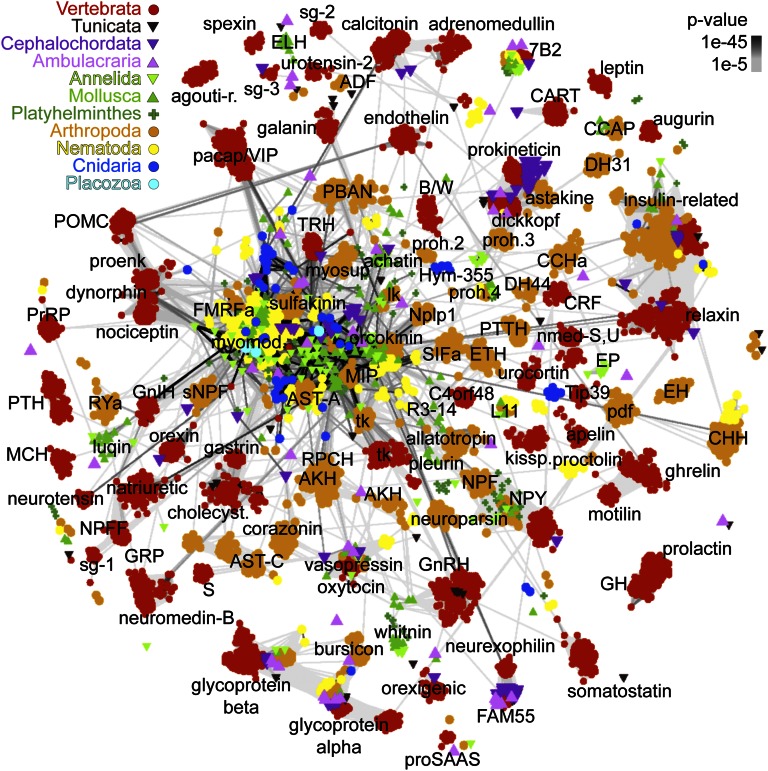
A nonredundant dataset of 6,225 pNPs (Methods and Dataset S1) from 10 animal phyla belonging to ∼80 families were clustered based on all-against-all sequence similarity [Basic Local Alignment Search Tool (BLAST) P values] (32) using either the BLOSUM62 or the PAM30 matrix. Clustering recovered all known families (Fig. 1), several of them unique with no connections to other families [e.g., prolactin, galanin, and cocaine- and amphetamine-regulated transcript (CART)], or only few spurious hits to unrelated clusters [e.g., crustacean hyperglycemic hormone (CHH) to relaxin]. However, 22 of 80 families were strongly connected to form one large central cluster (CC; Figs. 1 and 2 and Fig. S1). In the CC some sequences were only indirectly connected via a network of transitive BLAST connections. The core of the CC contained repetitive pNPs that give rise to short, amidated neuropeptides [e.g., FMRFamides (Phe-Met-Arg-Phe-NH2), myoinhibitory peptide (MIP), and LWamide (Leu-Trp-NH2)]. Several peripheral groups [e.g., neuropeptide FF (NPFF), and gonadotropin-inhibitory hormone (GnIH)] were connected to the core, but not to other derived families, representing independent divergences from the more ancestral sequences of the core (Figs. 1 and 2 and Fig. S1).
Fig. 1.
BLOSUM62 cluster map of metazoan pNP families. Nodes correspond to pNPs and are colored based on taxonomy. Edges correspond to BLAST connections of P value >1e-5.
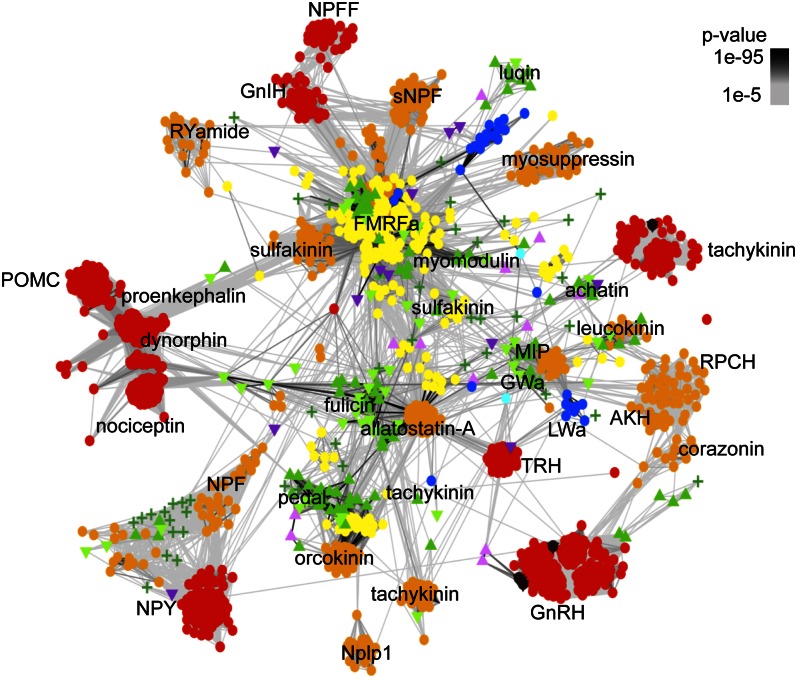
Fig. 2.
PAM30 map of the CC. The largest cluster in the PAM30 map was defined using linkage clustering and optimized separately. Nodes correspond to pNPs and are colored based on taxonomy. Edges represent BLAST connections of P value > 1e-5. Color code is the same as in Fig. 1.
The clustering of repetitive pNPs may not reflect evolutionary relatedness but rather spurious BLAST matches due to identical repeat length and reoccurring dibasic cleavage sites. To exclude this possibility, I mapped the length of the neuropeptide repeats of a subset of pNPs to the cluster map. Several pNPs with the same repeat length were far from each other in the map, indicating that repeat length alone does not explain the observed clustering (Fig. S1C). To test whether clustering correlates with short terminal amidated motifs I mapped the occurrence of such motifs. I identified 32 3-aa amidated motifs specific to a certain cluster (Figs. S1D and S2A).
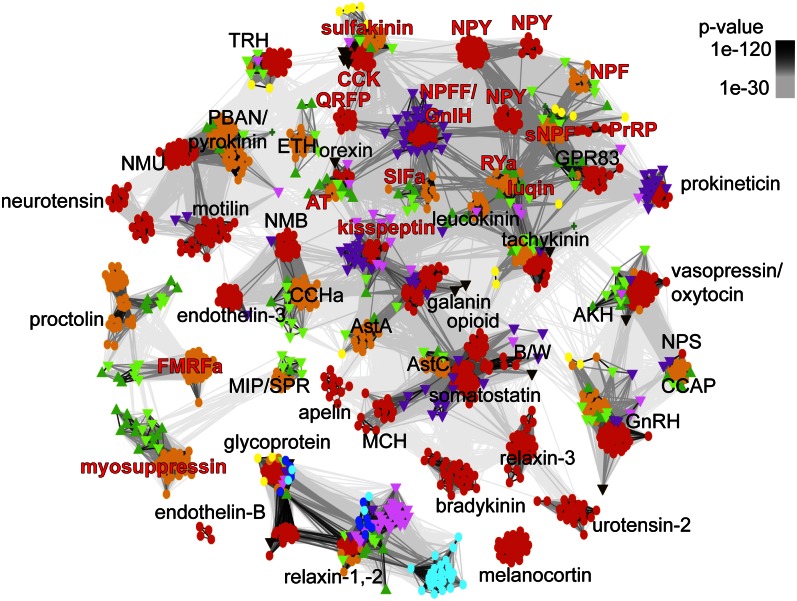
To analyze the phyletic distribution of pNP families, I projected taxonomic information onto the cluster map, distinguishing 11 metazoan clades (Figs. 1 and 2 and Fig. S3A). I also performed sensitive similarity searches to detect distant homologs, allowing the reconstruction of pNP repertoire at key nodes of the metazoan tree (Fig. S3A). I also clustered class-A (rhodopsin) and class-B (secretin) neuropeptide GPCRs, revealing several orthologous clusters (P value < 1e-50, Fig. 3, Fig. S2B, and Datasets S2 and S3), and the phyletic breadth and time of origin of different families (Fig. S3B). The combined analyses of pNP and GPCR distribution allowed a detailed reconstruction of the evolution of neuropeptide signaling in metazoa.
Placozoan Sequences Reveal the Deep Origin of CC pNPs.
Database searches identified three pNPs in the neuronless placozoan Trichoplax adhaerens (Fig. S4A). These pNPs have a SP and repetitive short sequences flanked by dibasic cleavage sites, preceded by the amidation signature glycine. They showed BLAST hits to repetitive pNPs from various bilaterians, including Famides (Phe-NH2), mollusk PRQFVamide (Pro-Arg-Gln-Phe-Val-NH2), or Saccoglossus kowalevskii Samide (Ser-NH2) and mapped to the CC (Fig. 1). The T. adhaerens genome contains enzymes for pNP processing and several GPCRs. Orthologs of the relaxin and glycoprotein hormone GPCRs could be identified (Fig. 3), but no relaxin- or glycoprotein-hormone-like pNP was found. No pNP sequence or neuropeptide GPCR could be identified in the sponge Amphimedon queenslandica despite the fact that sponges contain pNP processing enzymes.
Fig. 3.
BLOSUM62 cluster map of class-A neuropeptide GPCRs. Nodes correspond to class-A GPCRs and are colored based on taxonomy. Edges represent BLAST connections of P value > 1e-30. Color code is the same as in Fig. 1.
Ancient Eumetazoan pNP Families.
The last common ancestor of eumetazoans had an extended pNP repertoire with short amidated peptides [Wamide (Trp-NH2) and R[FY]amide (Arg-[PheTyr]-NH2)], insulin-related peptide, prokineticin (33, 34), and glycoprotein hormone. FMRFamide-like peptides (FLP) form a heterologous group of F/Yamides (Phe/Tyr-NH2) with many families of unclear relatedness (35, 36). The pNP map clarifies the relationships of FLPs (Figs. 1 and 2 and Fig. S1 B and D). Cnidarian RFamides map near the repetitive bilaterian R[FY]amides in the CC, revealing an ancestral eumetazoan R[FY]amide orthology group (Fig. 2 and Fig. S1D). FLPs with several repeats could also be identified in the cephalochordate Branchiostoma floridae and the hemichordate S. kowalevskii, but not in vertebrates, indicating that vertebrates have a structurally more derived pNP complement. [LV]Wamides ([LeuVal]Trp-NH2) constitute another eumetazoan orthology group within the CC (Figs. 1 and 2 and Fig. S1D). This group contains cnidarian GLWamides and diverse protostome Wamides (GWamides and MIPs), which cluster together and share an amidated Trp residue preceded by a small aliphatic residue. Protostome MIPs have another conserved Trp (W-X6–8-Wamide motif) that is lacking from LWamides and GWamides (Gly-Trp-NH2) (Dataset S4). Wamides at the core of the CC connect to peripheral families. GWamides connect to adipokinetic hormone (AKH) and red pigment-concentrating hormone (RPCH) (Fig. 2). These three families are present in arthropods, mollusks, and annelids and share a C-terminal segment with a disulphide bond (37). These pNPs in turn connect to the ancestral bilaterian gonadotropin-releasing hormone (GnRH)/corazonin family, suggesting a complex scenario of duplication and gene loss for the origin of these families from repetitive Wamides (38). A relationship between GnRH and AKH has been proposed based on shared sequence features (38) and the relatedness of the receptors (39, 40). The clustering of GnRH/AKH receptors in the GPCR map confirms this (Fig. 3).
Glycoprotein hormones and prokineticins also trace back to the stem eumetazoan. Glycoprotein hormones have two related subunits belonging to the Cys-knot superfamily that also includes TGF-β, NGF, PDGF, and the bone morphogenetic protein (BMP) antagonists gremlin/DAN (41). Cys-knot domains are also present in several multidomain proteins (e.g., mucins). Position-specific iterated BLAST (PSI-BLAST) searches identified a glycoprotein hormone in the sea anemone Nematostella vectensis, showing highest similarity to arthropod bursicons (Fig. 1 and Dataset S4). This is consistent with the presence of glycoprotein hormone receptor-like sequences in cnidarians (42)(Fig. 3). A bursicon-like sequence is also present in the sea urchin Strongylocentrotus purpuratus, but not in other deuterostomes. Prokineticins/astakines consist of a Cys-rich domain that is also found in colipases (34) and as a C-terminal domain in the Cys-rich Wnt antagonists, the dickkopf-related proteins. Dickkops have an additional N-terminal domain. The prokineticin/colipase domain is also present in Hydra magnipapillata (e.g., XP_002160463.2) and N. vectensis (XP_001641384.1) independently of the dickkopf N-terminal domain and is potentially a precursor of the cognate domain in prokineticins (Fig. S2C).
The presence of two insulin-related peptides in N. vectensis indicates that this family is also ancestral to eumetazoans. A group of cnidarian GPCRs was also identified that clustered with receptors for the chordate insulin-like peptide relaxin (Fig. 3), suggesting that relaxin receptors may be ancestrally involved in insulin-related peptide signaling.
Greatly Extended pNP Repertoire in Urbilateria.
Several pNP families have previously been shown to be ancestral to bilaterians including the tachykinins (43), corticotropin-releasing factors (CRFs) (44), calcitonin (45), neuromedin-U/pyrokinin (46), allatostatin-C/somatostatin (47), cholecystokinin/sulfakinin (48), pedal peptide/orcokinin (49), vasopressin/oxytocin (50), GnRH/corazonin/AKH (40, 51), 7B2 (52), and neuropeptide Y and F (NPY/NPF) (53). Many of these families form well-connected clusters in the pNP map with both protostome and deuterostomes sequences (Fig. 1), with some exceptions, where sequence conservation is limited (neuromedin-U/pyrokinin, calcitonin/diuretic hormone 31 (DH31), cholecystokinin/sulfakinin, allatostatin-C/somatostatin; Dataset S4).
The GPCR map revealed several orthologous clusters of protostome and deuterostome sequences with orthologous neuropeptide ligands, confirming the above relationships. These included the class-A receptors for tachykinin, neuromedin-U/pyrokinin, vasopressin/oxytocin, GnRH/corazonin/AKH, allatostatin-C/somatostatin, and cholecystokinin/sulfakinin (Fig. 3). The class B GPCRs for calcitonin/DH31 and CRF/diuretic hormone 44 (DH44) also formed bilaterian-wide clusters (Fig. S2B). Members of several of these families and their putative receptors could also be identified in S. kowalevskii and B. floridae (Figs. 1 and 3, Fig. S2B, and Datasets S2, S3, and S5).
The urbilaterian origin of four further pNP families was revealed by sequence searches and was partly supported by GPCR clustering. The evidence for opioid-like peptides in protostomes has been controversial (54), and no pNP has yet been described in any invertebrate. Database searches identified proenkephalin-like pNPs in annelid and mollusk expressed sequence tags (EST). HHpred searches with the Lottia gigantea and Platynereis dumerilii pNPs identified the nociceptin/proenkephalin family as homologs (probability 94.6%, P = 6.5e-07 and 81%, P = 1.8e-05, respectively). The opioid peptides produced from these pNPs are longer than their vertebrate counterparts, are often amidated, and share the N-terminal motif YGx[FL]+ (+ is a hydrophobic residue; Fig. S4B and Dataset S4). Vertebrate opioid pNPs have a segment after the SP with six Cys (22). Six to eight Cys following the SP are also present in the annelid and mollusk opioid pNPs, although not in conserved positions. These similarities establish the homology of the lophotrochozoan pNPs with the vertebrate opioid family. No protostome opioid receptor could be identified.
Sensitive similarity searches also identified deuterostome and ecdysozoan orthologs of lophotrochozoan luqins (55). Luqins retrieved a S. purpuratus sequence that identified a S. kowalevskii pNP. Both ambulacrarian sequences have an RWamide (Arg-Trp-NH2) motif and a proline-rich C-terminal peptide with two conserved Cys residues (Dataset S4). PSI-BLAST searches also revealed the homology of luqins and insect RYamide (Arg-Tyr-NH2) pNPs. Luqin and RYamide pNPs have two R[YF]amide peptides directly following the SP and also share the C-terminal peptide with the two conserved Cys residues (56). In the GPCR map mollusk luqin and insect RYamide receptors cluster together with two GPCRs from S. purpuratus that likely represent deuterostome luqin receptor orthologs, confirming the ancestral presence of luqin signaling in bilaterians. Another ancestral bilaterians family, first described in lophotrochozoans, is achatin (57). Homologs of mollusk achatin could be identified in annelids, in S. kowalevskii, and B. floridae (Dataset S5). Achatins share the GF[GAF][DNG] motif (Dataset S4). No achatin receptor has yet been described.
Database searches and the GPCR map also revealed the orthology of allatotropin and orexin pNPs (Fig. 3). A S. kowalevskii sequence (Dataset S5), identified by PSI-BLAST using arthropod allatotropin queries, shares a conserved C-terminal domain with protostome allatotropins. This sequence also contains an orexin peptide directly after the SP (Dataset S4) (58). The allatotropin and orexin receptors cluster together, and this cluster also contains a S. kowalevskii receptor. These results establish allatotropin/orexin and their receptors as orthologs representing an ancient bilaterian family.
Several other orthologous GPCR pairs for seemingly unrelated peptide families were also recovered, suggesting that the corresponding pNPs also represent orthologous, ancestral bilaterian families. These include the CCHamide (Cys-Cys-His-NH2)/neuromedin-B, neuropeptide-S/crustacean cardioactive peptide (CCAP), and allatostatin-A/galanin receptors. Of these families the orthology of CCAP/neuropeptide S (NPS) is supported by a shared N-terminal K-R-x-F-x-N motif (Dataset S4). CCHamides (59, 60) are related to annelid and mollusk excitatory peptide pNPs (61). This family is also related to the L11 pNPs of annelids, mollusks, nematodes (62), and crustaceans, as shown by PSI-BLAST. L11/CCHamide/polychaete excitatory peptides (EP) share the signature sequence Cys-x6,8-Cys-X-Gly-X2,3 with an internal disulphide bond (Dataset S4) and represent two paralogous families, ancestrally present in protostomes. The orthology of L11/CCHamide/EP and neuromedin-B is not recognizable at the pNP level but they are listed as orthologs based on the receptor evidence. Likewise, an orthologous relationship between allatostatin-A and galanin is not evident at the pNP level, but is supported by the orthology of their receptors and a similar role in the regulation of lipid storage (63), among other functions. Neuromedin-B and galanin receptors are present in B. floridae, but no pNP orthologs were found. Identifying the ligands for these receptors may reveal intermediate sequences that could reinforce these relationships.
Other GPCRs also formed bilaterian orthologous clusters, even though their known peptide ligands have a more limited distribution. Leucokinins were first described in arthropods, and sequence searches identified homologs in nematodes, mollusks, and annelids. These peptides share the FxxW[GA]-NH2 motif (Dataset S4) and connect to the CC (Fig. 2). The GPCR map revealed the presence of an ambulacrarian leucokinin receptor, indicating that leucokinin-like peptides may also be present in ambulacraria. GPCR clustering revealed another bilaterian GPCR family, the orphan GPR83 receptors. These may be receptors for an as-yet-unidentified ancient bilaterian peptide. Thyrotropin-releasing hormone (TRH) receptor orthologs are also present in protostomes (Fig. 3 and Fig. S3B). The ancestry of TRH has been traced to the stem deuterostome (49) but may be urbilaterian. Among the class-B GPCRs, the vertebrate parathyroid hormone (PTH) receptor has orthologs in invertebrates (Fig. S2B). PTH could only be identified in vertebrates, but the broader distribution of the receptors indicates that this family also has urbilaterian origin. Likewise, a S. purpuratus ortholog of protostome pigment-dispersing factor (Pdf) receptors suggests a deeper origin for this family (Fig. S2B).
GPCRs for most F/Yamide peptide families cluster together, including sulfakinin/cholecystokinin (CCK), pyroglutamylated RFamide peptide (QRFP), NPFF/GnIH, SIFa, NPY, NPF, short neuropeptide F (sNPF), prolactin-releasing peptide (PrRP), RYamide, luqin, kisspeptin, and allatotropin receptors (Fig. 3). These GPCRs likely represent stem bilaterian duplicates. Within this large GPCR cluster, bilaterian orthologous groups can be recognized for sulfakinin/CCK, orexin/allatotropin, and RYamide/luqin pNP pairs, in agreement with pNP classification. GPCR orthologies further suggest that NPFF/GnIH/SIFamide (Ser-Ile-Phe-NH2) and PrRP/sNPF pNPs likewise represent ancestral bilaterian families, and that vertebrate kisspeptins may have invertebrate orthologs. Vertebrate NPFF and GnIH pNPs are paralogs as indicated by sensitive similarity searches, an intermediate sequence from the cyclostome Paramyxine atami, and a shared PQRFamide motif. A B. floridae NPFF/GnIH could also be identified. NPFF and GnIH receptors cluster with several B. floridae GPCRs and are connected to protostome SIFamide receptors, indicating that these Famide families are likely orthologs. The receptors for PrRP and sNPF pNPs also cluster together, in the vicinity of NPY/NPF receptors (Fig. 3), in agreement with a distant relationship between the sNPF, NPF, and NPY pNP families (64).
Protostome- and Deuterostome-Specific pNPs.
Some pNPs are restricted to protostomes, either tracing back to the protostome stem or present in one or two phyla only (Fig. S3A). Ancient protostome pNPs include proctolin (65), prohormone-2 and -4, and myomodulin/myosuppressin. The protostome origin of proctolin and myomodulin/myosuppressin is also supported by the GPCR map (prohormone-2 and -4 receptors are not known). Arthropod myosuppressins are muscle inhibitory peptides (66). Their receptors cluster with mollusk and annelid receptors, indicating that orthologous peptides are present in lophotrochozoans. The likely orthologs are the myomodulins (67), myoactive peptides found in mollusks, annelids, and nematodes (68) that share an LR[MLF]-NH2 motif with myosuppressins (Dataset S4). The biochemical characterization of these lophotrochozoan receptors could confirm this connection. Some pNPs are more restricted phyletically, including arthropod antidiuretic factor (ADF) (15), neuroparsin, and prothoracicotropic hormone (PTTH), mollusk R3-14, ecdysozoan CHH/ion transport peptide, and lophotrochozoan fulicin. Cnidarians, platyhelminthes, echinoderms, and nematodes also have several pNPs with no similarity to sequences from other phyla (not all listed in Fig. S3A).
The arthropod PTTHs are related to the extracellular signaling molecule trunk (69) that is a member of an ancient bilaterian family; trunk orthologs could be identified in annelids, mollusks, and in B. floridae. Trunk is distantly related to the TGF-β inhibitors, noggins, which are also present in placozoans and sponges (Fig. S2D). PTTH represents an arthropod-specific paralog of trunk and is listed as an arthropod-specific pNP (Fig. S3A). The Cys-rich neuroparsins show similarity to the insulin-like growth factor binding protein (HHpred probability 99.5% and P = 1e-18), a domain found in various multidomain proteins (e.g., HTRA serine proteases), and represent an arthropod-specific duplication and stand-alone version of this widespread domain (Fig. S2E).
Several pNP families are only found in the deuterostomes. Some originated either in the chordate or the deuterostome stem lineage (Fig. S3A). Ambulacrarian orthologs reveal the stem deuterostome origin of some pNPs. Secretogranin-3 could be identified in S. kowalevskii, S. purpuratus, and B. floridae. Several members of the FAM55/neurexophilin family, first described in vertebrates (70), could also be identified in S. purpuratus, S. kowalevskii, and B. floridae (Fig. 1 and Fig. S3A). B. floridae orthologs revealed the stem chordate origin of orexigenic neuropeptide QRFP-like/26RFa and CART (CART, Datasets S4 and S5). GPCR clustering suggests a stem-chordate origin of further families, where the pNPs are only known in vertebrates. These include motilin/ghrelin, melanin-concentrating hormone (MCH)/MCH gene-related peptide (Mgrp), and endothelin. The presence of glucagon and gastric inhibitory peptide receptor orthologs in Ciona intestinalis suggests that these families originated before the vertebrate–urochordate divergence. Other vertebrate-specific pNPs show no resemblance to any family and represent likely vertebrate innovations (Fig. 1 and Fig. S3). The history of the GPCRs supports this for urotensin, adrenomedullin, pituitary adenylate cyclase-activating polypeptide (PACAP)/vasoactive intestinal peptide (VIP), neuropeptide-B/-W, and neurotensin. Growth hormone, VIP, glucagon, and adrenomedullin could also be identified in the lamprey Petromyzon marinus, indicating that these originated along the vertebrate stem.
Discussion
A striking pattern in the evolution of pNPs is the interrelatedness of several pNPs in the CC and the independent derivation of several families from the CC. This indicates that a large fraction of metazoan pNP families are deep paralogs. Importantly, only a clustering approach could reveal this pattern, because many of the derived families are not similar to each other, and the homologies are only revealed by indirect links in a network of BLAST interactions. The broad network of interactions in the highly diverse CC demonstrates the unique pattern of sequence evolution of pNPs. pNPs may evolve more freely in sequence space than globular proteins, where key conserved residues can be identified across very distant homologs. pNPs at two sides of the CC may not show any sequence similarity, except for the SP and cleavage sites, yet be related transitively, via a network of strong sequence similarity. The paralogous nature of several Y/Famide pNPs in the CC is further supported by the close relationship of their receptors.
The finding of repetitive pNPs belonging to the CC in the placozoan T. adhaerens indicates that this class of pNPs was present in the common ancestor of placozoans and eumetazoans and a large diversity of pNPs evolved from such ancestral sequences via successive phases of clade-specific derivations. The presence of pNPs in placozoans indicates that neuropeptide signaling may predate the origin of nervous systems. Although the phylogenetic position of placozoans is not fully resolved, they may be the sister group to the eumetazoans, potentially representing a primitive neuronless organism (71). If this is the case, then the study of placozoan neuropeptidergic cells may give unique insights into the evolutionary origin of neuronal signaling. The role of neuropeptide signaling in T. adhaerens is unclear but may involve paracrine communication between sensory cells and effector cells to regulate ciliary crawling, or digestive enzyme secretion.
The last common ancestor of eumetazoans had various small amidated peptides (RFamide, RYamide, and Wamide), a glycoprotein hormone, prokineticin, and insulin-related peptide. In vertebrates, the glycoprotein hormones of the pituitary are under the control of short, amidated peptides (TRH and GnRH) and regulate sexual development, reproduction, growth, and metabolism. Insulin-related peptides are conserved regulators of growth and metabolism. In cnidarians, external stimuli are directly translated into neuroendocrine signals by chemosensory–neurosecretory cells releasing small amidated peptides to regulate growth and metamorphosis (72). Wamides may have been ancestrally involved in mediating life-cycle transitions triggered by chemosensory cues (73). RFamides may have ancient roles in muscle control (74), ciliary locomotion (27, 62), and food intake (75). The role of glycoprotein hormones and insulin-like peptides in cnidarians is unclear, but they may be part of a small-peptide/glycoprotein/insulin module in the regulation of growth, metabolism, or sexual maturation.
The combined analysis of pNPs and neuropeptide GPCRs identified 27 ancestral urbilaterian pNP-receptor families pointing at a hitherto unknown sophistication of neuropeptidergic systems in the urbilaterian. These pNPs regulate several aspects of physiology, including sexual behavior and reproduction (GnRH, achatin, oxytocin, and GnIH/SIFamide), diuresis (CRF/diuretic hormone, calcitonin, and vasopressin), gut and heart activity (achatin, luqin, and orcokinin), pain perception (opioid), and food intake (NPY, kinins, neuromedin-U, galanin/allatostatin-A, and orexin/allatotropin). These pNPs may have originated concomitantly with the origin of a complex bilaterian body plan having a through gut with novel controls for food intake and digestion, excretory and circulatory systems, light-controlled reproduction (50), a centralized nervous system (76), complex reproductive behavior (77, 78), and learning (79). The stable association of receptor–ligand families across bilateria revealed the long-term coevolution of receptor–ligand pairs. This pattern suggests the deep conservation of neuropeptidergic regulation between different metazoan phyla, the extent of which will be revealed by further comparative functional studies in a broader selection of taxa.
Methods
pNPs and neuropeptide GPCRs were retrieved using a combination of strategies. Curated lists of 6225 pNPs, 1465 Class A and 547 Class B receptor sequences were clustered using CLANS2 (32). To read the Clans files (Datasets S1–S3) install CLANS2 and run the command line command: java -Xmx4000m -jar /your_install_directory/CLANS.jar -load Clans_file. Detailed procedures are described in SI Methods.
Supplementary Material
Acknowledgments
I thank Elizabeth Williams and Markus Conzelmann for helpful comments. The research leading to these results received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013)/European Research Council Grant Agreement 260821.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KC708483).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221833110/-/DCSupplemental.
References
- 1.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11(6):1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rechler MM, Nissley SP. The nature and regulation of the receptors for insulin-like growth factors. Annu Rev Physiol. 1985;47:425–442. doi: 10.1146/annurev.ph.47.030185.002233. [DOI] [PubMed] [Google Scholar]
- 3.Leung DW, et al. Growth hormone receptor and serum binding protein: Purification, cloning and expression. Nature. 1987;330(6148):537–543. doi: 10.1038/330537a0. [DOI] [PubMed] [Google Scholar]
- 4.Lowe DG, et al. Human atrial natriuretic peptide receptor defines a new paradigm for second messenger signal transduction. EMBO J. 1989;8(5):1377–1384. doi: 10.1002/j.1460-2075.1989.tb03518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tartaglia LA, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83(7):1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 6.Yamauchi T, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423(6941):762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 7.Chang JC, Yang RB, Adams ME, Lu KH. Receptor guanylyl cyclases in Inka cells targeted by eclosion hormone. Proc Natl Acad Sci USA. 2009;106(32):13371–13376. doi: 10.1073/pnas.0812593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378(6558):730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 9.Marder E. Neuromodulation of neuronal circuits: Back to the future. Neuron. 2012;76(1):1–11. doi: 10.1016/j.neuron.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y-J, Zitňan D, Galizia CG, Cho K-H, Adams ME. A command chemical triggers an innate behavior by sequential activation of multiple peptidergic ensembles. Curr Biol. 2006;16(14):1395–1407. doi: 10.1016/j.cub.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Hartenstein V. The neuroendocrine system of invertebrates: A developmental and evolutionary perspective. J Endocrinol. 2006;190(3):555–570. doi: 10.1677/joe.1.06964. [DOI] [PubMed] [Google Scholar]
- 12.Hook V, et al. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: Peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 14.Dircksen H, et al. Genomics, transcriptomics, and peptidomics of Daphnia pulex neuropeptides and protein hormones. J Proteome Res. 2011;10(10):4478–4504. doi: 10.1021/pr200284e. [DOI] [PubMed] [Google Scholar]
- 15.Li B, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2008;18(1):113–122. doi: 10.1101/gr.6714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hummon AB, et al. From the genome to the proteome: Uncovering peptides in the Apis brain. Science. 2006;314(5799):647–649. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 17.Mirabeau O, et al. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 2007;17(3):320–327. doi: 10.1101/gr.5755407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser F, et al. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J Proteome Res. 2010;9(10):5296–5310. doi: 10.1021/pr100570j. [DOI] [PubMed] [Google Scholar]
- 19.Predel R, et al. Neuropeptidomics of the mosquito Aedes aegypti. J Proteome Res. 2010;9(4):2006–2015. doi: 10.1021/pr901187p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie F, et al. The zebra finch neuropeptidome: Prediction, detection and expression. BMC Biol. 2010;8:28. doi: 10.1186/1741-7007-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Pérez F, et al. Loss of DNA: A plausible molecular level explanation for crustacean neuropeptide gene evolution. Peptides. 2007;28(1):76–82. doi: 10.1016/j.peptides.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Dores RM, Lecaude S. Trends in the evolution of the proopiomelanocortin gene. Gen Comp Endocrinol. 2005;142(1-2):81–93. doi: 10.1016/j.ygcen.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Wegener C, Gorbashov A. Molecular evolution of neuropeptides in the genus Drosophila. Genome Biol. 2008;9(8):R131. doi: 10.1186/gb-2008-9-8-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Baggerman G, Schoofs L, Wets G. Uncovering conserved patterns in bioactive peptides in Metazoa. Peptides. 2006;27(12):3137–3153. doi: 10.1016/j.peptides.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Lindemans M, et al. A neuromedin-pyrokinin-like neuropeptide signaling system in Caenorhabditis elegans. Biochem Biophys Res Commun. 2009;379(3):760–764. doi: 10.1016/j.bbrc.2008.12.121. [DOI] [PubMed] [Google Scholar]
- 26.Veenstra JA. Neuropeptide evolution: Neurohormones and neuropeptides predicted from the genomes of Capitella teleta and Helobdella robusta. Gen Comp Endocrinol. 2011;171(2):160–175. doi: 10.1016/j.ygcen.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Conzelmann M, Jékely G. Antibodies against conserved amidated neuropeptide epitopes enrich the comparative neurobiology toolbox. Evodevo. 2012;3(1):23. doi: 10.1186/2041-9139-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen T, Lindemans M, Meelkop E, Temmerman L, Schoofs L. Coevolution of neuropeptidergic signaling systems: From worm to man. Ann N Y Acad Sci. 2010;1200:1–14. doi: 10.1111/j.1749-6632.2010.05506.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y-J, et al. MIPs are ancestral ligands for the sex peptide receptor. Proc Natl Acad Sci USA. 2010;107(14):6520–6525. doi: 10.1073/pnas.0914764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park Y, Kim Y-J, Adams ME. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci USA. 2002;99(17):11423–11428. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Baggerman G, Schoofs L, Wets G. The construction of a bioactive peptide database in Metazoa. J Proteome Res. 2008;7(9):4119–4131. doi: 10.1021/pr800037n. [DOI] [PubMed] [Google Scholar]
- 32.Frickey T, Lupas A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20(18):3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 33.Watthanasurorot A, Söderhäll K, Jiravanichpaisal P, Söderhäll I. An ancient cytokine, astakine, mediates circadian regulation of invertebrate hematopoiesis. Cell Mol Life Sci. 2011;68(2):315–323. doi: 10.1007/s00018-010-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravind L, Koonin EV. A colipase fold in the carboxy-terminal domain of the Wnt antagonists—the Dickkopfs. Curr Biol. 1998;8(14):R477–R478. doi: 10.1016/s0960-9822(98)70309-4. [DOI] [PubMed] [Google Scholar]
- 35.Espinoza E, Carrigan M, Thomas SG, Shaw G, Edison AS. A statistical view of FMRFamide neuropeptide diversity. Mol Neurobiol. 2000;21(1-2):35–56. doi: 10.1385/MN:21:1-2:035. [DOI] [PubMed] [Google Scholar]
- 36.Walker RJ, Papaioannou S, Holden-Dye L. A review of FMRFamide- and RFamide-like peptides in metazoa. Invert Neurosci. 2009;9(3-4):111–153. doi: 10.1007/s10158-010-0097-7. [DOI] [PubMed] [Google Scholar]
- 37.Martínez-Pérez F, Becerra A, Valdés J, Zinker S, Aréchiga H. A possible molecular ancestor for mollusk APGWamide, insect adipokinetic hormone, and crustacean red pigment concentrating hormone. J Mol Evol. 2002;54(6):703–714. doi: 10.1007/s00239-001-0036-7. [DOI] [PubMed] [Google Scholar]
- 38.Lindemans M, et al. Gonadotropin-releasing hormone and adipokinetic hormone signaling systems share a common evolutionary origin. Front Endocrinol (Lausanne) 2011;2:16. doi: 10.3389/fendo.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Asadulina A, Panzera A, Veraszto C, Liebig C, Jékely G (2012) Whole-body gene expression pattern registration in Platynereis larvae. EvoDevo 3:1–33. [DOI] [PMC free article] [PubMed]
- 40.Lindemans M, et al. Adipokinetic hormone signaling through the gonadotropin-releasing hormone receptor modulates egg-laying in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106(5):1642–1647. doi: 10.1073/pnas.0809881106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1(5):673–683. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- 42.Anctil M. Chemical transmission in the sea anemone Nematostella vectensis: A genomic perspective. Comp Biochem Physiol Part D Genomics Proteomics. 2009;4(4):268–289. doi: 10.1016/j.cbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Van Loy T, et al. Tachykinin-related peptides and their receptors in invertebrates: A current view. Peptides. 2010;31(3):520–524. doi: 10.1016/j.peptides.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Patel M, Hayes T, Coast G. Evidence for the hormonal function of a CRF-related diuretic peptide (Locusta-DP) in Locusta migratoria. J Exp Biol. 1995;198(Pt 3):793–804. doi: 10.1242/jeb.198.3.793. [DOI] [PubMed] [Google Scholar]
- 45.Furuya K, et al. Cockroach diuretic hormones: Characterization of a calcitonin-like peptide in insects. Proc Natl Acad Sci USA. 2000;97(12):6469–6474. doi: 10.1073/pnas.97.12.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melcher C, Bader R, Walther S, Simakov O, Pankratz MJ. Neuromedin U and its putative Drosophila homolog hugin. PLoS Biol. 2006;4(3):e68. doi: 10.1371/journal.pbio.0040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veenstra JA. Allatostatin C and its paralog allatostatin double C: The arthropod somatostatins. Insect Biochem Mol Biol. 2009;39(3):161–170. doi: 10.1016/j.ibmb.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Janssen T, et al. Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrinology. 2008;149(6):2826–2839. doi: 10.1210/en.2007-1772. [DOI] [PubMed] [Google Scholar]
- 49.Rowe ML, Elphick MR. The neuropeptide transcriptome of a model echinoderm, the sea urchin Strongylocentrotus purpuratus. Gen Comp Endocrinol. 2012;179(3):331–344. doi: 10.1016/j.ygcen.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 50.Tessmar-Raible K, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell. 2007;129(7):1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 51.Roch GJ, Busby ER, Sherwood NM. Evolution of GnRH: Diving deeper. Gen Comp Endocrinol. 2011;171(1):1–16. doi: 10.1016/j.ygcen.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 52.Hwang JR, Siekhaus DE, Fuller RS, Taghert PH, Lindberg I. Interaction of Drosophila melanogaster prohormone convertase 2 and 7B2. Insect cell-specific processing and secretion. J Biol Chem. 2000;275(23):17886–17893. doi: 10.1074/jbc.M000032200. [DOI] [PubMed] [Google Scholar]
- 53.Rajpara SM, et al. Identification and molecular cloning of a neuropeptide Y homolog that produces prolonged inhibition in Aplysia neurons. Neuron. 1992;9(3):505–513. doi: 10.1016/0896-6273(92)90188-j. [DOI] [PubMed] [Google Scholar]
- 54.Danielson PB, Dores RM. Molecular evolution of the opioid/orphanin gene family. Gen Comp Endocrinol. 1999;113(2):169–186. doi: 10.1006/gcen.1998.7206. [DOI] [PubMed] [Google Scholar]
- 55.Aloyz RS, DesGroseillers L. Processing of the L5-67 precursor peptide and characterization of LUQIN in the LUQ neurons of Aplysia californica. Peptides. 1995;16(2):331–338. doi: 10.1016/0196-9781(94)00140-5. [DOI] [PubMed] [Google Scholar]
- 56.Li L, et al. Mass spectrometric survey of interganglionically transported peptides in Aplysia. Peptides. 1998;19(8):1425–1433. doi: 10.1016/s0196-9781(98)00094-1. [DOI] [PubMed] [Google Scholar]
- 57.Kamatani Y, et al. Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Férussac containing a D-amino acid residue. Biochem Biophys Res Commun. 1989;160(3):1015–1020. doi: 10.1016/s0006-291x(89)80103-2. [DOI] [PubMed] [Google Scholar]
- 58.Sakurai T, et al. Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92(4):573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 59.Roller L, et al. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1147–1157. doi: 10.1016/j.ibmb.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Hansen KK, Hauser F, Williamson M, Weber SB, Grimmelikhuijzen CJP. The Drosophila genes CG14593 and CG30106 code for G-protein-coupled receptors specifically activated by the neuropeptides CCHamide-1 and CCHamide-2. Biochem Biophys Res Commun. 2011;404(1):184–189. doi: 10.1016/j.bbrc.2010.11.089. [DOI] [PubMed] [Google Scholar]
- 61.Oumi T, et al. The GGNG peptides: Novel myoactive peptides isolated from the gut and the whole body of the earthworms. Biochem Biophys Res Commun. 1995;216(3):1072–1078. doi: 10.1006/bbrc.1995.2730. [DOI] [PubMed] [Google Scholar]
- 62.Conzelmann M, et al. Neuropeptides regulate swimming depth of Platynereis larvae. Proc Natl Acad Sci USA. 2011;108(46):E1174–E1183. doi: 10.1073/pnas.1109085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bendena WG, et al. A Caenorhabditis elegans allatostatin/galanin-like receptor NPR-9 inhibits local search behavior in response to feeding cues. Proc Natl Acad Sci USA. 2008;105(4):1339–1342. doi: 10.1073/pnas.0709492105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: Any similarities to vertebrate neuropeptide Y signaling? Peptides. 2011;32(6):1335–1355. doi: 10.1016/j.peptides.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 65.Veenstra JA. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen Comp Endocrinol. 2010;167(1):86–103. doi: 10.1016/j.ygcen.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 66.Holman GM, Cook BJ, Nachman RJ. Isolation, primary structure and synthesis of leucomyosuppressin, an insect neuropeptide that inhibits spontaneous contractions of the cockroach hindgut. Comparative Biochemistry and Physiology Part C: Comparative Pharmacology. 1986;85:329–333. doi: 10.1016/0742-8413(86)90077-0. [DOI] [PubMed] [Google Scholar]
- 67.Cropper EC, Tenenbaum R, Kolks MA, Kupfermann I, Weiss KR. Myomodulin: A bioactive neuropeptide present in an identified cholinergic buccal motor neuron of Aplysia. Proc Natl Acad Sci USA. 1987;84(15):5483–5486. doi: 10.1073/pnas.84.15.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditiselegans and other species. Proc Natl Acad Sci USA. 2001;98(24):14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rewitz KF, Yamanaka N, Gilbert LI, O’Connor MB. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326(5958):1403–1405. doi: 10.1126/science.1176450. [DOI] [PubMed] [Google Scholar]
- 70.Missler M, Südhof TC. Neurexophilins form a conserved family of neuropeptide-like glycoproteins. J Neurosci. 1998;18(10):3630–3638. doi: 10.1523/JNEUROSCI.18-10-03630.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19(8):706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 72.Gajewski M, Leitz T, Schloßherr J, Plickert G. LWamides from Cnidaria constitute a novel family of neuropeptides with morphogenetic activity. Rouxs Arch Dev Biol. 1996;205:232–242. doi: 10.1007/BF00365801. [DOI] [PubMed] [Google Scholar]
- 73.Conzelmann M, et al. Conserved MIP receptor–ligand pair regulates Platynereis larval settlement. Proc Natl Acad Sci USA. 2013;110(20):8224–8229. doi: 10.1073/pnas.1220285110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McFarlane ID, Graff D, Grimmelikhuijzen CJ. Excitatory actions of Antho-RFamide, an anthozoan neuropeptide, on muscles and conducting systems in the sea anemone Calliactis parasitica. J Exp Biol. 1987;133:157–168. [Google Scholar]
- 75.Dockray GJ. The expanding family of -RFamide peptides and their effects on feeding behaviour. Exp Physiol. 2004;89(3):229–235. doi: 10.1113/expphysiol.2004.027169. [DOI] [PubMed] [Google Scholar]
- 76.Denes AS, et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129(2):277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 77.Wagenaar DA, Hamilton MS, Huang T, Kristan WB, French KA. A hormone-activated central pattern generator for courtship. Curr Biol. 2010;20(6):487–495. doi: 10.1016/j.cub.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garrison JL, et al. Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science. 2012;338(6106):540–543. doi: 10.1126/science.1226201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beets I, et al. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science. 2012;338(6106):543–545. doi: 10.1126/science.1226860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.