Abstract
A great challenge in development biology is to understand how interacting networks of regulatory genes can direct the often highly complex patterning of cells in a 3D embryo. Here, we detail the gene regulatory network that describes the distribution of ciliary band-associated neurons in the bipinnaria larva of the sea star. This larva, typically for the ancestral deuterostome dipleurula larval type that it represents, forms two loops of ciliary bands that extend across much of the anterior-posterior and dorsal-ventral ectoderm. We show that the sea star first likely uses maternally inherited factors and the Wnt and Delta pathways to distinguish neurogenic ectoderm from endomesoderm. The broad neurogenic potential of the ectoderm persists throughout much of gastrulation. Nodal, bone morphogenetic protein 2/4 (Bmp2/4), and Six3-dependent pathways then sculpt a complex ciliary band territory that is defined by the expression of the forkhead transcription factor, foxg. Foxg is needed to define two molecularly distinct ectodermal domains, and for the formation of differentiated neurons along the edge of these two territories. Thus, significantly, Bmp2/4 signaling in sea stars does not distinguish differentiated neurons from nonneuronal ectoderm as it does in many other animals, but instead contributes to the patterning of an ectodermal territory, which then, in turn, provides cues to permit the final steps of neuronal differentiation. The modularity between specification and patterning likely reflects the evolutionary history of this gene regulatory network, in which an ancient module for specification of a broad neurogenic potential ectoderm was subsequently overlaid with a module for patterning.
Keywords: echinoderm, neural patterning, evolution
The fascinating challenge of developmental biology is to determine how gene regulatory networks (GRNs), which are encoded within the genome and initiated by anisotropies in the zygote, can orchestrate the complex final pattern of appropriately differentiated cells in time and 3D space. The specification and patterning of the nervous system has received particular attention from developmental biologists, as it is such a defining feature of the body plan. Many animals have diffuse nervous systems that form a neural net (1). Other animals have highly patterned centralized nervous systems (CNSs), of which the best characterized are the ventral nerve cord of Drosophila and the dorsal hollow neural tube of vertebrates. In both of these very disparate taxa, the position of the neuroectoderm is established by gradients of bone morphogenetic protein (BMP) and its antagonist, Chordin/short gastrulation (Sog), along the dorsal-ventral (DV) axis (2, 3). High concentrations of BMP promote the formation of nonneurogenic ectoderm, whereas neurogenic ectoderm forms where BMP concentration is low (4). Although there are other types of nervous system localizations (1), much less is known of the GRNs that lead to their final pattern. We were especially intrigued by the particularly distinctive localized pattern of neurons associated with the ciliary bands of the bipinnaria larva of the sea star, Patiria miniata. The sea star (Phylum Echinodermata) bipinnaria has two loops of ciliary bands: one that loops above the mouth and one below it, which extends from the ventral surface to the anterior, dorsal margins of the ectoderm (Fig. 1A). As there are two loops of ciliary bands, a particular anterior-posterior (AP) coordinate may have neurons along two locations within the DV axis and vice versa. These neurons coordinate the action of the cilia to enable the larvae to swim and feed in response to environmental cues within the water column (5). This design is very different from the DV-restricted patterning of the CNS in vertebrates and Drosophila, and even the echinoderm pluteus larva, which develops neurons associated with a single ciliary band that is restricted along the DV axis (Fig. 1A). Indeed, in sea stars there seems to be no simple AP and DV positioning mechanism that could establish the pattern of these neurons. However, unlike other well-characterized deuterostome embryos, this distinctive bipinnaria pattern of neurons is a common feature of the larvae of members of several classes of echinoderms [i.e., the auricularia of sea cucumbers (6) and the larvae of some basal crinoids (7)] and also some groups of hemichordates (i.e., tornaria larvae). This larval type has therefore arguably been defined as the ancestral deuterostome form and, as such, has featured strongly in evolutionary hypotheses of vertebrate CNS origins (8–10). However, very little is known of the mechanisms of development of this larva, particularly of the ectoderm. Thus, the motivation of this study was to understand the GRN that explains a distinctive nervous system pattern in what might be the basal representative of a large clade of animals.
Fig. 1.
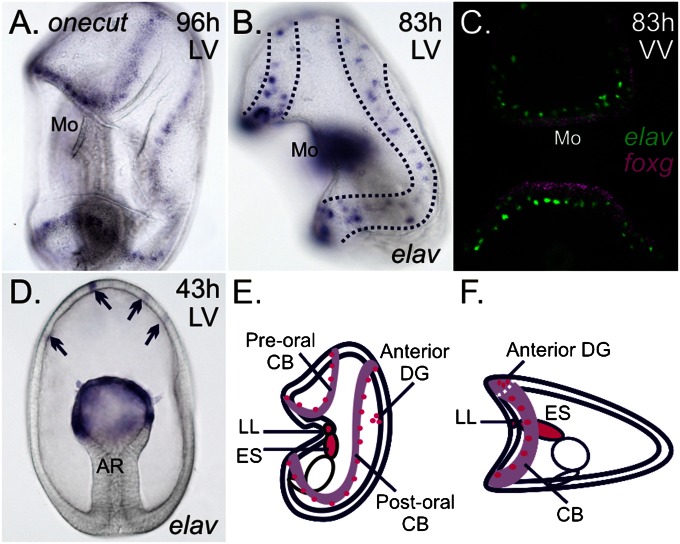
Expression of elav in P. miniata. In all figure plates, unless noted, sea star embryos are oriented with the anterior pole up and with ventral to the left: h, hours postfertilization; LV, lateral view; VV, ventral view. (A) Expression of onecut within the ectoderm marks the ciliary bands that loop above and below the mouth (Mo) of 96-h-old larvae (WMISH). (B) Dotted lines highlight elav-expressing ectodermal cells in two rows above and below the mouth. elav-expressing cells are also detected within the mesoderm and mesenchyme (not in plane of focus in B) (WMISH). (C) Two rows of elav-expressing cells (green) form near the ciliary bands, marked by foxg (purple) (FISH). (D) Transcripts of elav are first observed within cells of the anterior ectoderm (arrows) and in the mesodermal bulb of the archenteron (AR) of gastrulae (WMISH). Schematic of sea star (E) and sea urchin (F) larvae, viewed laterally, depicts expression of elav (magenta) in all known neural territories; the larval ciliary bands (CB) are shown in light purple. DG, dorsal ganglion; ES, esophagus; LL, lower lip. (Magnification: 200×.)
The initial placeholders of the GRN for this developmental process are the signaling events that first establish the AP and DV axes and distinguish ectoderm from endoderm and mesoderm. Sea star embryos undergo equal cleavage and hatch as a blastula at around 24 h after fertilization. The endoderm and mesoderm form at the vegetal pole and invaginate during gastrulation, leaving the ectoderm as a ciliated outer domain. Recent studies indicate that canonical Wnt (cWnt) pathways may have an ancestral role in establishing early animal (anterior)-vegetal (posterior) axes and distinguishing endomesoderm from ectoderm (11, 12). We therefore start our examination of the AP axis formation and the establishment of the ectoderm by considering the role of cWnt signaling in the sea star. The first morphological evidence of DV patterning in sea stars is the formation of an invagination (where the mouth will later from) on the ventral surface at around 72 h. This formation is preceded by the expression of the transcription factor foxa in this oral territory before gastrulation (13). In sea urchins, Nodal is needed to establish the mouth ectoderm and BMP gradients are used to establish territories along the DV ectodermal axis (14–17). These pathways therefore also serve as starting points for our investigations of DV patterning in the sea star.
Results and Discussion
Localized Patterns of Neurons in the Sea Star Larva.
We first characterized the pattern of neurons by examining the expression of the single P. miniata ortholog of elav (Fig. S1). Elav is an RNA binding protein that plays a role in the transition from neural progenitor to committed and differentiated states (18) and is a conserved molecular marker of postmitotic neurons (19, 20). Expression is detected in two rows of ectodermal cells along the pre- and postoral ciliary bands that surround the mouth (Fig. 1 B and C and Fig. S2A); additionally, elav expression is present in the lower lip of the mouth and esophagus (Fig. S2B) and within the bilateral anterior dorsal ganglia (Fig. S2C). Earlier in development, elav is first observed within several cells in the anterior pole ectoderm of 2-d-old (gastrula stage) embryos (Fig. 1D). We think that these elav-positive cells will later coalesce to form the bilateral, anterior dorsal ganglia of larvae, rather than neurons of the ciliary bands. Previous studies have characterized the distribution of neurons in the sea star bipinnaria based on immunoreactivity to the serotonin precursor, 5-hydroxytryptophan, and synaptotagmin-B (21). These studies identify neurons associated with the anterior ectoderm, larval ciliary bands, lower lip of the mouth, and esophagus (22, 23). Thus, we are confident that elav expression within the ectoderm serves as a marker of most, if not all, differentiated neurons. Expression of elav is observed additionally within the mesodermal bulb at the top of the archenteron of gastrulae (Fig. 1D), and later in these cells as they ingress into the blastocoel. It is not yet known if any of these elav-expressing mesenchymal cells are neurons.
This finding is very different from the pattern of neurons found in the two closest relatives to the sea star for which any mechanisms of neural specification are known (i.e., the sea urchin and the directly developing hemichordate Saccoglossus, which contrasts to the indirectly developing hemichordates that develop tornaria larvae). In sea urchins, a single ciliary band and its associated neurons (22) form at the boundary between the oral (ventral) and aboral (dorsal) ectoderm (14–17, 24) (Fig. 1F). In Saccoglossus, elav is expressed throughout the ectoderm with no apparent DV patterning (20).
Neurogenesis Occurs Broadly Throughout the Ectoderm During Gastrulation.
We previously identified many transcription factors expressed within the sea star ectoderm (25–27) that, based on their orthology to other taxa, likely have roles in neurogenesis. P. miniata orthologs of otx (in particular the ectodermally localized otxßb isoform) (26), onecut (see ref. 27 and Fig. 2A), and soxb1 (Fig. 2B) are expressed throughout the ectoderm through gastrulation. Quantitative reverse-transcription PCR (qPCR) reveals that transcripts of sea star orthologs of these genes, as well as other pan-ectodermal–expressed regulatory genes (25), are abundant within the fertilized eggs (Figs. S3 and S4), and therefore may operate near the top of a developmental GRN hierarchy for specification of the ectoderm. When cWnt signaling is blocked by injecting one-cell–stage embryos with a construct that blocks the nuclearization of β-catenin (Δ-cadherin) (28), the expression of these genes is found throughout the later embryo (Fig. S5 A and B). elav-expressing cells are also found scattered throughout this nβ-catenin–deficient ectoderm (Fig. S5C) rather than being restricted the anterior pole neural territory (e.g., compare with Fig. 1D). Thus, we suggest that similar to sea urchins and many other taxa, cWnt signaling is used to initially distinguish ectoderm from endomesoderm. In particular, cWnt signaling in sea stars segregates—as we show further below—the broad neurogenic potential ectoderm from the endomesoderm.
Fig. 2.
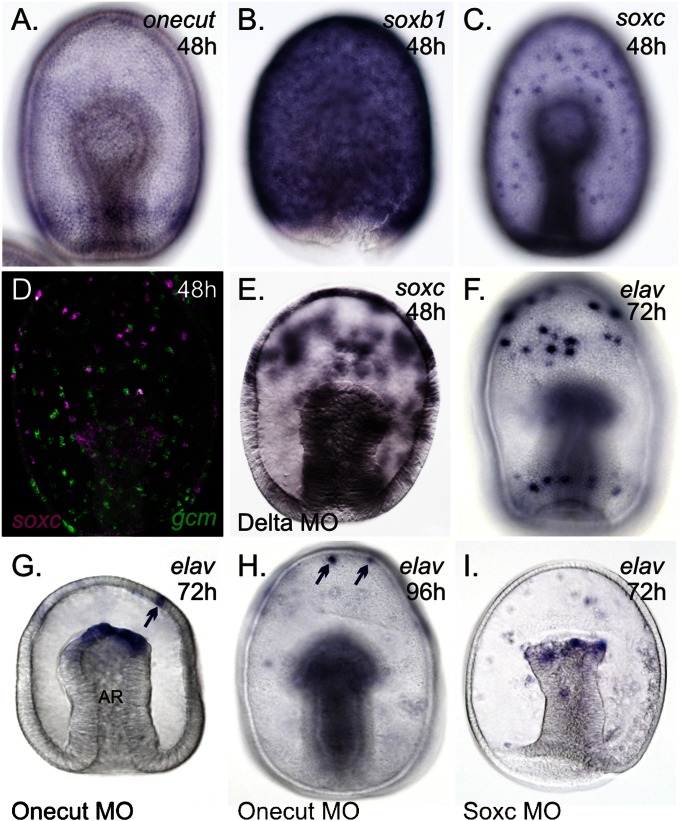
Sea star gastrulae develop a broad neurogenic potential ectoderm. Transcripts of onecut (A), soxb1 (B), and soxc (C) are found throughout the ectoderm of gastrulae. (D) soxc-expressing cells (purple) do not colocalize with those expressing glial cells missing (gcm; green) (32). (E) Ectopic expression of soxc within the ectoderm of Delta morphants (MO). (F) Transcripts of elav in the ciliary band neurons of an ∼72-h-old larva. In Onecut morphants at ∼72 h (G) or 96 h (H) postfertilization, elav-positive ciliary band neurons are not observed. (I) Both anterior and ciliary band elav-expressing neurons are lost in Soxc morphants around 72 h postfertilization. elav remains expressed in the mesoderm (G–I) at the tip of the archenteron (AR), later in mesenchyme (not in plane of focus in H and I) and in some anterior ectodermal cells (arrows in G and H). (Magnification: 200×.)
In vertebrates, and possibly protostomes (29), the expression of sox gene-family members marks a progression along a pathway toward neural commitment; soxb orthologs function during the early specification of neurons, and the soxc orthologs play a role in the commitment of neurons (30). We find here that in sea stars, soxb1 is expressed throughout the ectoderm of gastrulae (Fig. 2B), but transcripts of soxc are also found throughout the ectoderm, but only within distinct cells, giving the appearance of a spotted pattern of localization (Fig. 2 C and D). Delta Notch lateral inhibition is a conserved mechanism for distinguishing one cell type from another, particularly neuronal from nonneuronal, within an epithelium (31). In sea stars, delta is expressed in a spotted pattern throughout the ectoderm at the gastrulae stage (32) and, when disrupted, results in an expansion of soxc expression to adjacent cells within the ectoderm (compare Fig. C and E). This finding suggests that a lateral inhibition mechanism may also be used to segregate soxc-expressing cells within the ectoderm.
We next sought to determine whether the expression of elav depends on these pan-ectodermal–expressed genes. Perturbation, by injecting zygotes with a sequence-specific translation blocking morpholinos (hereafter referred to as a morphant) of onecut results in the loss of elav-expressing cells associated with the developing ciliary bands (compare Fig. 2 F and G). Some elav-expressing cells remain within the anterior pole territory in what we think are cells that will later become the dorsal ganglia (Fig. 2G). To confirm that the loss of ciliary band neurons in Onecut morphants is not because of a developmental delay resulting from the morpholino injection, we show that these neurons remain absent from the ectoderm of Onecut morphants even at 4 d postfertilization (early larval stage) (Fig. 2H). We additionally show that perturbing the function of Onecut does not broadly respecify the ectoderm, as another cell type that is marked by the expression of gcm is expressed normally in these morphants (Fig. S6). We also demonstrate that the sea star ortholog of Soxc is required for elav expression. In Soxc morphants at 3 d following fertilization, a loss of elav-positive cells was observed from both the anterior pole territory and ectoderm near the developing ciliary bands (Fig. 2I).
Taken together, these data suggest that a suite of genes, which we term the pan-neurogenic suite, are first expressed within the sea star fertilized egg, are restricted by cWnt signaling, and initiate a regulatory program for the specification of a broad neurogenic potential ectodermal territory. We do not know yet if these factors are directly involved in the neural differentiation. However, the pan-neurogenic suite, which includes at least onecut-, soxb1-, and soxc-expressing cells, forms throughout the ectoderm and are therefore not regulated by AP or DV patterning mechanisms within the ectoderm. In this respect, the sea star develops much like Saccoglossus, which maintains a pan-neurogenic ectoderm throughout gastrulation (20). It has been argued that the ectoderm of sea urchins will become broadly neurogenic in the absence of signaling (33); but, in normal development, this neurogenic potential is rapidly restricted to the anterior pole territory (34, 35) and ciliary band domain (16, 17). The presence of a broad neurogenic potential ectoderm, therefore, may be a basal feature of all echinoderms and hemichordates maintained in common for almost 800 million y (36), although the particular details of the GRN associated with this may differ. The striking difference is that during development, elav-expressing differentiated neurons become localized to ciliary bands and the anterior ganglia in echinoderms, but in Saccoglossus differentiated neurons remain throughout the ectoderm.
Patterning a Foxg-Defined Ciliary Band Domain.
To understand how the broad neurogenic potential ectoderm nonetheless leads to the localized expression of elav, we next considered the role of AP and DV patterning mechanisms in sea stars. We have shown previously that there are at least four molecularly distinct AP restricted domains within the sea star ectoderm that are similar to orthologous domains observed within the hemichordate ectoderm and the developing vertebrate nervous system (25). For example, we find concentric domains of expression of zic, foxq2/retinal homeobox (rx), six3, and nk1 along the AP axis (25). Although these domains are less apparent in the sea urchin, we have suggested previously that at least some of these AP domains may exist and that they are likely compressed into the anterior pole ectoderm much earlier in development (25).
In addition to AP domains, sea star gastrulae also have distinct domains of gene expression within the ectoderm along the DV axis. For example, we have previously shown that foxg and foxa are expressed on the ventral surface of gastrulae (13, 25). We also cloned nodal and bmp2/4 orthologs from P. miniata and examined their expression. Similarly to sea urchin, and as expected, the expression of nodal (Fig. S7A) and bmp2/4 (Fig. S7B) is restricted along the DV axis early in development, although by late gastrulation bmp2/4 expression is no longer detected in the ectoderm. We additionally examined the localization of activated Smad1/5/8, the transcriptional effector for BMP2/4 signaling. Just as has been shown in sea urchins (15), we find that phosphorylated Smad1/5/8 is found throughout the dorsal ectoderm of blastula (see, for example, Fig. 4). Again, by around 60 h of development (late gastrula) we no longer detect pSmad1/5/8 by immunohistochemistry. Thus, it is immediately evident that neurogenesis can occur broadly throughout the ectoderm without regard to the presence of active BMP signaling.
Fig. 4.
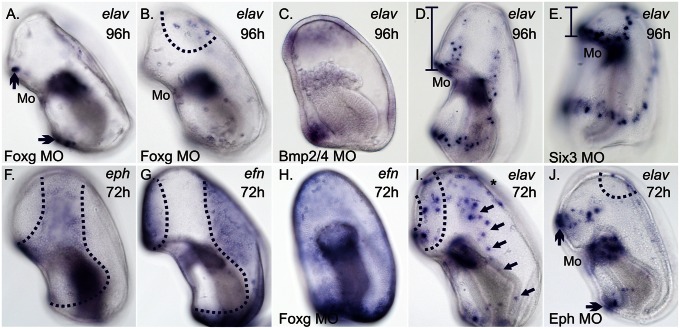
Linking the Foxg-CBD to localized neural patterning. (A) In Foxg morphants at about 96 h, elav-expressing ciliary band associated neurons are absent from the lateral surface; however, elav-expressing ciliary band neurons remain on the ventral surface (arrows in A) above and below the mouth (Mo). Expression of elav remains in the mesoderm (not in plane of focus). (B) A surface ectodermal view of A shows the presence of anterior dorsal elav-expressing neurons in Foxg morphants. (C) In Bmp2/4 morphants at ∼96 h, loops of elav expression within the ectoderm appear similar to foxg (see Fig. 3J). (D) elav expression within the ectoderm near the pre- and postoral ciliary bands, above and below the mouth. (E) Expression of elav within the preoral ciliary band above mouth shifts anteriorly in Six3 morphants similar to foxg (see Fig. 3O). Expression of elav remains unchanged in the mesoderm (not in plane of focus in D and E). Vertical bracketed bars in (D and E) highlight the reduction in the anterior oral hood ectoderm of Six3 morphants compared with controls. (F) Transcripts of eph are found throughout the ectoderm of the CBD around 72 h. (G) Transcripts of efn are found throughout the non-CBD ectoderm in normal embryos, but (H) throughout the ectoderm in Foxg morphants. (I) Preoral (dotted lines) and postoral (arrows) elav-expressing ciliary band neurons above and below the mouth, respectively, around 72 h. The asterisk marks elav-expressing anterior dorsal neurons. (J) In ∼72-h-old Eph morphants, elav-expressing ciliary band neurons are largely absent from the lateral surface; however, elav-expressing ciliary band neurons remain on the ventral surface (arrows) and within the anterior, dorsal ectoderm (dotted line). (Magnification: 200×.)
Of the DV restricted genes, the ectodermal expression of foxg is quite different from that of other sea star regulatory genes that are later expressed within the ectoderm of the ciliary bands. It is first observed relatively much later in development and only on the ventral side of early gastrulae (Fig. 3A) (25). As development proceeds, foxg expression clears from the stomodeal (mouth) ectoderm and remains expressed within a mostly concentric domain that appears not unlike the expression of its ortholog in the sea urchin (37) (Fig. S8). A critical difference, however, is that foxg expression in sea stars is detected shortly thereafter in a domain, which we term the ciliary band domain (CBD), that starts to extend dorsally and anteriorly over the anterior pole (Fig. 3B). By larval stage, foxg expression is restricted to both ciliary bands (Fig. 3C) in what we think is the edge of its former domain. We observe a loss of localized onecut expression within the ectoderm of the ciliary bands of Foxg morphants (compare Fig. 3 D and E). This finding suggests that one of the roles of Foxg is to organize gene expression within the ectoderm of the pre- and postoral ciliary bands. Therefore, it is possible that the observed role of Onecut in neurogenesis (Fig. 2 G and H) may occur only after its expression is restricted to the ciliary bands. The spatial relationships of foxg, onecut, and elav within the ectoderm are summarized in Fig. S8.
Fig. 3.
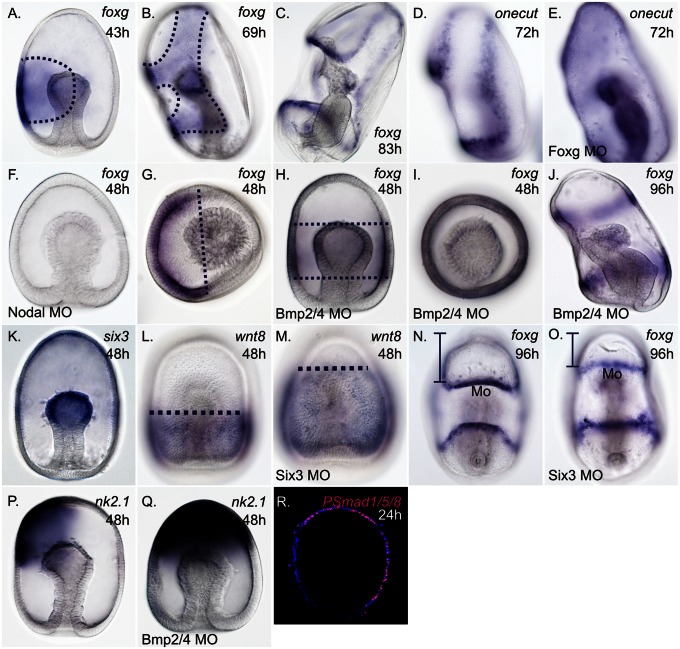
Patterning of a foxg-expressing CBD. (A) Transcripts of foxg are initially localized within one ectodermal domain on the ventral side of 43-h-old gastrulae. (B) By 69 h, foxg transcripts are cleared within the stomodeal (mouth) ectoderm and remain within in a single domain that forms around the mouth and loops over the anterior pole. (C) In 83-h-old larvae, transcripts of foxg are clearly localized within the ectoderm of both ciliated bands. (D) Transcripts of onecut localize to the ectoderm of the ciliary bands in ∼72-h-old larvae. (E) Around 72 h, transcripts of onecut are found throughout the ectoderm of Foxg morphants (MO). (F) Expression of foxg is lost from the ectoderm in Nodal morphants around 48 h. (G) Anterior pole view (APV) shows foxg transcripts within only the ventral ectoderm. (H and I) In Bmp2/4 morphants, foxg transcripts are detected within a concentric domain around the middle of gastrulae. (I) APV of foxg expression in Bmp2/4 morphants. (J) In Bmp2/4 morphants at around 96-h postfertilization, foxg is expressed in two concentric loops above and below the mouth that correspond to the pre- and postoral ciliary bands, respectively; foxg expression does not extend into the anterior pole ectoderm. (K) Transcripts of six3 localize within the anterior ectoderm at 48 h. (L) Transcripts of wnt8 localize within the posterior ectoderm at 48 h, and (M) expand anteriorly in Six3 morphants. (N) Ventral view of foxg expression within the ectoderm of the pre- and postoral ciliary bands, above and below the mouth (Mo), respectively. (O) Ventral view of foxg expression shows that preoral ciliary band above the mouth shifts anteriorly in Six3 morphants. Vertical bracketed bars in (N and O) highlight the reduction in the anterior oral hood ectoderm of Six3 morphants compared with controls. (P) Transcripts of nk2.1 within the ventral anterior pole ectoderm at the gastrula-stage. (Q) Expression of nk2.1 expands dorsally in Bmp2/4 morphants. (R) The transcriptional effector of BMP2/4 signaling, phosphorylated Smad1/5/8 (pSmad1/5/8) is localized throughout the dorsal ectoderm (pink); DAPI staining shows nuclei (blue). (Magnification: 200×.)
We next sought to understand how this foxg-CBD is shaped. Expression of foxg within the ectoderm is lost in Nodal morphants (compare Fig. 3 A and F, and Fig. S7), as is foxa expression within the stomodeal ectoderm (Fig. S7). Thus, Nodal signaling establishes two nested ectodermal domains on the ventral side of sea star gastrulae, the foxg-expressing domain that includes and surrounds the more restricted foxa-expressing stomodeal domain.
In addition, foxg is no longer polarized to the ventral surface of gastrulae when Bmp2/4 signaling is disrupted using a targeted morpholino. Expression of foxg is observed in a ring around the middle of Bmp2/4 morphants (compare Fig. 3 A and G to H and I, respectively); however, the AP boundary of foxg expression above and below the mouth does not appear to change (compare Fig. 3 A and H). This finding suggests that AP positioning of the foxg-CBD domain is established independently of DV patterning mechanisms. In 4-d-old Bmp2/4 morphants, expression of foxg is similarly radialized, although foxg expression has properly resolved into distinct pre- and postoral ciliary bands (Fig. 3J).
We next examined how AP patterning mechanisms may affect foxg expression. In vertebrates, Six3 gene products repress Wnt-mediated posteriorization of the neuroectoderm (38, 39). The sea urchin ortholog of Six3 also has the potential to repress Wnt signaling, because overexpression of Six3 represses the expression of several wnts, although their expression remains unchanged in Six3 morphants (35). In 2-d-old sea star gastrulae, six3 is expressed in the anterior ectoderm (Fig. 3K), in a domain that seemingly does not overlap with wnt8 expression within the posterior ectoderm (Fig. 3L). To determine if Six3 promotes anterior ectodermal identity by repression of wnt8, we blocked the translation of six3 using a specific translation blocking morpholino. When the function of Six3 is disrupted, the expression of wnt8 expands anteriorly, suggesting that Six3 antagonism of Wnt signaling establishes a boundary between the anterior and posterior ectoderm (compare Fig. 3 L and M) and that this mechanism is conserved between these echinoderms and vertebrates. Consistent with this finding, in Six3 morphants at 4 d postfertilization (early larval stage) there is a morphologically obvious reduction in the anterior region of larvae (i.e., the oral hood ectoderm above the mouth is reduced). In addition, the archenteron, a structure that derives from vegetal (posterior)-most epithelium and forms the endoderm and mesoderm, appears elongated. Importantly, expression of foxg indicates that the position of the preoral ciliary band above the mouth shifts anteriorly in Six3 morphants; the position postoral ciliary band below the mouth, as expected, remains relatively unchanged (compare Fig. 3 N and O). Thus, shifting the Wnt8-Six3 boundary that respectively demarcates the posterior versus anterior ectoderm, results in an anterior shift in foxg expression.
Although the finer details of how the foxg-CBD extends up through the anterior pole ectoderm are not yet understood, we suggest that a change in Bmp2/4-mediated gene expression within the anterior pole ectoderm might also be involved in the shaping of the CBD in this territory. We find that nk2.1 normally is expressed within the anterior pole ventrally to the domain of foxg expression in this territory. In Bmp2/4 morphants, nk2.1 is now expressed throughout the anterior pole (compare Fig. 3 P and Q) and foxg expression concomitantly fails to move into the anterior (Fig. 3). It is possible that DV restriction of the gene expression within the anterior pole ectoderm permits the extension of foxg expression that is observed in late gastrulae. Thus, the normal processes of ventralization and dorsalization of the anterior pole ectoderm may allow for the extension of the CBD into this territory.
In summary, we have shown that the sea star first produces a broad neurogenic potential ectoderm that is not initially patterned along the DV or AP ectoderm axis. This broad neurogenic potential ectoderm is then overlaid with a highly patterned CDB. The AP and DV patterning mechanisms that shape the expression of this CBD and rely on interactions between Nodal, BMP2/4, Wnt and Six3, at least, to establish this foxg-expression territory.
Linking Patterning to the Final Steps of Neurogenesis.
What remains to be shown is how the final differentiation to elav-expressing neurons is achieved. Crucially, we show that elav-expressing ciliary band neurons, but not anterior neurons that will likely form the dorsal ganglia, are largely absent from the ectoderm of foxg morphants (Fig. 4 A and B). We note that this aspect is unlike Soxc morphants, in which both ciliary band and anterior dorsal neurons are absent (Fig. 2I). We find that the localization of elav-expressing ciliary band neurons within the ectoderm also shifts when the shape of the foxg-CBD is altered. The expression of elav is radialized in two distinct rings above and below the mouth in these Bmp2/4 morphants (Fig. 4C) and the positioning of the elav moves anteriorly in Six3 morphants (compare Fig. 4 D and E). Importantly, the presence of elav-expressing cells does not appear to change in response to these perturbations, rather only the location of elav-positive cells changes within the ectoderm.
Therefore, elav-expressing cells can form in many locations within the ectoderm as long as they are associated with foxg and only after foxg becomes localized into ciliary bands. Additionally these elav-expressing cells only appear after phosphorylated Smad1/5/8 is no longer detected within the ectoderm. However, elav-positive cells do not overlap with foxg expression and only form along the outer edge of its domain of expression and not, for instance, within the former foxg-CBD territory (Fig. 1C). An additional mechanism is required to allow final neural differentiation and this is likely controlled by the action of some transcription factor or signaling molecule that forms a boundary demarcated by foxg expression. We observed that the ephrin receptor (eph) is expressed throughout the foxg-CBD of late gastrulae (Fig. 4F), but the ephrin ligand (efn) is expressed exactly opposite to this, throughout the non-CBD ectoderm (Fig. 4G). The expression of these genes therefore forms a border at the edge of the foxg domain. In Foxg morphants, we observed a nearly ubiquitous expression of efn throughout the ectoderm (Fig. 4H). Therefore, an additional function of Foxg is to segregate out two molecularly distinct ectodermal domains, such that there is a boundary of gene-expression profiles; this is represented by, at least, the apposed expression of eph and efn at the edge of its territory. In vertebrates, interactions of Eph and Efn can mediate directed cell migrations (40). Therefore, we suspect that one of the roles of the Eph/Efn boundary in the sea star is to guide cells to a closer association with the ciliary bands. In sea stars, Eph morphants have a reduction of differentiated neurons along the lateral parts of the ciliary bands (compare Fig. 4 I and J), which could suggest that cell migration is needed to bring predifferentiated cells from the pan-neurogenic ectoderm to a closer association with the foxg-CBD before they can express elav. It is also possible that the Eph and Efn are simply involved in establishing two distinct territories that are needed to permit final differentiation and have no direct role in neural migrations. At present, we do not know how many times soxc-expressing neural precursor cells divide and thus, for now, cannot compare numbers of soxc versus elav-expressing cells to help distinguish between these alternatives. This is a direction of further study.
Conclusions
This study was driven by the important need to understand the developmental mechanisms that lead to a complex distribution of neurons across the ectoderm of a dipleurula-like larva, which is considered the basal larval type of deuterostomes. The data presented here clearly explain how elav-expressing cells are patterned across the AP and DV axes in association with the two ciliary bands in sea stars (summarized in Fig. 5 and Fig. S9). The sea star first likely uses maternally inherited anisotropies and the cWnt pathway to distinguish endomesoderm from neurogenic ectoderm that persists through gastrulation. This endomesoderm is then overlaid with a foxg-patterning module, which is shaped by Nodal signaling from the mouth and is patterned by Bmp2/4 and Six3. Foxg, in turn, is needed to form two distinct ciliary bands at the edge of this domain and to distinguish a molecular boundary between the CBD and other ectoderm. Thus, significantly, Bmp2/4 signaling in sea stars does not distinguish differentiated neurons from nonneuronal ectoderm but instead contributes to the patterning of the CBD, which then in turn provides cues to permit the final steps of neuronal differentiation. The final fate commitment to neuronal cell type in sea star may instead be governed by a temporal loss of BMP signaling rather than a spatial gradient as observed in many other taxa. This functional modulation between specification and patterning likely reflects the evolutionary history of this GRN, in which an ancestral module for specification of a broad neurogenic potential ectoderm was subsequently overlaid with a GRN for patterning. Such patterning could allow a swimming larval form to more efficiently coordinate ciliary beating.
Fig. 5.
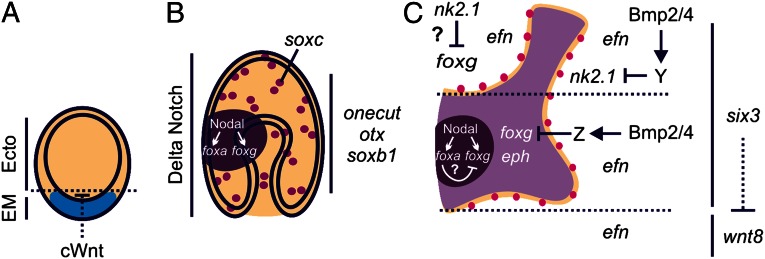
Model for localized patterning of neurons within a broad neurogenic potential ectoderm. (A) Canonical Wnt (cWnt) signaling initially establishes endomesoderm (EM, blue) from ectoderm (Ecto, tan). A suite of neurogenic genes including onecut-, otxßb-, and soxb1 are expressed throughout the ectoderm of (A) blastulae and (B) gastrulae. (B) Delta Notch signaling patterns distinct soxc-expressing cells within the broad neurogenic potential ectoderm. Nodal signaling establishes foxg and foxa expression within the ventral ectoderm (dark purple). (C) During late gastrulation, foxg expression clears from the stomodeal (mouth) ectoderm and a foxg-CBD (light purple) is shaped by Bmp2/4 signaling and Six3. The creation of this foxg-CBD, distinguishes between eph- and efn-expressing ectodermal territories. Differentiated neurons (magenta) form along this boundary. Dotted lines in C denote the position of the foxg-CBD within the ectoderm along the AP axis; six3-expressing anterior ectoderm extends upward from the bottom dotted line and includes the anterior pole ectoderm, above the top dotted line.
Experimental Procedures
Embryo Culture and Characterization of P. miniata Gene Expression.
Embryos were cultured in seawater at 15 °C and fixed for in situ hybridization, as previously described (26). Partial sequences of P. miniata orthologs were obtained via screening a P. miniata arrayed cDNA library (26) or were obtained from the publically available transcriptome. Whole-mount in situ hybridization (WMISH) and FISH was performed as previously described (25, 26).
qPCR and NanoString nCounter Assay.
RNA from developmentally staged P. miniata embryos was extracted using GenElute Mammalian Total RNA Kit (Sigma-Aldrich). cDNA was synthesized using iScript Select cDNA Synthesis Kit (Bio-Rad). Abundance of gene transcripts was determined using the nCounter Gene Expression Assay (NanoString Technologies) as described by ref. 41 and using custom P. miniata Reporter CodeSet and Capture ProbeSet.
Microinjection.
Microinjection was performed as described previously (26). Morpholino antisense oligonucleotides were designed by GeneTools. Approximately 20–40 injected embryos were used in spatial analyses of gene expression. Experiments were repeated at least twice. Similar phenotypes were observed when a second morpholino targeted to the same transcript was used. The embryos presented are representative of changes observed in greater than 90% of injected embryos.
Supplementary Material
Acknowledgments
We thank Smadar Ben-Tabou de-Leon and Lynne Angerer for critical reading of this manuscript and fruitful discussions; James Fitzpatrick and Haibing Teng for technical advice; David McClay for kindly providing the dominant-negative cadherin construct; two anonymous reviewers for their thoughtful comments; Brenna McCauley for sharing data about the temporal expression of several Patiria miniata orthologs; Adam Foote for assistance with phylogenetic analysis; and Marinus Inc. and Pete Halmay and Pat Leahy for animal collection. This work was partially supported by National Science Foundation Grant 0844948 (to V.F.H.) and Howard Hughes Medical Institute Undergraduate Education Grant 52006917 (C.S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Patiria miniata cDNA sequences reported in this paper have been deposited in the GenBank database [accession nos.: JX844799 (ephrin), JX844800 (ephrin receptor), JX844801 (elav), JX844802 (soxb1), JX844804 (soxc), JX844803 (wnt8), KC669537 (nodal), and KC669538 (bmp2/4)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220903110/-/DCSupplemental.
References
- 1.Moroz LL. On the independent origins of complex brains and neurons. Brain Behav Evol. 2009;74(3):177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Robertis EM, Sasai Y. A common plan for dorsoventral patterning in Bilateria. Nature. 1996;380(6569):37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 3.Arendt D, Nübler-Jung K. Comparison of early nerve cord development in insects and vertebrates. Development. 1999;126(11):2309–2325. doi: 10.1242/dev.126.11.2309. [DOI] [PubMed] [Google Scholar]
- 4.Mizutani CM, Bier E. EvoD/Vo: The origins of BMP signalling in the neuroectoderm. Nat Rev Genet. 2008;9(9):663–677. doi: 10.1038/nrg2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strathmann RR, Grünbaum D. Good eaters, poor swimmers: Compromises in larval form. Integr Comp Biol. 2006;46(3):312–322. doi: 10.1093/icb/icj031. [DOI] [PubMed] [Google Scholar]
- 6.Hyman LH. The Invertebrates: Echinodermata, the coelomate Bilateria. McGraw-Hill, vol IV; 1955. [Google Scholar]
- 7.Nakano H, Hibino T, Oji T, Hara Y, Amemiya S. Larval stages of a living sea lily (stalked crinoid echinoderm) Nature. 2003;421(6919):158–160. doi: 10.1038/nature01236. [DOI] [PubMed] [Google Scholar]
- 8.Garstang W. Preliminary note on a new theory of the phylogeny of the Chordata. Zool Anz. 1894;17:122–125. [Google Scholar]
- 9.Nielsen C. Origin of the chordate central nervous system—And the origin of chordates. Dev Genes Evol. 1999;209(3):198–205. doi: 10.1007/s004270050244. [DOI] [PubMed] [Google Scholar]
- 10.Lacalli TC. Apical organs, epithelial domains, and the origin of the chordate central nervous system. Am Zool. 1994;34(4):533–541. [Google Scholar]
- 11.Range RC, Angerer RC, Angerer LM. Integration of canonical and noncanonical Wnt signaling pathways patterns the neuroectoderm along the anterior-posterior axis of sea urchin embryos. PLoS Biol. 2013;11(1):e1001467. doi: 10.1371/journal.pbio.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Röttinger E, Dahlin P, Martindale MQ. A framework for the establishment of a cnidarian gene regulatory network for “endomesoderm” specification: The inputs of ß-catenin/TCF signaling. PLoS Genet. 2012;8(12):e1003164. doi: 10.1371/journal.pgen.1003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100(23):13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duboc V, Röttinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell. 2004;6(3):397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- 15.Lapraz F, Besnardeau L, Lepage T. Patterning of the dorsal-ventral axis in echinoderms: Insights into the evolution of the BMP-chordin signaling network. PLoS Biol. 2009;7(11):e1000248. doi: 10.1371/journal.pbio.1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saudemont A, et al. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 2010;6(12):e1001259. doi: 10.1371/journal.pgen.1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, Burke RD. TGFβ signaling positions the ciliary band and patterns neurons in the sea urchin embryo. Dev Biol. 2010;347(1):71–81. doi: 10.1016/j.ydbio.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascale A, Amadio M, Quattrone A. Defining a neuron: Neuronal ELAV proteins. Cell Mol Life Sci. 2008;65(1):128–140. doi: 10.1007/s00018-007-7017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koushika SP, Lisbin MJ, White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6(12):1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- 20.Lowe CJ, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113(7):853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- 21.Burke RD, et al. Neuron-specific expression of a synaptotagmin gene in the sea urchin Strongylocentrotus purpuratus. J Comp Neurol. 2006;496(2):244–251. doi: 10.1002/cne.20939. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima Y, Kaneko H, Murray G, Burke RD. Divergent patterns of neural development in larval echinoids and asteroids. Evol Dev. 2004;6(2):95–104. doi: 10.1111/j.1525-142x.2004.04011.x. [DOI] [PubMed] [Google Scholar]
- 23.Chee F, Byrne M. Development of the larval serotonergic nervous system in the sea star Patiriella regularis as revealed by confocal imaging. Biol Bull. 1999;197(2):123–131. doi: 10.2307/1542609. [DOI] [PubMed] [Google Scholar]
- 24.Bradham CA, et al. Chordin is required for neural but not axial development in sea urchin embryos. Dev Biol. 2009;328(2):221–233. doi: 10.1016/j.ydbio.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yankura KA, Martik ML, Jennings CK, Hinman VF. Uncoupling of complex regulatory patterning during evolution of larval development in echinoderms. BMC Biol. 2010;8:143. doi: 10.1186/1741-7007-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinman VF, Nguyen AT, Davidson EH. Expression and function of a starfish Otx ortholog, AmOtx: A conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mech Dev. 2003;120(10):1165–1176. doi: 10.1016/j.mod.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Otim O, Hinman VF, Davidson EH. Expression of AmHNF6, a sea star orthologue of a transcription factor with multiple distinct roles in sea urchin development. Gene Expr Patterns. 2005;5(3):381–386. doi: 10.1016/j.modgep.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126(2):345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 29.Kerner P, Simionato E, Le Gouar M, Vervoort M. Orthologs of key vertebrate neural genes are expressed during neurogenesis in the annelid Platynereis dumerilii. Evol Dev. 2009;11(5):513–524. doi: 10.1111/j.1525-142X.2009.00359.x. [DOI] [PubMed] [Google Scholar]
- 30.Bergsland M, et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25(23):2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis J. Neurogenic genes and vertebrate neurogenesis. Curr Opin Neurobiol. 1996;6(1):3–10. doi: 10.1016/s0959-4388(96)80002-x. [DOI] [PubMed] [Google Scholar]
- 32.Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA. 2007;104(49):19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: Insights from sea urchin development. Development. 2011;138(17):3613–3623. doi: 10.1242/dev.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yaguchi S, Yaguchi J, Burke RD. Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development. 2006;133(12):2337–2346. doi: 10.1242/dev.02396. [DOI] [PubMed] [Google Scholar]
- 35.Wei Z, Yaguchi J, Yaguchi S, Angerer RC, Angerer LM. The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development. 2009;136(7):1179–1189. doi: 10.1242/dev.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22(11):2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 37.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev Biol. 2006;300(1):49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 38.Lagutin OV, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17(3):368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavado A, Lagutin OV, Oliver G. Six3 inactivation causes progressive caudalization and aberrant patterning of the mammalian diencephalon. Development. 2008;135(3):441–450. doi: 10.1242/dev.010082. [DOI] [PubMed] [Google Scholar]
- 40.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6(6):462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 41.Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr Patterns. 2010;10(4-5):177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.