Abstract
Berninamycin is a member of the pyridine-containing thiopeptide class of antibiotics that undergoes massive posttranslational modifications from ribosomally generated preproteins. Berninamycin has a 2-oxazolyl-3-thiazolyl-pyridine core embedded in a 35-atom macrocycle rather than typical trithiazolylpyridine cores embedded in 26-atom and 29-atom peptide macrocycles. We describe the cloning of an 11-gene berninamycin cluster from Streptomyces bernensis UC 5144, its heterologous expression in Streptomyces lividans TK24 and Streptomyces venezuelae ATCC 10712, and detection of variant and incompletely processed scaffolds. Posttranslational maturation in S. lividans of both the wild-type berninamycin prepeptide (BerA) and also a T3A mutant generates macrocyclic compounds as well as linear variants, which have failed to form the pyridine and the macrocycle. Expression of the gene cluster in S. venezuelae generates a variant of the 35-atom skeleton of berninamycin, containing a methyloxazoline in the place of a methyloxazole within the macrocyclic framework.
Keywords: heterologous expression, linear precursors, prepeptide gene replacement
The thiazolyl peptide (thiopeptide) antibiotics, first discovered in 1948, now number almost one hundred members and encompass a wide range of chemical diversity (1–8). They can be classified into subgroups by several criteria, including the size of the macrocyclic ring(s), the oxidation state and substitution pattern of the central pyridine core, and also by the nature of the target in susceptible bacteria. These antibiotics are effective against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), and function by disrupting one of two steps in bacterial protein biosynthesis: (i) blockade of elongation factor binding to the 50S ribosomal subunit (9–11) or (ii) by binding to the conditional GTPase elongation factor Tu (EF-Tu) and inhibiting its aminoacyl-tRNA chaperone activity (12–15). There are crystal structures of thiostrepton or nosiheptide bound to the 50S ribosomal subunit (10) and of GE2270 bound to EF-Tu (12), giving detailed information about interactions of these constrained antibiotic scaffolds with their respective targets.
Thiostrepton (tetrahydropyridine core) and nosiheptide (hydroxypyridine core), two thiopeptides that exemplify the 26-membered ring class, have a central nitrogen heterocycle substituted at positions 2, 3, and 6 with three thiazoles. This tetracyclic core is embedded within a peptidyl macrocycle, which forms a rigid pharmacophore that contributes to tight target binding. Both of these antibiotics also have a second macrocyclic ring that adds additional conformational constraint to the scaffold. Thiocillins have a trithiazolylpyridine core embedded within a single 26-membered macrocycle. We have recently shown that the peptide macrocycle of thiocillins is produced late in the maturation process, in the same posttranslational cyclization step that builds the central pyridine ring (16). All these thiazolyl peptide antibiotics with a 26-member macrocycle bind to the 50S ribosome.
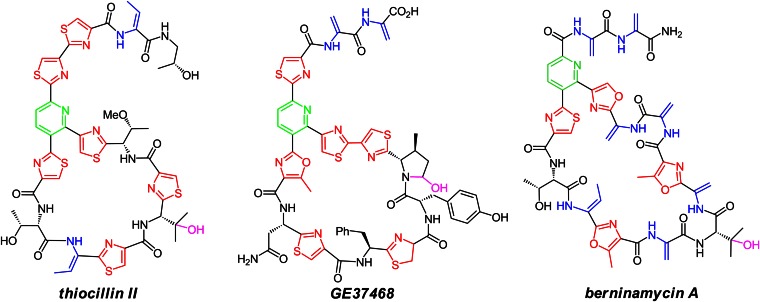
On the other hand, three 29-membered macrocyclic family members, GE2270, GE37468, and thiomuracin A, all inhibit EF-Tu but not the 50S ribosome (12–15, 17, 18). The molecular basis of thiazolylpyridine macrocyclic ring size as determinant of ribosomal versus elongation factor targeting is not yet known. One additional macrocyclic ring size in the thiopeptide antibiotic class is represented by the berninamycins, sulfomycin (19), thioplabin (20), and TP-1161A (21, 22). This class has a unique oxazolyl-thiazolyl-pyridine core embedded in a 35-membered (13-residue) peptidyl macrocycle (Fig. 1) (23–25). The antibacterial mechanism of TP-1161, recently isolated from a marine Nocardiopsis species (21, 22), has not been disclosed, whereas berninamycin is reported to target the 50S ribosome, similar to thiostrepton (26, 27).
Fig. 1.
Macrocyclic thiopeptide antibiotics: 26-membered ring thiocillin II, 29-membered ring GE37468, and 35-membered ring berninamycin A.
In recent years it has become clear that the various thiazolylpyridine antibiotic scaffolds arise from extensive posttranslational modification (PTM) of ribosomally generated preproteins of approximately 50–60 residues (14, 17, 21, 22, 28–31), where the core 14–16 residues that end up in the mature antibiotic are at the C termini, downstream of leader peptide sequences of the protein precursors (6, 32). We have been among the groups that have reported gene clusters for the ribosome-targeting 26-membered macrocycle class (thiocillins) (28) and the EF-Tu-targeting 29-membered macrocycles (GE37468) (17, 18). In this study, we have turned to the 35-membered macrocycle scaffold and identified a candidate biosynthetic gene cluster for berninamycin from a draft genome sequence of the known producer Streptomyces bernensis UC 5144. The cluster was validated by heterologous expression from both Streptomyces lividans and Streptomyces venezuelae. Examination of the product distribution in S. lividans reveals linear intermediates, which give insights into the timing of various posttranslational modifications during the maturation process. We also observe an altered macrocyclic variant from S. venezuelae expression, where a methyloxazole residue within the 35-atom ring has been left at a methyloxazoline stage.
Results
Identification of a Candidate Berninamycin Gene Cluster from S. bernensis UC 5144 Through Genome Mining.
The genome of S. bernensis UC 5144 (S. bernensis) has been sequenced by next-generation Illumina sequencing technology and a portion of the data assembled into two contigs, comprising the suite of genes normally associated with thiopeptide production as well as the predicted serine-rich structural gene of berninamycin A. The two contigs were connected through PCR amplification of genomic DNA and traditional Sanger sequencing. Finally, a primer-walking strategy was adopted to extend the Illumina sequencing coverage and identify the boundaries of the cluster (SI Appendix).
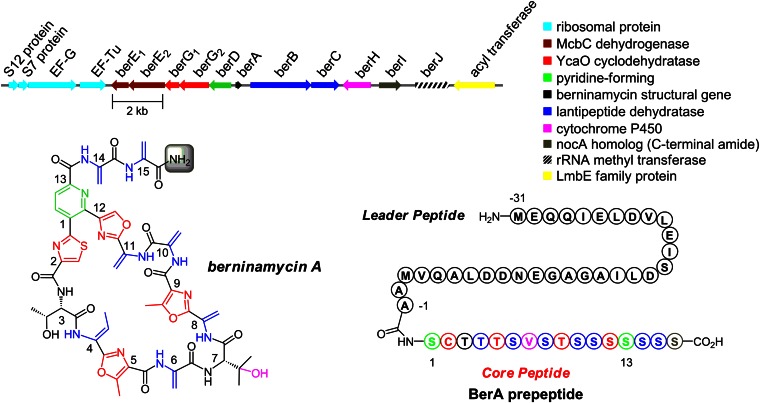
The candidate cluster consists of 11 ORFs (berA–J) spanning 12.9 kb, bordered at one end by ribosomal and translation-associated proteins and at the other by an acyl transferase, presumably not involved in berninamycin production (Fig. 2). The berninamycin structural gene (berA) encompasses a 31-aa leader peptide and a C-terminal 16-aa core peptide, where 10 of the 16 amino acids are serine. Ser16 itself is not found in the mature scaffold but is instead cleaved to reveal the C-terminal amide, presumably by the action of BerI, which is homologous to NocA and NosA from nocathiacin and nosiheptide biosynthesis, respectively (33). The remainder of the cluster contains many of the hallmarks of thiopeptide synthesis: lantipeptide-like dehydratases BerB-C for installing the dehydroalanine (Dha) and dehydrobutyrine (Dhb) functional groups; the purported pyridine-forming BerD; and McbC-like dehydrogenases BerE1 and BerE2 and YcaO-type cyclodehydratases BerG1 and BerG2, together responsible for thiazole, oxazole, and methyloxazole synthesis. Additionally, cytochrome P450 BerH is likely involved in hydroxylation at Val7 and berJ encodes for a 23S-rRNA methyl transferase, a well known mechanism in establishing resistance to thiostrepton (27, 34, 35). Overall, the cluster bears a great deal of resemblance to the reported gene cluster for 35-membered ring thiopeptide TP-1161A (SI Appendix, Fig. S1 and Table S1) (21, 22).
Fig. 2.
The berninamycin A gene cluster and the BerA prepeptide.
Expression of the Cluster in the Heterologous Hosts S. lividans TK24 and S. venezuelae ATCC 10712 and Evaluation of the BerA Prepeptide Conversion to the Mature Antibiotic.
We next focused our attention to transferring the cluster to a model host for heterologous expression, which would have two distinct advantages: (i) expression of berninamycin in a heterologous host would confirm the identity and completeness of the gene cluster and (ii) it would allow us to test whether berninamycin analogs could be prepared through simple mutations of the BerA prepeptide. The complete gene cluster was isolated through screening of a genomic fosmid library followed by restriction digest, and was inserted into the pSET152 shuttle vector to yield the 22.7-kb pSET152+bern plasmid (SI Appendix, Fig. S2).
The plasmid was then integrated into the genome of S. lividans TK24 (S. lividans) and S. venezuelae ATCC 10712 (S. venezuelae) by conjugative transfer to produce S. lividans/pSET152+bern and S. venezuelae/pSET152+bern, respectively. As controls, the unmodified pSET152 plasmid (lacking the presumed berninamycin gene cluster) was transferred to S. lividans and S. venezuelae genomes to furnish S. lividans/pSET152 and S. venezuelae/pSET152, respectively (SI Appendix).
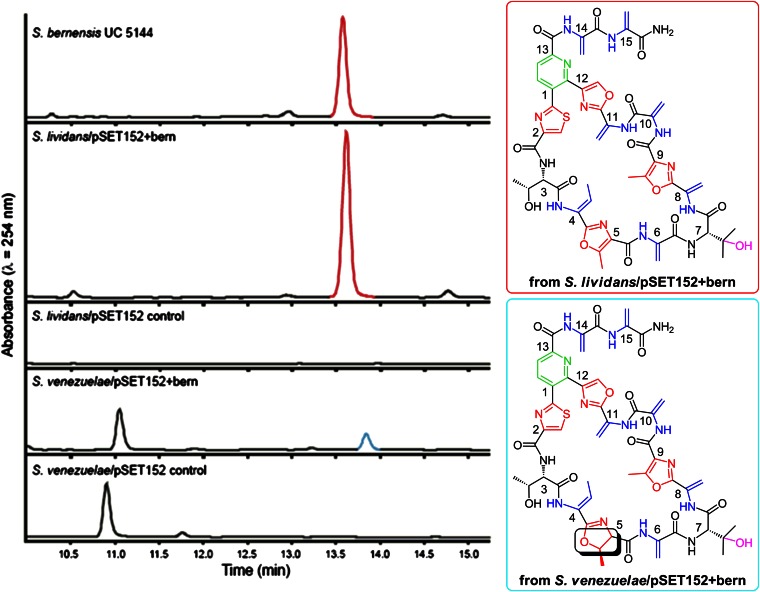
Small-scale cultures revealed that S. lividans/pSET152+bern is capable of producing berninamycin A at a similar level to the wild-type S. bernensis (2.4 times the quantity produced by S. bernensis, Fig. 3); however, the level of expression was variable (SI Appendix). [Berninamycin B (1.5% of berninamycin A), a small amount of berninamycin D (approximately 1.0% of berninamycin A), resulting from hydrolysis of the C-terminal tail, as well as a trace amount of berninamycin C (obscured by berninamycin A), from partial hydrolysis of the C-terminal tail, were obtained from S. bernensis cultures, as initially reported by Rinehart and coworkers (ref. 24; SI Appendix).] The macrocycle could not be detected from the S. lividans/pSET152 under identical conditions, demonstrating that the collection of 11 genes (berA–J) is sufficient for producing berninamycin.
Fig. 3.
Liquid chromatography traces (Left) of berninamycin A (Upper Right) expression from the natural producer S. bernensis UC 5144 and the heterologous host S. lividans/pSET152+bern (red peak); synthesis of an altered 35-membered ring macrocycle (Lower Right) from heterologous host S. venezuelae/pSET152+bern (blue peak). The stereochemistry shown for this latter metabolite is assumed based on the structure of berninamycin A and its prepeptide and has not been confirmed spectroscopically.
Similar to the S. bernensis expression, S. lividans/pSET152+bern also delivers a substantially reduced amount of berninamycin B (3.9% of berninamycin A), which lacks hydroxylation at Val7 (24). [A small amount of berninamycin D (<1.0% the quantity of berninamycin A) could be detected in 50-mL expression cultures of S. lividans/pSET152+bern; however, berninamycin C, was not observed (SI Appendix).] A small quantity of a compound bearing two hydroxylations at Val7 (approximately 1.0% of berninamycin A), a metabolite that has not been observed previously (SI Appendix, Fig. S3) but that has an analogous double hydroxylation of an isoleucine residue in thiostrepton and thiomuracin maturation (13, 29), is also produced in both S. bernensis and S. lividans/pSET152+bern cultures. The site of the second hydroxylation was determined by MS/MS fragmentation studies (SI Appendix).
S. venezuelae/pSET152+bern expression also results in macrocycle production, albeit at a reduced level (21% the quantity produced by S. bernensis); however, high-resolution LCMS analysis showed that rather than producing berninamycin A (M+H+ = 1,146.3483 m/z), a compound with a 2-Da increase in mass is formed (M+H+ = 1,148.3639 m/z). Subsequent MS/MS fragmentation analysis and chemical derivitization studies (SI Appendix) revealed that the metabolite contains a methyloxazoline, rather than the normal methyloxazole of berninamycin, at position 5 (Fig. 3). Similarly, thiazolines, rather than thiazoles, are naturally found at or near this position of the macrocycle in GE37468 and thiostrepton. Whereas berninamycin displays modest activity against Gram-positive bacteria such as Bacillus subtilis [minimum inhibitory concentration (MIC) = 6.3 μM] and MRSA (MIC = 10.9 μM), the analog produced by S. venezuelae is inactive as an antibiotic (MIC > 200 μM against B. subtilis), suggesting the importance of structural rigidity at this fifth residue in binding to the 50S ribosome. Neither berninamycin nor the M+2 compound were detected in the S. venezuelae/pSET152 control.
Partial Processing of BerA in S. lividans/pSET152+bern Results in Linear Intermediates in both Wild-Type and Mutant Strains.
Although the core peptide of BerA is dominated by the presence of serines, the differential processing of Thr3–5 (Thr3 is unmodified, Thr4 is dehydrated to a dehydrobutyrine, and Thr5 is cyclodehydrated and oxidized to a methyloxazole) provides an opportunity for understanding the role of each residue in ultimately forming the macrocyclic ring of berninamycin (Fig. 2) and its binding to its biological target. Therefore, T3A, T4A, and T5A mutations were individually introduced into the berA gene through a PCR-directed mutagenesis (SI Appendix, Fig. S4) and incorporated into S. lividans TK24 via conjugative transfer (SI Appendix).
Whereas the T4A and T5A mutants do not generate any detectable macrocyclic products, the T3A BerA prepeptide is converted to the fully processed 35-membered ring along with the metabolites lacking Val7 hydroxylation and with double hydroxylation (at Cβ and Cγ) at that residue in direct analogy to the wild-type expression. The T3A variant of berninamycin does not show antimicrobial activity against B. subtilis (MIC > 400 μM); mutation to this residue in thiocillin has similarly abolished the compound’s ability to act as an antibiotic (36, 37).
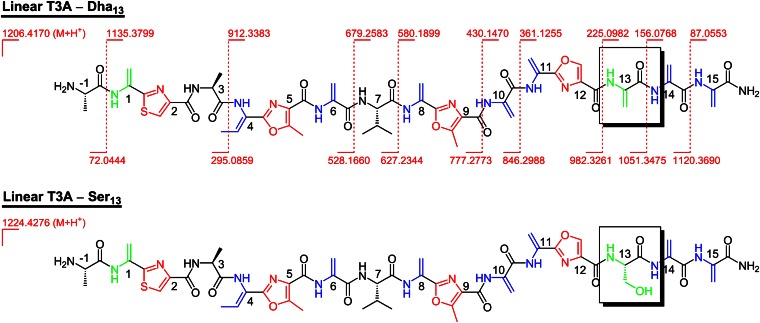
Further examination of the S. lividans/pSET152+bern T3A mutant expression revealed the presence of two additional major metabolites that elute before the 35-membered macrocycle (SI Appendix). High-resolution mass spectrometry determined the more polar of the two to have a 1,224.4276-Da mass (M+H+) and the other compound to have a 1,206.4170-Da mass (M+H+), suggesting a loss of water.
The masses, taken together with MS/MS fragmentation of each compound, indicate that the two metabolites are linear precursors to the macrocyclic structure, where almost all of the leader peptide has been truncated (Fig. 4); fragmentation sites for the dehydrated compound are shown and are identical to those found in the more polar hydrate. Both compounds contain a C-terminal amide, an unmodified Val7 residue, and an N-terminal Ala−1 as the sole remaining amino acid of the leader peptide (as the free amine). A linear intermediate, where the majority of the leader peptide has been cleaved, has been observed for thiocillin production in Bacillus cereus, but only in a mutant where tclM, the gene encoding for the pyridine-forming enzyme, has been knocked out (16). Furthermore, analysis of the MS/MS data restricts the hydration site to Ser13 in the 1,224.4276-Da compound (linear T3A–Ser13), one of the residues that takes part in the pyridine-forming event to make the 35-membered ring macrocycle. For the dehydrated compound (linear T3A–Dha13), all PTMs of the core peptide en route to the macrocycle have taken place apart from pyridine formation and valine hydroxylation.
Fig. 4.
Linear metabolites from S. lividans/pSET152+bern (T3A) expression, resulting from interruption of macrocycle production by proteolysis of the leader peptide; observed MS/MS fragmentation sites are highlighted for the linear T3A–Dha13 compound and are identical in the linear T3A–Ser13 metabolite. The stereochemistry shown for these linear metabolites is based on the structure of berninamycin A and its prepeptide and has not been confirmed spectroscopically.
The two linear metabolites are also observed in S. lividans/pSET152+bern expression of wild-type berninamycin and in the expression of the T4A mutant of S. lividans/pSET152+bern (SI Appendix, Fig. S5); the linear compounds have not been observed for the T5A mutant. Additionally, the linear precursors have not been found in the S. venezuelae heterologous expressions; however, when fermentations of the native producer, S. bernensis, were reexamined for these specific linear metabolites, we discovered that the linear precursor to berninamycin, bearing a Dha13, is detectable, but the corresponding compound with an unprocessed Ser13 has not been found.
Discussion
The isolation of linear precursors to the berninamycin macrocycle from the S. lividans heterologous expressions provides a glimpse into the possible timing of late-stage events in the many posttranslational maturation steps and also into the importance of key structural residues for mature scaffold assembly. Both linear compounds have undergone nearly all of the PTMs required for berninamycin synthesis, including all thiazole and oxazole formations, Dha and Dhb formations, and C-terminal amide formation by BerI. It has been demonstrated that NosA, the BerI homolog involved in nosiheptide biosynthesis, is capable of acting on a macrocyclized substrate to generate the C-terminal amide (33); however, these data indicate that BerI reacts with a linear precursor in berninamycin construction.
Val7 lacks hydroxylation in both linear compounds; taken together with the presence of mature macrocycle lacking hydroxylation at that residue (berninamycin B) along with singly hydroxylated (berninamycin A), and doubly hydroxylated (a previously uncharacterized compound) macrocycle, the data indicate that cytochrome P450 BerH acts after scaffold macrocyclization. [Lack of valine hydroxylation was also observed in the linear compound obtained from the tclM knockout in the thiocillin gene cluster (16).]
More important is the difference between the two linear compounds, the dehydration of Ser13, one of the two Dha moieties to take part in pyridine formation, or the lack thereof. With all other essential PTMs in place, the observation of the dehydrated as well as unprocessed residue 13 in the two compounds suggests that dehydration at this amino acid occurs just before pyridine formation, even though there are several serines and one threonine that are dehydrated both upstream and downstream of Ser13. This finding could be explained by the presence of two lantipeptide dehydratases in the berninamycin gene cluster, BerB and -C. One of the enzymes may carry out the phosphorylation/elimination sequence at several serines and Thr4, whereas the other dehydratase may subsequently act on the remaining unmodified residues. In one scenario, one dehydratase might process Ser1,6,8,10–11,14–16 and Thr4 in an N-to-C direction, whereas the other may be dedicated to reaction at Ser13 (SI Appendix, Fig. S6). In that case, the presence of oxazole12 may assist in orchestrating the position of enzymatic modification to the berninamycin prepeptide, halting the action of one dehydratase at Ser13 and directing the action of the other. All of the reported thiazolyl peptide gene clusters contain two lantipeptide dehydratases, perhaps indicating that separation of dehydration events might be a general phenomenon in thiopeptide biosynthesis. Future gene deletion studies may allow evaluation of such regioselective dehydrative functions in this and other thiopeptide gene clusters.
The presence of the two linear compounds in the S. lividans expressions with wild-type and T3A berA in the presence of a functional pyridine-forming enzyme, BerD, is likely due to an imbalance in expression of Ber proteins in that heterologous host. Underexpression of (active) BerD may lead to early adventitious proteolysis of the leader peptide, thus prohibiting BerD from recognizing its substrate. Similarly, the loss of the leader peptide may compete with dehydration at Ser13 by the required dehydratase, again due to a change in rates through underexpression of the enzyme. (Lantibiotic-type dehydratases such as BerB and BerC are known to require the upstream leader sequences for downstream Ser and Thr phosphorylation/elimination chemistry; ref. 32.) For the T4A mutant, however, only the two linear compounds can be detected, which probably does not reflect competitive proteolysis of the leader region but rather the inability for BerD to cyclize a substrate bearing an sp3-hybridized residue at position 4 instead of the normally sp2-hybridized Dhb4. For the T5A mutant, neither linear nor cyclized metabolites are observed, demonstrating the need for a rigidifying methyloxazole (or methyloxazoline as observed in the S. venezuelae heterologous expression) at that site and implying the early action of the cyclodehydratase enzymes in the berninamycin pathway. The incomplete processing of berninamycin in the S. bernensis strain, although minor, suggests a partial failure of posttranslational processing late in the PTM cascade even in the native producer.
Conclusions
We have identified the biosynthetic gene cluster for the production of thiopeptide berninamycin A in S. bernensis through genome sequencing and confirmed the assignment by heterologous expression of the antibiotic in S. lividans TK24 and S. venezuelae ATCC 10712. Mutations to the berA structural gene have given insight into the structural requirements for posttranslational modifications of the BerA prepeptide. A number of metabolites have been observed, including linear precursors to berninamycin from S. lividans heterologous expressions, which have provided insight into the possible late-stage timing of events in berninamycin synthesis and perhaps suggest a general strategy in thiopeptide production.
Materials and Methods
Additional and more detailed procedures and data analysis can be found in the SI Appendix.
Identification of a Candidate Berninamycin Gene Cluster and Isolation Through Construction of a Genomic Library.
S. bernensis UC 5144 (NRRL 3575) was grown in LB media for 3 d, and genomic DNA was isolated by chloroform extraction followed by isopropanol precipitation. A portion of the genomic DNA was used for next-generation Illumina sequencing, and the portion of the data that related to thiopeptide production (including the serine-rich BerA prepeptide) was assembled. Another portion of genomic DNA was used to generate a fosmid library in the pCC2FOS vector according to the CopyControl Fosmid Library Production kit protocol (Epicentre Biotechnologies). The library was screened according to established procedures (17), and a single clone identified that contained the entire candidate berninamycin gene cluster (established by PCR and traditional sequencing). The gene cluster was isolated from the fosmid through restriction digest (SbfI-HF and NotI-HF).
Synthesis of the pSET152+bern Plasmid and Conjugative Transfer to S. lividans and S. venezuelae.
To introduce the cluster into the pSET152 vector, two SbfI restriction sites were first removed from the vector and SbfI and NotI restriction sites introduced into the lacZ gene by site-directed PCR mutagenesis (SI Appendix). The vector was then subjected to restriction digest (SbfI-HF and NotI-HF) and ligated to the berninamycin gene cluster, obtained from the fosmid library screen, to yield the pSET152+bern plasmid. The plasmid was integrated into the genomes of S. lividans TK24 and S. venezuelae ATCC 10712 through conjugative transfer from ET12567/pUZ8002 E. coli according to an established procedure (38) to furnish S. lividans/pSET152+bern and S. venezuelae/pSET152+bern (SI Appendix, Fig. S2).
The T3A, T4A, and T5A mutations to the berA gene in the pSET152+bern plasmid were introduced through site-directed mutagenesis (SI Appendix, Fig. S4). Restriction digest of pSET152+bern with SpeI and NsiI yielded the pSET152+bern vector, lacking the berD, berA, and berB genes. Additionally, two PCRs were run with the pSET152+bern plasmid as template to introduce the mutation into the berA gene. The products from these two reactions were linked through another PCR and the product of this overlap PCR (comprised of the berD, mutant berA, and berB genes) was then subjected to restriction digest (SpeI and NsiI). These thus-generated mutant inserts and the pSET152+bern vector were then ligated to furnish the mutant pSET152+bern plasmids. The gene cluster with the mutant berA gene was introduced to S. lividans through conjugative transfer.
Expression and Isolation of Berninamycin and Analogs.
Starter cultures (25 mL of Seed media) were begun from spores of Streptomyces taken from soy flour mannitol plates (3–4 d, 30 °C); cultures were grown for 3 d at 30 °C [250 rpm shaking in 250-mL baffled flasks with ColiRollers glass beads (Novagen)]. At that time, 50-mL expression cultures (AF/MS or GYM media) were initiated by inoculation with 0.5 mL of starter culture and were grown for 4 d at 30 °C [250 rpm shaking in 250-mL baffled flasks with ColiRollers glass beads (Novagen)]. At that time, 10 mL of each expression culture was pelleted by centrifugation and the supernatant discarded. The cell pellet was extracted with acetone containing anhydrous Na2SO4 by shaking and vortexing vigorously for 30 min. The mixture was then filtered through a cotton plug, and the volatiles removed in vacuo. The residue was dissolved in 0.5 mL of 50:50 acetonitrile:water and filtered through a 0.45-μM filter for high-resolution LC-MS analysis.
Preparative amounts of berninamycin A and its analogs were obtained from large scale fermentations at the NERCE/BEID core facility. Starter cultures were initiated as described above and were used to inoculate 5 L of media in fermentors, maintaining oxygen in excess and the pH at 7.0 for the duration of the fermentation (SI Appendix). The cell pellets obtained from these fermentation cultures were extracted analogously to the procedure described above. The unpurified cell extract was dissolved in 50:50 acetonitrile:water and purified by preparative scale HPLC to obtain analytically pure macrocyclic thiopeptides (SI Appendix, Fig. S2).
Supplementary Material
Acknowledgments
We thank Dr. Timothy Wencewicz for careful reading of this manuscript, Dr. Robin Ross and Mr. Benjamin Seiler (Harvard Medical School, New England Regional Center of Excellence/Biodefense and Emerging Infectious Diseases Biomolecule Production Core Facility) for large-scale fermentations of heterologous expression hosts, Dr. Albert Bowers for NMR characterization of berninamycin A, and Prof. Mervyn Bibb (John Innes Centre) for helpful suggestions and donation of the S. venezuelae ATCC 10712 strain. This work was supported by New England Regional Center of Excellence Grant NIAID057159 (to C.T.W.) and National Institutes of Health (NIH) Grant GM20011 (to C.T.W.). S.J.M. and T.S.Y. were supported by NIH NRSA postdoctoral Fellowships F32AI098147 and F32GM098051, respectively.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KC894738).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307111110/-/DCSupplemental.
References
- 1.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105(2):685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 2.Arndt H-D, Schoof S, Lu J-Y. Thiopeptide antibiotic biosynthesis. Angew Chem Int Ed Engl. 2009;48(37):6770–6773. doi: 10.1002/anie.200901808. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat Prod Rep. 2010;27(2):153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT, Acker MG, Bowers AA. Thiazolyl peptide antibiotic biosynthesis: A cascade of post-translational modifications on ribosomal nascent proteins. J Biol Chem. 2010;285(36):27525–27531. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh CT, Malcolmson SJ, Young TS. Three ring posttranslational circuses: Insertion of oxazoles, thiazoles, and pyridines into protein-derived frameworks. ACS Chem Biol. 2012;7(3):429–442. doi: 10.1021/cb200518n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Liu W. Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat Prod Rep. 2013;30(2):218–226. doi: 10.1039/c2np20107k. [DOI] [PubMed] [Google Scholar]
- 8.Dunbar KL, Mitchell DA. Revealing Nature’s synthetic potential through the study of ribosomal natural product biosynthesis. ACS Chem Biol. 2013;8(3):473–487. doi: 10.1021/cb3005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron DM, Thompson J, March PE, Dahlberg AE. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J Mol Biol. 2002;319(1):27–35. doi: 10.1016/S0022-2836(02)00235-8. [DOI] [PubMed] [Google Scholar]
- 10.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Mikolajka A, et al. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem Biol. 2011;18(5):589–600. doi: 10.1016/j.chembiol.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heffron SE, Jurnak F. Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 A resolution: Atomic basis for GE2270A inhibition of EF-Tu. Biochemistry. 2000;39(1):37–45. doi: 10.1021/bi9913597. [DOI] [PubMed] [Google Scholar]
- 13.Parmeggiani A, et al. Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu. Biochemistry. 2006;45(22):6846–6857. doi: 10.1021/bi0525122. [DOI] [PubMed] [Google Scholar]
- 14.Morris RP, et al. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc. 2009;131(16):5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 15.LaMarche MJ, et al. Antibiotic optimization and chemical structure stabilization of thiomuracin A. J Med Chem. 2012;55(15):6934–6941. doi: 10.1021/jm300783c. [DOI] [PubMed] [Google Scholar]
- 16.Bowers AA, Walsh CT, Acker MG. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J Am Chem Soc. 2010;132(35):12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young TS, Walsh CT. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci USA. 2011;108(32):13053–13058. doi: 10.1073/pnas.1110435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young TS, Dorrestein PC, Walsh CT. Codon randomization for rapid exploration of chemical space in thiopeptide antibiotic variants. Chem Biol. 2012;19(12):1600–1610. doi: 10.1016/j.chembiol.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egawa Y, Umino K, Tamura Y, Shimizu M, Kaneko K. Sulfomycins, a series of new sulfur-containing antibiotics. I. Isolation, purification and properties. J Antibiot (Tokyo) 1969;22(1):12–17. doi: 10.7164/antibiotics.22.12. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama S, Wada Y, Hasumi K. Antibiotic A10255 (thioplabin) enhances fibrin binding and activation of plasminogen. J Antibiot (Tokyo) 2002;55(1):83–91. doi: 10.7164/antibiotics.55.83. [DOI] [PubMed] [Google Scholar]
- 21.Engelhardt K, et al. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl Environ Microbiol. 2010;76(15):4969–4976. doi: 10.1128/AEM.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelhardt K, Degnes KF, Zotchev SB. Isolation and characterization of the gene cluster for biosynthesis of the thiopeptide antibiotic TP-1161. Appl Environ Microbiol. 2010;76(21):7093–7101. doi: 10.1128/AEM.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe H, Kushida K, Shiobara Y, Kodama M. The structures of sulfomycin I and berninamycin A. Tetrahedron Lett. 1988;29:1401–1404. [Google Scholar]
- 24.Lau RCM, Rinehart KL. Berninamycins B, C, and D, minor metabolites from Streptomyces bernensis. J Antibiot (Tokyo) 1994;47(12):1466–1472. doi: 10.7164/antibiotics.47.1466. [DOI] [PubMed] [Google Scholar]
- 25.Lau RCM, Rinehart KL. Biosynthesis of berninamycin: Incorporation of 13C-labeled amino acids. J Am Chem Soc. 1995;117:7606–7610. [Google Scholar]
- 26.Reusser F. Mode of action of berninamycin. An inhibitor of protein biosynthesis. Biochemistry. 1969;8(8):3303–3308. doi: 10.1021/bi00836a026. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J, Cundliffe E, Stark MJR. The mode of action of berninamycin and mechanism of resistance in the producing organism, Streptomyces bernensis. J Gen Microbiol. 1982;128(4):875–884. doi: 10.1099/00221287-128-4-875. [DOI] [PubMed] [Google Scholar]
- 28.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci USA. 2009;106(8):2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: Prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131(12):4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 30.Liao R, et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16(2):141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, et al. Identification and analysis of the biosynthetic gene cluster encoding the thiopeptide antibiotic cyclothiazomycin in Streptomyces hygroscopicus 10-22. Appl Environ Microbiol. 2010;76(7):2335–2344. doi: 10.1128/AEM.01790-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oman TJ, van der Donk WA. Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6(1):9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y, et al. NosA catalyzing carboxyl-terminal amide formation in nosiheptide maturation via an enamine dealkylation on the serine-extended precursor peptide. J Am Chem Soc. 2010;132(46):16324–16326. doi: 10.1021/ja106571g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cundliffe E. Mechanism of resistance to thiostrepton in the producing-organism Streptomyces azureus. Nature. 1978;272(5656):792–795. doi: 10.1038/272792a0. [DOI] [PubMed] [Google Scholar]
- 35.Cundliffe E, Thompson J. Ribose methylation and resistance to thiostrepton. Nature. 1979;278(5707):859–861. doi: 10.1038/278859a0. [DOI] [PubMed] [Google Scholar]
- 36.Acker MG, Bowers AA, Walsh CT. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J Am Chem Soc. 2009;131(48):17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: Structure, conformation, and activity of heterocycle substitution mutants. J Am Chem Soc. 2010;132(21):7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kieser T, Bibb MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.