Abstract
The regulated binding of effector proteins to the nucleosome plays a central role in the activation and silencing of eukaryotic genes. How this binding changes the properties of chromatin to mediate gene activation or silencing is not fully understood. Here we provide evidence that association of the budding yeast silent information regulator 3 (Sir3) silencing protein with the nucleosome induces a conformational change in the amino terminus of histone H4 that promotes interactions between the conserved H4 arginines 17 and 19 (R17 and R19) and nucleosomal DNA. Substitutions of H4R17 and R19 with alanine abolish silencing in vivo, but have little or no effect on binding of Sir3 to nucleosomes or histone H4 peptides in vitro. Furthermore, in both the previously reported crystal structure of the Sir3-bromo adjacent homology (BAH) domain bound to the Xenopus laevis nucleosome core particle and the crystal structure of the Sir3-BAH domain bound to the yeast nucleosome core particle described here, H4R17 and R19 make contacts with nucleosomal DNA rather than with Sir3. These results suggest that Sir3 binding generates a more stable nucleosome by clamping H4R17 and R19 to nucleosomal DNA, and raise the possibility that such induced changes in histone–DNA contacts play major roles in the regulation of chromatin structure.
Keywords: gene silencing, histone H4 tail
Assembly of eukaryotic DNA into chromatin plays a central role in the regulation of gene expression and genome stability. The fundamental unit of chromatin folding, the nucleosome, is composed of 147 bp of DNA wrapped twice around an octamer of histones H2A, H2B, H3, and H4 (1). Posttranslational modifications of histones play important roles in the regulation of chromatin structure by affecting the interaction of histones with nucleosomal DNA and by recruiting effector molecules that perform downstream functions (2–6). Although many different types of chromatin domains have been defined based on histone modification patterns (7), the active gene-rich and inactive, repetitive, gene-poor chromosome regions are commonly referred to as euchromatin and heterochromatin, respectively.
In the budding yeast Saccharomyces cerevisiae, the silent mating type cassettes and telomeric DNA regions are assembled into heterochromatin-like structures that display epigenetic inheritance patterns and regional effects on gene expression (8–10). Studies by Grunstein and coworkers (11, 12) provided the first evidence of a specific role for histones in silencing. The conserved amino terminus of histone H4 is dispensable for growth but is required for repression of the silent mating type loci (11, 12). In the H4 amino terminus, substitutions within a basic patch region, composed of lysine 16 (K16), arginine 17 (R17), histidine 18 (H18), and arginine 19 (R19), abolish silencing, whereas substitution of lysine 20 (K20) has a partial silencing defect (12). Furthermore, H4K16 is hypoacetylated within silent domains, and although its substitution to arginine is tolerated, substitutions to alanine and glutamine abolish silencing, providing evidence that H4K16 acetylation regulates silencing in vivo (12, 13). In addition to histone H4, the globular domain of histone H3 surrounding lysine 79 (K79), termed the loss of rDNA silencing (LRS) surface, is required for silencing (14–17).
The establishment and maintenance of silent domains at telomeres and the mating type loci also requires the silent information regulator (Sir) 2, 3, and 4 proteins (18, 19). The Sir2 and Sir4 proteins form a subcomplex that associates with Sir3 into the silent infomation regulator (SIR) complex (20–23). Sir2 is a NAD-dependent deacetylase with preference for H4K16 (24-26), whereas Sir3 is a histone H4 and nucleosome binding protein that displays a strong preference for histone H4 peptides and nucleosomes that contain unacetylated H4K16 (22, 27, 28). The association of Sir3 with chromatin is also inhibited by dimethylation or trimethylation of histone H3K79 (29, 30) occurring in transcribed genomic regions (14, 16).
Sir3 associates with nucleosomes via a conserved N-terminal domain, the bromo adjacent homoloy (BAH) domain, as well as a less well-characterized C-terminal domain with similarities to AAA ATPases (AAL domain) but lacking ATP binding or hydrolysis activity (27, 29, 31–33). The recently solved 3-Å resolution crystal structure of the BAH–nucleosome complex reveals how the BAH domain binds to the nucleosome and how this binding is controlled by acetylation and methylation (34). H4K16 and H18 make multiple hydrogen-bonding interactions with a deep pocket in the BAH domain, which would be disrupted by acetylation of the H4K16 epsilon amino group. The importance of this BAH binding pocket is supported by studies indicating that mutations within the binding pocket and surrounding residues disrupt silencing in vivo (32, 33, 35). The H4 tail exits the nucleosome near the LRS regions, and within this region, H3K79 and its surrounding residues also have multiple bonding interactions with the BAH domain (34). Thus, although the BAH–nucleosome structure provides clear evidence of the participation of H3K79, H4K16, and H4H18 in Sir3–nucleosome binding, it does not address how this binding mediates the silencing function of Sir3. In particular, the roles of H4R17 and R19, two residues in the basic patch region that are as critical for silencing as H4K16 (12), remain unclear.
In this study, we examined the roles of amino acids in the histone H4 basic patch in histone peptide and nucleosome binding. As expected, we found that substitutions of H4K16 and H18 with alanine abolished histone H4 peptide and nucleosome binding. However, substitutions of H4R17 and R19, which disrupted silencing to the same extent as K16 or H18 substitutions, had no effect on Sir3 binding in our in vitro assays. Consistent with the biochemical binding data, both H4R17 and R19 point away from the BAH domain in the crystal structure of the BAH domain in complex with the yeast nucleosome core particle (reported here), as well as the BAH domain in complex with the Xenopus laevis nucleosome core particle reported by Armache et al. (34). Rather than making contact with the BAH domain, these arginines make contacts with phosphates in the nucleosomal DNA backbone. Based on the foregoing findings, we propose that binding of Sir3 to the nucleosome induces a conformational change that clamps H4R17 and R19 onto nucleosomal DNA to create a silenced nucleosome.
Results and Discussion
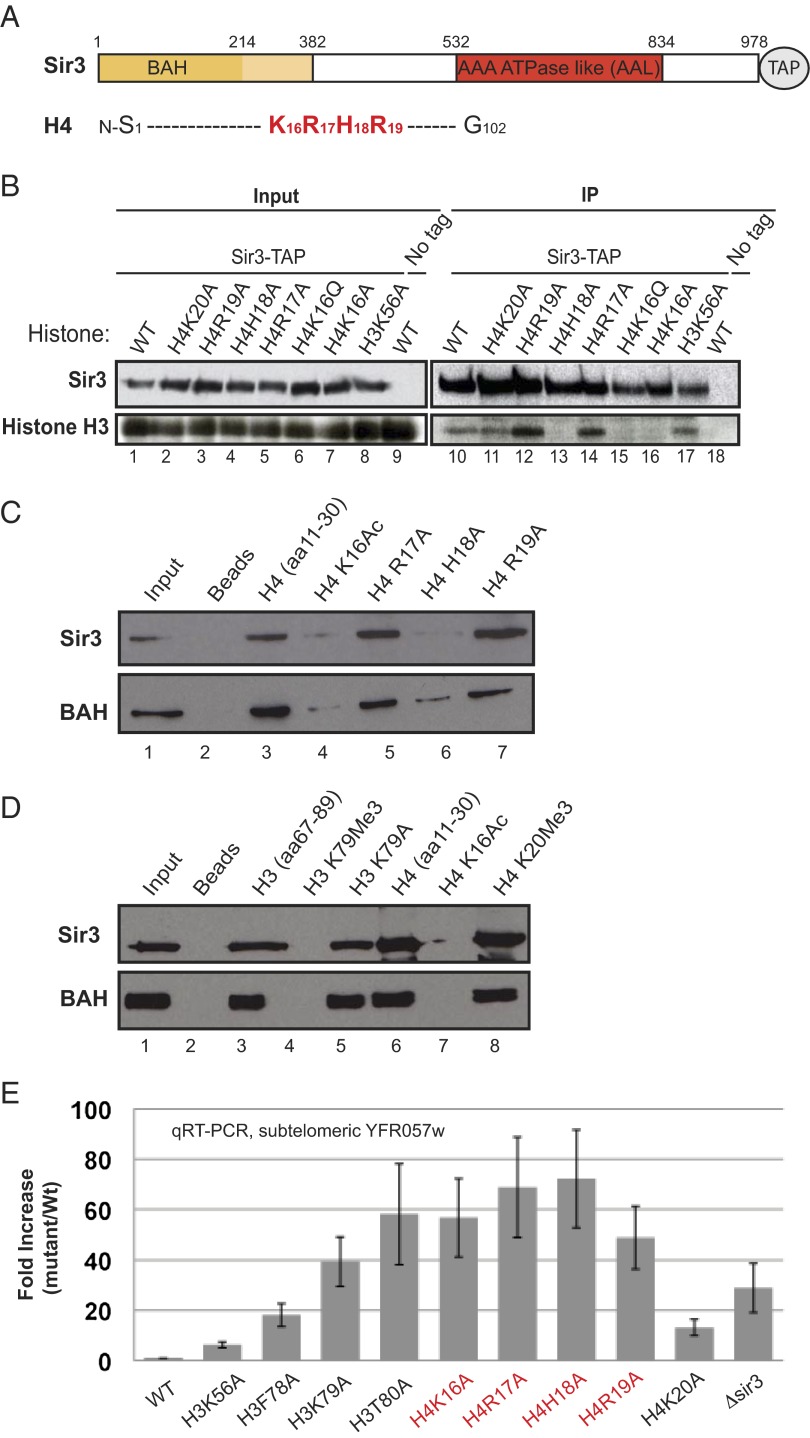
We used a pull-down assay that examines the association of nucleosomes from a solubilized chromatin extract with tandem affinity purification (TAP)-tagged Sir3 to determine the contribution of H4R17, R19, and other amino acids in the H4 basic patch region to the binding of Sir3 to nucleosomes (Fig. 1A; see Tables S1 and S2 for yeast strains and plasmids). As a control and consistent with previous results (29), the substitution of H4K16 with either glutamine or alanine (H4K16Q and K16A, respectively) abolished the association of nucleosomes with Sir3 (Fig. 1B, compare lanes 10, 15, and 16; Fig. S1). Similarly, consistent with its interaction with the BAH domain (34), the substitution of H4H18 with alanine (H4H18A) abolished the binding of nucleosomes to Sir3 (Fig. 2B, compare lanes 10 and 13). In contrast, the substitution of either H4R17 or R19 with alanine (H4R17A and R19A, respectively) resulted in increased binding of nucleosomes to Sir3 (Fig. 1B, compare lanes 10, 12, and 14). As further controls, substitutions at H4K20 and K56 (H4K20A and K56A, respectively), which have weak silencing defects, had no effect on the Sir3–nucleosome interaction in this assay (Fig. 1B, lanes 11 and 17).
Fig. 1.
Histone H4R17 and R19 are critical for silencing but do not affect the association of Sir3 with the nucleosome. (A) Schematic diagram highlighting the boundaries of the BAH and AAA ATPase-like (AAL) domains of Sir3, and the basic patch region of histone histone H4 (in red). (B) Sir3-TAP pull-downs showing the effect of histone H4 N-terminal basic patch and H3K56 mutations on the nucleosome–Sir3 association. Sir3-TAP was expressed in yeast under the control of its own promoter. (C and D) Peptide pull-down assays showing the association of Sir3 and the BAH domain (N-terminal subfragment, Sir3-BAH381) with WT histone peptides and histone peptides containing the indicated amino acid substitutions. In addition, the data show that trimethylation of the H3, amino acid 67–89 peptide at K79 abolishes its binding to Sir3 and its BAH-containing subfragment. H4K16Ac and K20me3 serve as positive and negative controls, respectively. Full-length Sir3 and Sir3-BAH381 (Sir3 amino acids 1–381) were purified from yeast. (E) qRT-PCR data comparing the silencing of the subtelomeric YFR057w, located on chromosome VI-R in WT and the indicated histone mutant cells. sir3∆ serves as a control.
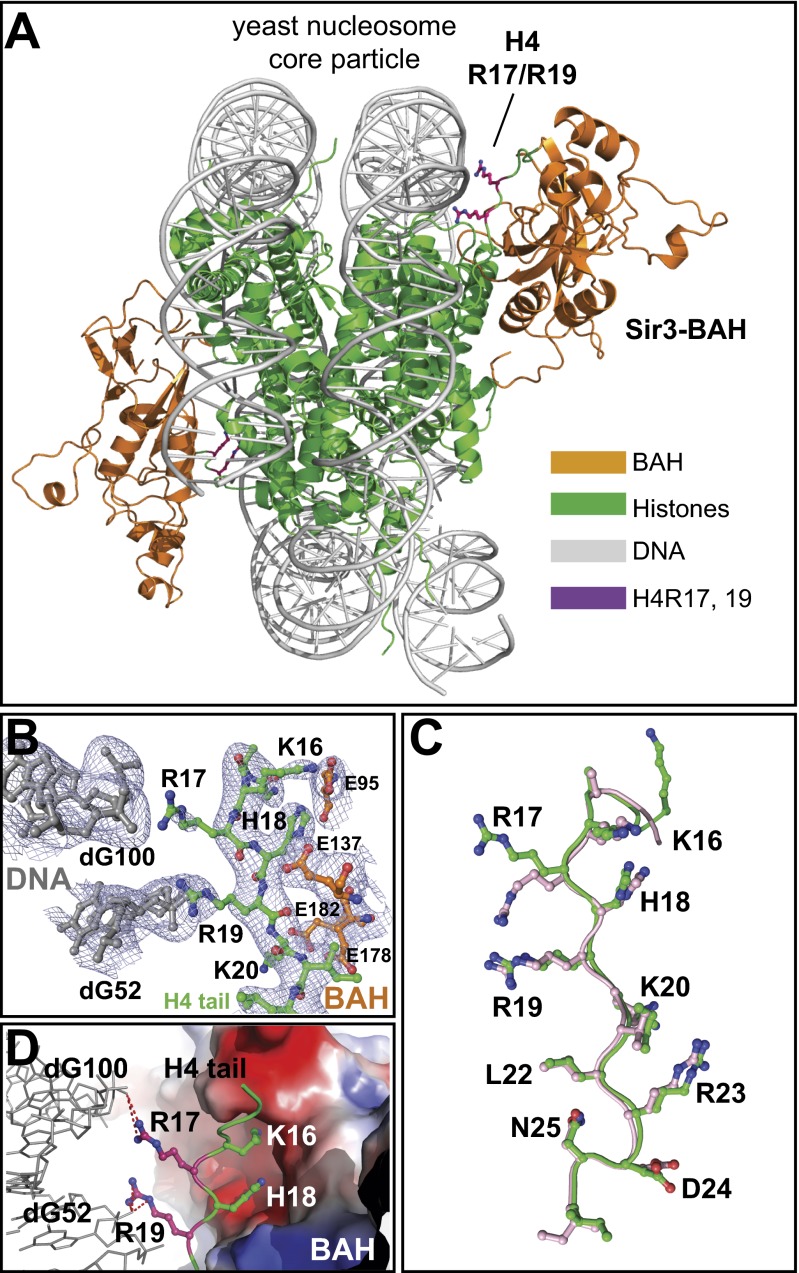
Fig. 2.
Structure of the Sir3-BAH in complex with the yeast nucleosome. (A) Overall view of the structure highlighting interactions between histone H4R17 and R19 and nucleosomal DNA. The BAH domain is in orange; H4R17 and R19 are in magenta; histones H2A, H2B, H3, and H4 are in green; and the DNA is in gray. (B) Electron density map (σ = 1.0) showing the H4 basic patch region (green) and its interactions with DNA (gray) and the BAH domain (orange). (C) Overlay of the H4 tail in the BAH–XlNCP (pink; ref. 34) and BAH–ScNPC (green; this study) complexes. (D) High-magnification view of the interactions involving the H4 basic patch region. H4K16 and H18 interact with a deep pocket in the BAH domain, shown in electrostatic surface representation, whereas H4R17 and R19 point away from the BAH domain and make salt bridges with the phosphates of different strands of nucleosomal DNA at nucleotides 52 and 100.
We next tested the importance of the H4 basic patch residues in Sir3 binding using a biotinylated peptide pull-down assay. Consistent with the nucleosome binding data, the acetylation of H4K16 (H4K16Ac) or the substitution of H4H18 with alanine diminished the ability of these residues to bind to either full-length Sir3 or the BAH domain (Fig. 1C, compare lanes 3, 4, and 6; Fig. 1D, lanes 6 and 7). On the other hand, the substitution of either H4R17 or R19 with alanine had little or no effect on the binding of full-length Sir3 or the BAH domain (Fig. 1C, compare lanes 3, 5, and 7). Thus, H4R17 and R19 are not required for the association of Sir3 with either nucleosomes or histone H4 N-terminal peptides.
Consistent with previous nucleosome-binding studies (29) and extensive contacts between the BAH domain and H3K79 (34), trimethylation of H3K79 abolished the binding of full-length Sir3 and the BAH domain to a peptide spanning amino acids 67–89 of histone H3 (Fig. 1D, lanes 3 and 4). Moreover, consistent with nucleosome-binding results (28), the substitution of H3K79 with alanine had no effect on binding (Fig. 1D, lane 5). This suggests that the methylation of H3K79 by Dot1 inhibits Sir3 binding via a steric hindrance mechanism that prevents interactions between Sir3 and the surrounding LRS region, rather than with H3K79 itself.
To verify that the point mutations used in our studies had the expected loss of silencing defects, we performed quantitative RT-PCR (qRT-PCR) to measure RNA levels for the subtelomeric YFR057w open reading frame on the right arm of chromosome VI, which is silenced in a Sir3-dependent manner. Substitutions of R17 and R19 with alanine displayed a similar increase in YFR057w RNA levels as seen with substitutions of H4K16 and H18, which are Sir3-contacting amino acids (Fig. 1E). However, ChIP experiments showed that H4R17A and R19A mutations disrupted the association of Sir3 with YFR057w in vivo (Fig. S2), likely owing to disruption of the Sir3–nucleosome interaction in cells carrying the arginine mutations during chromatin remodeling and transcription activation.
We note that the basic patch region of H4 also provides a binding site for the H3K79 methyltransferase disruptor of telomeric silencing 1 (Dot1), an event required for efficient silencing (30, 36). However, the complete loss of Dot1-mediated H3K79 methylation requires substitutions of both H4R17 and R19 with alanine (30, 36), and dot1∆ cells have near-WT silencing at the homothalic left (HML) mating type locus (37, 38), whereas the substitution of either H4R17 or R19 results in loss of silencing at HML and subtelomeric regions (12) (Fig. 1E). Thus, the silencing defects of H4R17 and R19 cannot be explained by a loss of Dot1 binding. In fact, the increase in Sir3 binding in the H4R17A and R19A mutations noted above (Fig. 1B) is similar to what we had previously observed comparing WT with ∆dot1 cells using the same nucleosome pull-down assay (29), and suggests that increased Sir3 binding results from weaker Dot1 binding to the H4 basic patch region and reduced H3K79 methylation.
The crystal structure of the BAH domain (Sir3 amino acids 1–214) in complex with the heterologous X. laevis nucleosome core particle suggests that H4R17 and R19 do not contact the BAH domain (34). To determine whether these arginines occupy similar positions when the BAH domain is bound to the yeast nucleosome, we solved the crystal structure of a Sir3 N-terminal fragment from amino acids 2–382 (Sir3-382), which included the conserved BAH domain, in complex with the nucleosome core particle (NCP), reconstituted using the 147-bp Widom 601 positioning DNA and bacterially produced yeast histones. We used the Sir3-382 protein because it binds to the nucleosome with greater affinity than the smaller BAH domain (Sir3-214), and its binding is sensitive to substitutions at either H4K16 or H3K79 (Figs. S1 and S3). We further introduced a point mutation at position 205 (E205N), which was previously shown to increase the affinity of Sir3 for the nucleosome (34, 39). The 3.1-Å resolution structure of Sir3-382–ScNCP shows two BAH domains bound symmetrically to each side of the nucleosome (Fig. 2A and Table S3). Most of the Sir3–nucleosome interaction interface in our structure was identical to the Sir3-214–XlNCP complex (34) (Fig. S4), showing extensive contacts between the BAH domain and the globular domain of histone H3 surrounding H3K79 and the amino terminus of histone H4. Amino acids 216–382, beyond the BAH domain, displayed discontinuous electron density, which is consistent with predictions that this low-complexity region is unstructured, and are not presented in the structure.
We observed clear electron density for the H4R19 side chain and weaker electron density for the H4R17 side chain, but both arginines clearly pointed away from the BAH domain and were in a position to make contact with DNA (Fig. 2 A and B). H4R19 was located in a similar position as that previously described for the Sir3-214–XlNCP structure (Fig. 2C). However, whereas in the Sir3-214–XlNCP structure, both arginines were located close to the phosphate of nucleotide 100, in the Sir3-382–ScNCP structure, H4R17 was located closer to the phosphate of nucleotide 52 (Fig. 2 C and D). Thus, H4R17 and R19 interact with phosphates 100 and 52, respectively, which are located across the minor groove on opposite strands of the DNA double helix in the BAH–nucleosome complex (Fig. 2 C and D). The different positions of H4R17 in the two structures may be related to alternative conformations or flexibility of R17, the location of which may be stabilized by additional Sir3 sequences or the other subunits of the SIR complex. Unlike H4K16 and H18, which penetrate binding pockets in the BAH domain, H4R17 and R19 make salt bridges with phosphates of nucleosomal DNA (Fig. 2D). Nucleotides 52 and 100, which contact H4R17 and R19, respectively, are located on complementary strands across the DNA minor groove (Fig. 2D). In addition, the contacts are symmetrical for the two histone H4 chains (Fig. S5 A and B). Thus, the locations of H4R17 and R19 in BAH complexes with yeast and frog nucleosomes are consistent, and support the idea that the silencing function of these amino acids involves their interaction with nucleosomal DNA rather than with Sir3.
To determine whether silencing requires H4R17 and R19 under conditions in which Sir3 is present on chromatin, we took advantage of the observation that overexpressed Sir3 associates with the HML locus and silences the expression of the α1 and α2 genes even in sir4I1311N mutant cells, in which it cannot interact with the Sir2/Sir4 deacetylase complex (Fig. S2). This silencing is not accompanied by the spread of the Sir2/Sir4 deacetylase and requires the substitution of H4K16 with arginine (H4K16R), which mimics deacetylated lysine. To determine whether the silencing mediated by this Sir3 binding requires H4R17 or R19, we constructed Sir3-overexpressing (SIR3OE) cells with H4R17A and R19A substitutions alone or in combination with H4K16R substitutions. Sir3 bound to the HML locus at near-WT levels in H4R17A and R19A cells, but this binding did not correlate with silencing of the α1 and α2 genes (Fig. S2 B and C). Furthermore, no silencing was observed even in cells in which the foregoing mutations were combined with H4K16R, indicating that the roles of H4R17 and R19 in silencing did not involve a contribution to the binding of overexpressed Sir3 or K16 deacetylation (Fig. S2 B and C). We emphasize that, as shown in Fig. S2A for the subtelomeric YFR057W gene, in the absence of overexpression, the H4R17A and R19A mutations disrupted Sir3 binding to chromatin, suggesting that even though they are not Sir3 contact residues, H4R17 and R19 affect the stability of Sir3 on chromatin in vivo.
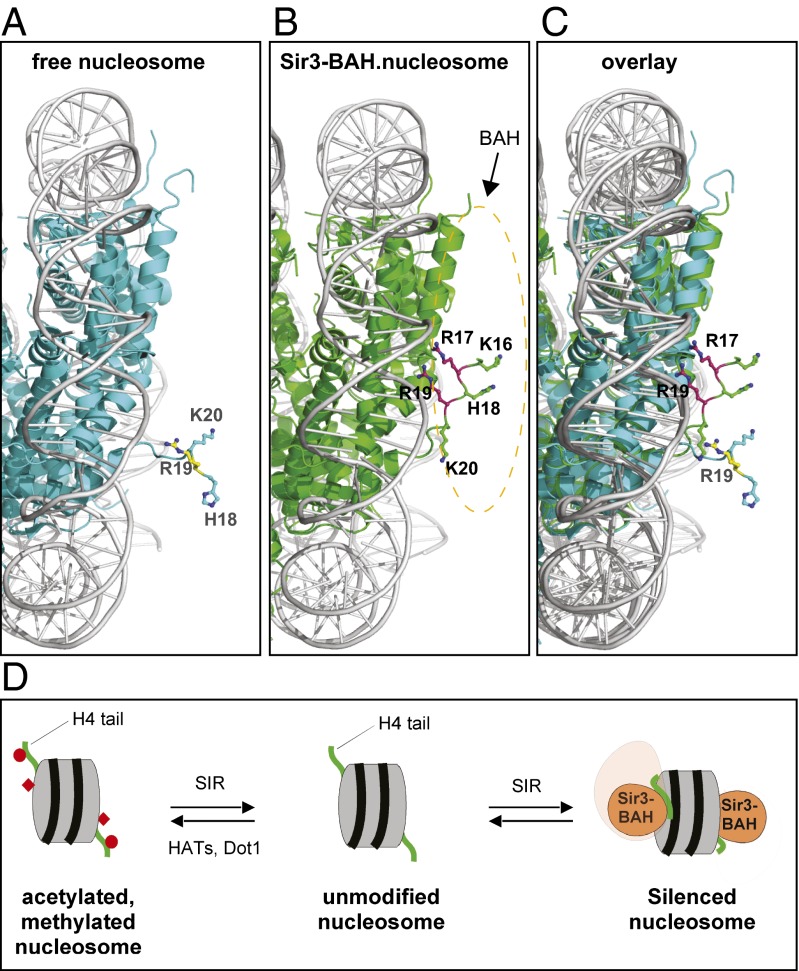
Taken together, the biochemical and structural evidence presented here indicate that H4R17 and R19 play a role in silencing that is distinct from the role of H4K16 and H18. Histone tails generally display relatively weak electron density in the crystal structures of free nucleosomes reported to date (for examples, see refs. 1 and 40). Nonetheless, the conformation of the H4 tail and H4R17 and R19 observed in the Sir3–NCP complexes are unique. Comparison of the H4 tail conformation in the free yeast nucleosome (40) and the Sir3-382–ScNCP complex indicates that the association of Sir3 with the nucleosome results in a conformational change in the H4 N-terminal tail involving a rotation around the main chain of asparagine 25 (Fig. 3 A–C). This rotation allows H4K16, H18, K20, L22, and R23 to interact with the BAH domain and brings the R17 and R19 side chains in close proximity to nucleosomal DNA. The locations of the R17 and R19 side chains may be further constrained by interactions of their backbone carbonyl groups with E137 in the BAH domain (Fig. 2B and Fig. S6). We propose that the salt bridges between these H4 arginines and the phosphates in the DNA backbone clamp the nucleosomal DNA to the histone octamer and help create a “silenced nucleosome” (Fig. 3D). This silenced nucleosome is likely to be more resistant to unwinding by chromatin remodeling complexes, a required step for DNA accessibility and transcription. The H4 amino terminus is likely to interact with nucleosomal DNA even in the absence of Sir3, such as in the high-resolution Xenopus nucleosome crystal structure (Fig. S4 B–D) (41). Previous studies have suggested that interactions between histone tails and DNA contribute to nucleosome stability and are regulated by acetylation (42). Thus, Sir3 may shift the equilibrium toward a conformation that has evolved to contribute to nucleosome stability.
Fig. 3.
Association of Sir3-BAH with the nucleosome induces a conformational change in histone H4. Comparison of the structure of the free yeast nucleosome (A; cyan, 1ID3) with the Sir3-BAH382–nucleosome complex (B; green) highlighting the basic patch region of histone H4. (C) Overlay of the two structures. The dotted oval indicates the location of the BAH domain, but the BAH atoms have been removed for clarity. The basic patch region in the free yeast nucleosome and the BAH–nucleosome complex are in yellow and magenta, respectively. H4K16 and R17 are absent in the free nucleosome structure. (D) Model for formation of the silenced nucleosome. Recruitment of the SIR complex results in the generation of nucleosomes containing deacetylated H4K16 and unmethylated H3K79. The SIR complex binds to such unmodified nucleosomes, and the association of the Sir3-BAH domain induces a conformational change in the H4 tail (in green; red circle, acetylated H4K16; red diamond, methylated H3K79) that clamps H4R17 and R19 to DNA to create a silenced nucleosome.
Our findings suggest that silencing requires a specific mode of Sir3–nucleosome association and provide a possible explanation for previous reports suggesting that SIR complex binding does not correlate with silencing (28, 43, 44). The formation of a silenced nucleosome most likely requires multiple bonding interactions between Sir3 and the nucleosome that together stably clamp nucleosomal DNA to the histone octamer. The loss of a subset of the bonding interactions may allow for a more dynamic form of Sir3 binding that is insufficient to silence transcription. The functions of H4R17 and R19 proposed herein are reminiscent of the histone sin mutations that suppress defects in chromatin remodeling (45, 46). A large fraction of these mutations affect the interaction of histone H4R45 with a minor groove in nucleosomal DNA (46), providing another example of profound effects on chromatin structure resulting from histone point mutations that affect contacts with DNA.
Materials and Methods
Protein Expression and Purification.
A DNA fragment encoding residues 2–382 of Sir3 was cloned into a modified pET28b vector with a His6-Sumo tag fused at the N terminus. The D205N point mutation was introduced using the QuikChange II Site-Directed Mutagenesis Kit (Agilent). The Sir3 (2-382, D205N) plasmid was transformed and expressed in Escherichia coli Rosetta DE3 strain. After induction for 16 h with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 25 °C, the cells were harvested by centrifugation, and the resulting pellet was resuspended in lysis buffer [50 mM Tris⋅HCl (pH 8.0), 50 mM NaH2PO4, 400 mM NaCl, 3 mM imidazole, 10% glycerol, 1 mM PMSF, 0.1 mg/mL lysozyme, 2 mM 2-mercaptoethanol, and complete protease inhibitor mixture tablets (Roche)]. The cells were then lysed by sonication, and the cell debris was removed by ultracentrifugation. The supernatant was mixed with Ni-nitriloacetic acid (NTA) agarose beads (Qiagen) and rocked for 4 h at 4 °C, then washed twice with 20 column volumes of 10 mM imidazole in lysis buffer before elution with 250 mM imidazole. The ubiquitin-like specific protease 1 (ULP1) protease was added to remove the His6-Sumo tag. Finally, the protein was further purified by passage through a Mono-Q ion-exchange column (GE Healthcare) and by gel-filtration chromatography on a Hiload Superdex 75 (GE Healthcare) equilibrated with 25 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 5 mM DTT. The purified Sir3 (2-382, D205N) protein was concentrated to 25 mg/mL using centrifugal filters (Millipore) and stored at −80 °C. Budding yeast S. cerevisiae histones H2A, H2B, H3, and H4 were expressed in E. coli; purified; renatured in 2 M NaCl; and assembled into octamers through stepwise salt dialysis. The histone octamer core was assembled with the 601 Widom positioning sequence into the nucleosome core particle as described previously (1).
Sir3–NCP Complex Preparation and Crystallization.
The complex of the Sir3-382 and the nucleosome core particle was assembled by mixing the Sir3 protein with nucleosome core particle in a 2:1 molar ratio before a final size-exclusion chromatography step (Superose 6; GE Healthcare). The assembled complex was concentrated to 8 mg/mL for crystallization screening trials. The crystals were grown by sitting drop vapor diffusion in 0.05 M sodium cacodylate (pH 7.5) and 32% (vol/vol) 2-methyl-2,4-pentanediol (MPD) at 4 °C. Crystals were transferred to 40% (vol/vol) MPD in 1% increments with 10 min between each step, and then flash-frozen in liquid nitrogen.
Data Collection and Crystallographic Analysis.
Diffraction data were collected at the Advance Photon Source The Northeastern Collaborative Access Team (NE-CAT) beamline 24-ID-C, and processed using XDS and the CCP4 package (47, 48). The structure was solved by molecular replacement using Phaser (49), and a search model containing the budding yeast histone octamer core and the 147-bp human α-satellite DNA (PDB ID code 1ID3), with the DNA bases manually changed to the 601 Widom sequence. A clearly positive density for Sir3-382 was present on both sides of the nucleosome in the difference electron density map. Crystallographic refinement was carried out using PHENIX with manual model building in Coot (50, 51). All graphic presentations were prepared in PyMOL (www.pymol.org/). Secondary structural prediction was performed using the PredictProtein server (www.predictprotein.org).
Yeast Strains and Protein Purification.
The strains and plasmids used in this study are listed in Tables S1 and S2. Yeast strains were made by a PCR-based gene targeting procedure (52–55). Histone mutant strains were constructed by introducing individual histone mutations by plasmid shuffle into the background strain of DMY3903 or DMY3985, as described previously (12, 56). Plasmids carrying histone mutations were kindly provided by Dr. Jef Boeke (57) and Dr. Sharon Dent (56). FLAG-tagged Sir3 proteins expressed in yeast were purified as described previously (22, 35).
TAP Protein Purification and Western Blot Analysis.
TAP-pull downs were performed as described previously (29, 55). In brief, cells were resuspended in equal volumes of lysis buffer [50 mM Hepes KOH (pH 7.6), 10 mM magnesium acetate; 5 mM EGTA, 0.1 mM EDTA, 150 mM potassium chloride, 0.2% Nonidet P-40, 5% glycerol, 2 mM phenylmethylsulfonylfluoride, 1 mM benzamidine, and 1 mg/mL each leupeptin, bestatin, and pepstatin] and lysed by bead beating. The resulting lysate was incubated with IgG-coupled M-270 Dynabeads (Invitrogen) for 90–120 min at 4 °C. Beads were washed four times with lysis buffer and resuspended in SDS sample buffer. Samples were loaded on SDS/PAGE, transferred to nitrocellulose membranes, and probed with peroxidase anti-peroxidase (Sigma-Aldrich) and anti-H3 (Abcam) antibodies.
Quantitative RT-PCR.
Yeast cultures were grown in yeast extract peptone dextrose (YEPD) medium at 30 °C to an OD600 of 0.5. Total RNA was isolated by the hot phenol procedure and cleaned using the RNeasy Kit (Qiagen) to remove potential genomic DNA contamination. Gene-specific primers for YFR057W were used to prepare cDNA, followed by quantitative PCR using a LightCycler (Applied Biosystems). Relative RNA levels were calculated from CT values according to the ΔCT method and normalized to act1+ RNA levels.
Native Gel Shift and Peptide Pull-Down Assays.
Mononucleosomes were reconstituted as described previously (1, 28). Increasing molar ratios of full-length Sir3 protein or its subdomains were incubated with 8 nM mononucleosome, assembled on a 218-bp DNA fragment containing the 601 Widom positioning sequence (58) in binding buffer containing 20 mM Hepes (pH 7.5), 4 mM Tris (pH 7.5), 80 mM KCl, 0.1% Nonidet P-40, 0.2 mM EDTA, 2 mM DTT, 0.5 mg/mL BSA, and 5% glycerol. The binding reaction was carried out at 30 °C for 1 h, followed by chilling on ice for 15–20 min before loading onto the 3.5% native polyacrylamide gel. The native polyacrylamide gel was prerun at 50V for 1 h in 0.25× tris-borate EDTA (TBE), and then changed with fresh 0.25× TBE buffer. Samples were separated at 100V for 1.5–2 h at 4 °C or for 4–4.5 h in the presence of antibody. The gel was dried, analyzed by a storage phosphor screen, and quantified with Quantity One software (Bio-Rad). Saturation curves were analyzed using Kaleidograph software (Synergy Software). Peptide pull-down assays were carried out by incubating 1 μg of biotinylated histone peptides with Sir3 protein in 25 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, 100 μg/mL BSA, 0.5 mM DTT, 0.1% Tween-20, and 1 μg/mL leupeptin, pepstatin, and aprotinin for 1 h at room temperature in a rotary shaker. Peptides were then isolated with magnetic streptavidin beads for 30 min at room temperature. Beads were resuspended in SDS sample buffer and loaded onto 12% SDS/PAGE, followed by Western blot analysis with anti-Flag antibody.
Supplementary Material
Acknowledgments
We thank Drs. Stephen Harrison and Simon Jenni for helpful discussions and comments; Drs. Jef Boeke and Sharon Dent for gifts of histone plasmids; and Dr. Piotrek Sliz and the staff of the SBGrid Consortium and Drs. J. Schuermann, F. Murphy, S. Banerjee, K. Perry, and the staff of the NE-CAT Argonne National Laboratory for assistance with data collection and processing. This work was supported by the National Institutes of Health (Grant GM GM61641, to D.M., and Pathway to Independence Award GM094291, to A.J.). F.W. is a Fellow of the Howard Hughes Medical Institute Life Sciences Research Foundation, and D.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4JJN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300126110/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11(6):266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8(3):489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 9.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 10.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146(4):510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayne PS, et al. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 12.Johnson LM, Fisher-Adams G, Grunstein M. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992;11(6):2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7(4):592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 14.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109(6):745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 15.Ng HH, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16(12):1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc Natl Acad Sci USA. 2003;100(4):1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat Genet. 2002;32(2):273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 18.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klar AJS, Fogel S, Macleod K. MAR1 - a regulator of HMa and HMα loci in Saccharomyces cerevisiae. Genetics. 1979;93(1):37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: A SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94(6):2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanny JC, Kirkpatrick DS, Gerber SA, Gygi SP, Moazed D. Budding yeast silencing complexes and regulation of Sir2 activity by protein–protein interactions. Mol Cell Biol. 2004;24(16):6931–6946. doi: 10.1128/MCB.24.16.6931-6946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121(4):515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 23.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8(19):2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 24.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 25.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97(26):14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JS, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97(12):6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80(4):583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 28.Johnson A, et al. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol Cell. 2009;35(6):769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28(6):1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Altaf M, et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28(6):1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrentraut S, et al. Structural basis for the role of the Sir3 AAA+ domain in silencing: Interaction with Sir4 and unmethylated histone H3K79. Genes Dev. 2011;25(17):1835–1846. doi: 10.1101/gad.17175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampath V, et al. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol Cell Biol. 2009;29(10):2532–2545. doi: 10.1128/MCB.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norris A, Bianchet MA, Boeke JD. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 2008;4(12):e1000301. doi: 10.1371/journal.pgen.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334(6058):977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchberger JR, et al. Sir3–nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28(22):6903–6918. doi: 10.1128/MCB.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: Identification of a new trans-histone pathway. Genes Dev. 2007;21(16):2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osborne EA, Dudoit S, Rine J. The establishment of gene silencing at single-cell resolution. Nat Genet. 2009;41(7):800–806. doi: 10.1038/ng.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi YH, et al. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol Cell. 2011;42(1):118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87(16):6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20(18):5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent- mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 42.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 43.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27(6):890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang B, Britton J, Kirchmaier AL. Insights into the impact of histone acetylation and methylation on Sir protein recruitment, spreading, and silencing in Saccharomyces cerevisiae. J Mol Biol. 2008;381(4):826–844. doi: 10.1016/j.jmb.2008.06.059. [DOI] [PubMed] [Google Scholar]
- 45.Kruger W, et al. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9(22):2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 46.Muthurajan UM, et al. Crystal structures of histone Sin mutant nucleosomes reveal altered protein–DNA interactions. EMBO J. 2004;23(2):260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, et al. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20(20):2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 54.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 55.Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein–protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol. 2005;25(11):4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17(11):3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai J, et al. Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134(6):1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li G, Widom J. Nucleosomes facilitate their own invasion. Nature structural & molecular biology. 2004;11(8):763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.