Significance
Previous studies have shown that abscisic acid (ABA) has a positive effect on virus infection, and ethylene has a negative effect. However, the possible crosstalk between ABA and ethylene signaling during plants–virus interaction remains unclear. Our current results demonstrate that WRKY8 is involved in the defense response against crucifer-infecting tobacco mosaic virus (TMV-cg) through the direct regulation of the expression of ABI4, ACS6, and ERF104 and thus may mediate the crosstalk between ABA and ethylene signaling during the TMV-cg–Arabidopsis interaction.
Keywords: WRKY8 regulation, virus movement, ABA signaling, ET signaling
Abstract
WRKY transcription factors are key players in the plant immune response, but less is known about their involvement in antiviral defense than about their roles in defense against bacterial or fungi pathogens. Here, we report that Arabidopsis thaliana WRKY DNA-binding protein 8 (WRKY8) has a role in mediating the long-distance movement of crucifer-infecting tobacco mosaic virus (TMV-cg). The expression of WRKY8 was inhibited by TMV-cg infection, and mutation of WRKY8 accelerated the accumulation of TMV-cg in systemically infected leaves. Quantitative RT-PCR analysis showed that the expression of ABA insensitive 4 (ABI4) was reduced and the expression of 1-aminocyclopropane-1-carboxylic acid synthase 6 (ACS6) and ethylene response factor 104 (ERF104) was enhanced in the systemically infected leaves of wrky8. Immunoprecipitation assays demonstrated that WRKY8 could bind selectively to putative W-boxes of the ABI4, ACS6, and ERF104 promoters. Furthermore, TMV-cg infection enhanced WRKY8 binding to the ABI4 promoter but reduced the binding of WRKY8 to the ACS6 and ERF104 promoters, indicating that regulation of ABI4, ACS6, and ERF104 by WRKY8 is at least partially dependent on TMV-cg. Exogenous applications of abscisic acid (ABA) reduced the systemic accumulation of TMV-cg. Mutations in ABA deficient 1, ABA deficient 2, ABA deficient 3, or abi4 accelerated systemic TMV-cg accumulation. In contrast, exogenous application of aminocyclopropane-1-carboxylic acid enhanced the systemic accumulation of TMV-cg, but mutations in acs6, erf104, or an octuple acs mutant inhibited systemic TMV-cg accumulation. Our results demonstrate that WRKY8 is involved in the defense response against TMV-cg through the direct regulation of the expression of ABI4, ACS6, and ERF104 and may mediate the crosstalk between ABA and ethylene signaling during the TMV-cg–Arabidopsis interaction.
The interaction between plants and viral pathogens reflects a sophisticated coevolution of recognition, defense, and counter-defense mechanisms. Although plant viruses are among the least genetically complex pathogens, they use a variety of strategies to suppress or bypass host defense and then promote their infection of susceptible hosts. In plants, these strategies involve enhancing infection by manipulating host resources, such as the formation of replication complexes (1), enlargement of the plasmodesma size-exclusion limit (2, 3), evolution of viral suppressors of RNA silencing to counteract antiviral silencing (4), interference with regulation of the plant cell cycle (5, 6), and using host components for its own replication (7).
In turn, plants have evolved intricate mechanisms to fight viral infection, such as pathways mediated by gene silencing, hormone-mediated signaling pathways, and regulation of metabolism (8–11). In addition to a conserved sequence-specific system of gene regulation, recent research demonstrated that RNA silencing also functions as an adaptive inducible antiviral defense pathway (10, 11). In plants, several important RNA-silencing components, such as RNA-dependent RNA Polymerase 6 (RDR6), dicer-like 2/4 (DCL2/4), Argonaute 1 (AGO1), and double-stranded RNA-binding protein 4 (DRB4), constitute the host’s silencing machinery involved in antiviral defense, which involves the production of viral siRNA or the formation of RNA-induced silencing complexes that target viral RNAs for destruction (12–18). In addition to RNA silencing, several plant hormones are involved in plant basal defense responses in plant–virus interactions. For example, up-regulation of less susceptible to BSCTV 1 (LSB1) affects geminivirus Beet severe curly top virus (BSCTV) infection by activating the salicylic acid (SA) pathway (9). Research showed that abscisic acid (ABA) has a positive effect on virus infection by inhibiting the transcription of a basic β-1,3-glucanase (PR2) (19). For ethylene (ET), two ET-signaling mutants, constitutive triple response 1 (ctr1) and ethylene insensitive 2 (ein2), showed no obvious differences between the wild type and mutants in the GFP fluorescence of cells within local lesions but showed reduced susceptibility to cauliflower mosaic virus (CaMV) infection upon systemic infection of leaves, implying that ET-dependent responses play an important role in mediating long-distance virus movement (20, 21).
Infection of viruses induces the expression of host genes mainly at the level of transcription and may contribute to resistance against virus infection. Plants devote a large portion of their genome capacity to transcription, with the Arabidopsis (Arabidopsis thaliana) genome encoding more than 2,100 transcription factors (22). These transcription factor genes often belong to large gene families, which in some cases are plant specific. Among them, the WRKY transcription factors comprise one large family of regulatory proteins in plants. In Arabidopsis, the WRKY transcription superfamily consists of an estimated 74 members that fall into three major structural groups, based on both the number of WRKY domains and the features of their zinc finger-like motifs (23–25). Although discovered relatively recently, the WRKY transcription factors are becoming one of the best-characterized classes of plant transcription factors. Genetic and molecular studies during the past decade have demonstrated that WRKY transcription factors participate in various biotic/abiotic stress responses and several developmental and physiological processes, including embryogenesis, seed coat and trichome development, leaf senescence, regulation of biosynthetic pathways, and hormone signaling (25, 26). For example, disruption of WRKY33 or WRKY4 enhanced Botrytis cinerea’s susceptibility to necrotrophic fungal pathogens (27, 28), and several members of group III WRKY factors may function as positive regulators of basal resistance against the biotrophic pathogen Pseudomonas syringae (29–31). Our recent studies have shown that WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance, because mutations of these three genes render the mutant plants more sensitive to heat stress (32). On the other hand, WRKY34 negatively mediates the mature pollen-specific cold stress response by regulating the expression of C-repeat?DRE binding factor (CBF) transcriptional activators (33). Another study showed that tobacco transcription factor WRKY4 (NtWRKY4) is involved in antiviral defense and leaf morphogenesis in tobacco (34). Thus, we can deduce that WRKY transcription factors are significantly involved in the tight regulation and fine-tuning of the complex signaling and transcriptional networks of both plant stress responses and developmental processes.
Despite their functional diversity, WRKY proteins recognize and bind cis-acting DNA elements with the minimal consensus T/CTGACC/T (W-box) sequence, which often is found in the promoters of putative target genes (23, 35). However, little is known about the specific interaction of a given WRKY protein with a defined target gene. Recent studies using ChIP analysis showed that WRKY40 regulates the expression of several important ABA-responsive genes [such as ABRE binding factor 4 (ABF4), ABA insensitive 4 (ABI4), ABA insensitive 5 (ABI5), dehydration response element B1A (DREB1A), MYB domain protein 2 (MYB2), and RAB GTPase homolog B18 (RAB18)] in ABA signaling (36). WRKY40 also modifies the transcript level of several defense-related genes, including enhanced disease susceptibility 1 (EDS1), redox response transcription factor 1 (RRTF1), and jasmonate-ZIM-domain protein 8 (JAZ8), through direct in vivo interaction with the W-box sequence upstream of their promoters (37). Recently, Birkenbihl and coworkers (38, 39) showed that WRKY33 directly regulates the expression of various distinct components of defense pathways during B. cinerea infection. Our results also showed that activated expression of WRKY57 confers drought tolerance in Arabidopsis by directly binding the W-box of response to dessication 29A (RD29A) and nine-CIS-epoxycarotenoid dioxygenase 3 (NCED3) promoter sequences (40). Identification of important components that are regulated directly by WRKY transcription factors will add to our understanding of stress-induced signaling pathways.
Crucifer-infecting tobacco mosaic virus (TMV-cg) belongs to the crucifer-infecting Tobamovirus subgroup (41, 42) and can replicate and spread systemically in Arabidopsis. Thus, TMV-cg constitutes an excellent model with which to investigate host responses to infection by a compatible virus. In previous studies, we showed that WRKY8 is involved in regulating plant basal defense responses to both the necrotrophic fungal pathogen B. cinerea and the biotrophic bacterial pathogen P. syringae (43) and also plays a role in plant response to salt stress (44). Here, we report that WRKY8 also functions as a positive regulator during TMV-cg’s systemic infection. We demonstrate that mutation of WRKY8 makes plants more susceptible to TMV-cg infection. WRKY8 positively regulates ABI4 expression and negatively modulates ACS6 and ERF104 expression by directly binding to the W-box consensus motifs within their promoters. Thus, WRKY8 participates in plant–TMV-cg interaction by regulating both the ABA- and ET-signaling pathways.
Results
Mutation of WRKY8 Alters Responses to Viral TMV-cg Infection.
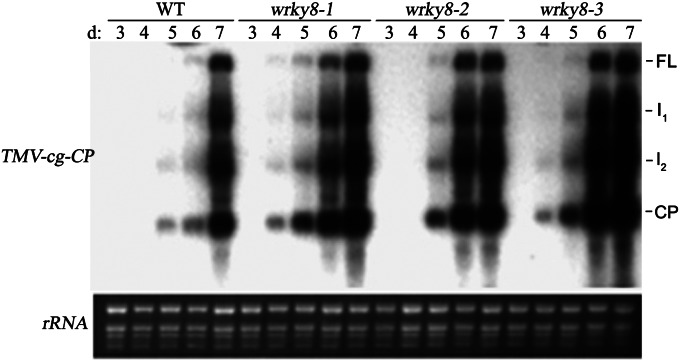
In a previous study, WRKY8 was demonstrated to be involved in regulating plant basal defense responses (43). Based on its induced expression by ABA and wounding treatments (see Fig. 2 A–C) (43), we hypothesized that WRKY8 also may be involved in antiviral defense responses. To clarify the potential functions of WRKY8, its roles in anti–TMV-cg responses were investigated. Three previously characterized wrky8 mutants were used (43). To assay plants’ responses to TMV-cg infection, we inoculated the third, fourth, and fifth true leaves of 26-d-old wrky8 mutants and wild-type plants with the virus. Viral RNA accumulated to similar levels in the inoculated leaves of wild-type and wrky8 mutant plants (Fig. S1). In upper systemic leaves of infected plants, viral RNAs [full-length viral genomic RNA and three subgenomic RNAs, i.e., I1, I2, and coat protein (CP)] were detected at 4 d postinfection (dpi) in wrky8 mutant plants but at 5 dpi in wild-type plants (Fig. 1). The highest levels of viral RNA were detected at 6 dpi in wrky8 mutant plants and at 7 dpi in wild-type plants (Fig. 1). Thus, TMV-cg virus accumulated more rapidly in the systemic leaves of infected wrky8 mutant plants than in the leaves of infected wild-type plants. These data indicate that WRKY8 might play an important role in preventing virus transmission from infected leaves to systemic leaves.
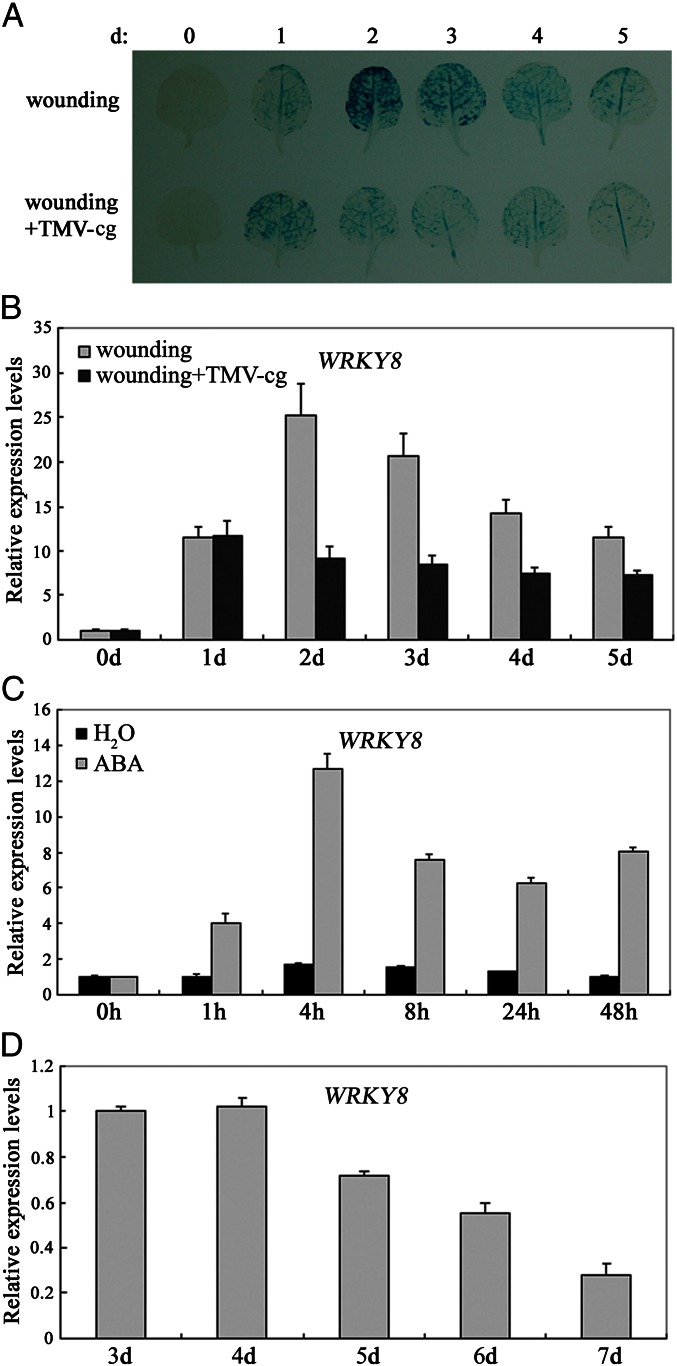
Fig. 2.
Induced expression of WRKY8. (A) GUS staining of WRKY8 in leaves locally infected with buffer or TMV-cg at 0, 1, 2, 3, 4, and 5 dpi. (B) Expression of WRKY8 in leaves locally infected with buffer or TMV-cg at 0, 1, 2, 3, 4, and 5 dpi. (C) Expression of WRKY8 after treatment with ABA (time course of 0, 1, 4, 8, 24, and 48 h). (D) Expression of WRKY8 in TMV-cg systemically infected leaves at 3, 4, 5, 6, and 7 dpi. These experiments were repeated three times with similar results.
Fig. 1.
Mutation of wrky8 affects the accumulation of TMV-cg in systemically infected leaves. The third, fourth, and fifth true leaves of 26-d-old wild-type and wrky8 mutants were inoculated with TMV-cg (5 µg/mL solution in 5 mM sodium phosphate, pH 7.5). RNA samples were prepared from systemically infected leaves of six plants inoculated with TMV-cg for 3, 4, 5, 6, and 7 d, respectively, and were probed with a TMV-cg coat protein (CP) cDNA fragment. Ethidium bromide-stained rRNA was used as a loading control. These experiments were repeated three times with similar results. FL, full length.
Temporal Expression of WRKY8.
WRKY8 appears to act as a positive regulator during the compatible interaction of Arabidopsis with TMV-cg. β-Glucuronidase (GUS) staining and quantitative real-time PCR (qRT-PCR) then was used to examine the inducibility and temporal kinetics of WRKY8 expression during infection. As shown in Fig. 2A, there was a stronger GUS signal in buffer-inoculated leaves than in leaves inoculated with TMV-cg. In buffer-inoculated leaves, the strongest GUS signal was observed at 2 dpi and decreased thereafter. In contrast, GUS expression was consistently lower and showed no peak of expression in leaves inoculated with TMV-cg. qRT-PCR results agreed with the results of GUS staining (Fig. 2B). Thus, both GUS staining and qRT-PCR showed that expression of WRKY8 was inhibited by TMV-cg infection in local infected leaves. Wounding and TMV-cg infection had opposite effects on the abundance of WRKY8 transcript in local incubated leaves.
TMV-cg infects Arabidopsis systemically and causes mild disease symptoms (45). The expression of WRKY8 was determined in systemically infected leaves. As shown in Fig. 2D, the expression of WRKY8 also was inhibited by TMV-cg infection in systemically infected leaves. The expression results showed that WRKY8 was inhibited during the course of TMV-cg infection and imply that WRKY8 may play an important role in anti–TMV-cg infection.
WRKY8 Responds to TMV-cg Infection Partially Through ABA Signaling.
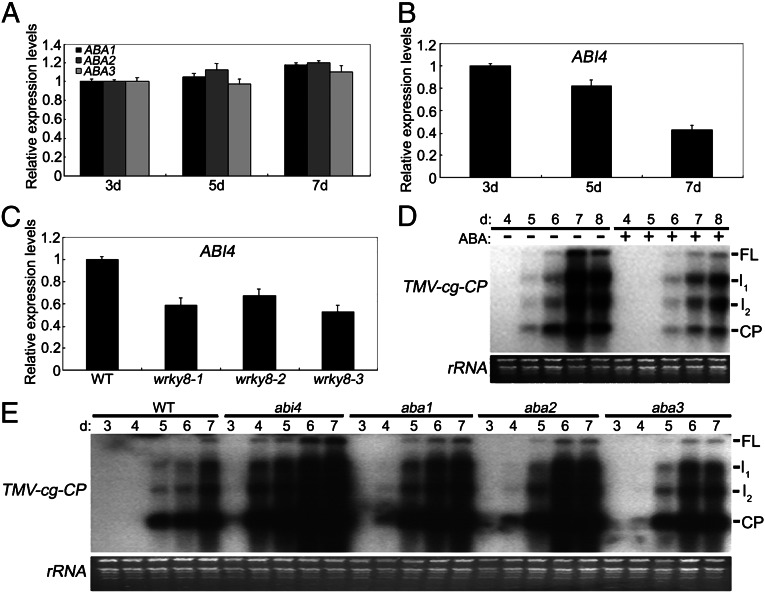
ABA plays a positive role in antiviral disease resistance, and treatment with ABA could increase resistance to a virus (46). Based on the induced expression of WRKY8 by ABA treatment (Fig. 2C) (43), we hypothesized that WRKY8’s function during TMV-cg infection may be mediated via ABA signaling. First, the expressions of ABA1, ABA2, ABA3, and ABI4 were investigated. As shown in Fig. 3 A and B, the expressions of ABA1, ABA2, and ABA3 were not significantly influenced by TMV-cg infection in wild-type plants, but the ABI4 expression was reduced during TMV-cg infection in the wild type. Furthermore, ABI4 was reduced more in systemically infected leaves in wrky8 mutant plants than in systemically infected leaves in wild-type plants (Fig. 3C). These results suggested that ABA signaling might be involved in TMV-cg defense responses.
Fig. 3.
The role of ABA in defense against TMC-cg infection. (A) Expression of ABA1, ABA2, and ABA3 in wild-type leaves systemically infected by TMV-cg at 3, 5, and 7 dpi. (B) Expression of ABI4 in wild-type leaves systemically infected by TMV-cg at 3, 5, and 7 dpi. (C) Expression of ABI4 in wild-type and wrky8 leaves systemically infected by TMV-cg at 6 dpi. (D) ABA delays the accumulation of TMV-cg in systemically infected leaves. Three leaves of 26-d-old wild-type plants treated or not treated with 100 µM ABA were inoculated with TMV-cg. Leaf collection, RNA isolation, and RNA blot analysis of TMV-cg CP were performed as in Fig. 1. (E) Mutation of abi4, aba1, aba2, or aba3 promotes the accumulation of TMV-cg in systemically infected leaves. Inoculation, leaf collection, RNA isolation, and RNA blot analysis of TMV-cg CP were performed as in Fig. 1. These experiments were repeated three times with similar results.
To determine the possible role of ABA in anti–TMV-cg resistance, the antiviral effect of ABA was investigated by infecting wild-type plants that were pretreated with ABA and plants that were not pretreated. As shown in Fig. 3D, the accumulation of TMV-cg in systemically infected leaves was both delayed and decreased in ABA-pretreated plants as compared with control plants (Fig. 3D). Thus, ABA contributes to virus resistance. The aba1, aba2, aba3, and abi4 mutants then were used to analyze their possible role in antiviral defenses. As shown in Fig. 3E, higher accumulation of TMV-cg in upper systemic leaves was observed in aba1, aba2, aba3, and especially in abi4 mutants as compared with wild-type plants. These results demonstrated that ABA signaling plays important roles during plant–TMV-cg interaction and that WRKY8 may participate in TMV-cg resistance, at least partially through ABA signaling.
Expression Profiling to Identify Putative WRKY8 Targets.
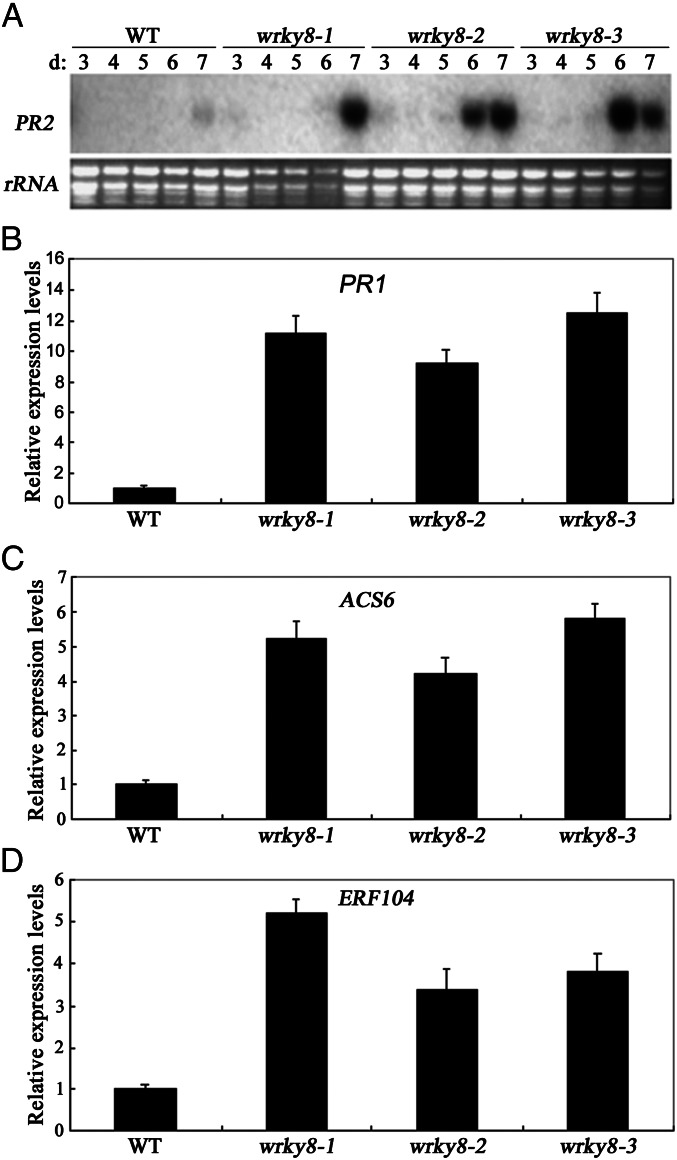
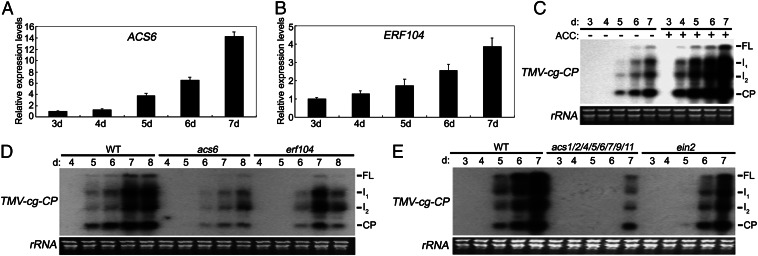
Infection by viruses causes the transcriptional reprogramming of their hosts, and the characterization of virally induced changes in host gene-expression patterns would provide valuable insight into the host’s response to viral activity. Thus, to dissect the role of the WRKY8 protein during TMV-cg infection further, we profiled the gene expression of the wrky8-1 mutant by microarray analysis (Dataset S1). Interestingly, when the microarray results in the wild type and wrky8-1 mutant after inoculation with TMV-cg for 6 d were compared, a number of defense-related genes (such as PR1 and PR2) and ET-synthesized or -responsive genes (such as ACS6 and ERF104) showed higher expression in the wrky8-1 mutant (Table S1). To confirm the validity of the microarray data, Northern blotting or qRT-PCR analysis was used to examine their expression levels. As shown in Fig. 4, the expression levels of these four genes were higher in wrky8-mutant plants than in wild-type plants; these results were in accordance with the microarray data. Based on these results, we deduced that both SA- and ET-signaling pathways might contribute to TMV-cg resistance.
Fig. 4.
Expression of PR2, ACS6, and ERF106. (A) Expression of PR2 in wild-type and wrky8 leaves systemically infected by TMV-cg at 3, 4, 5, 6, and 7 dpi. (B) Expression of PR1 in wild-type and wrky8 leaves systemically infected by TMV-cg at 6 dpi. (C) Expression of ACS6 in wild-type and wrky8 leaves systemically infected by TMV-cg a t6 dpi. (D) Expression of ERF104 in wild-type and wrky8 leaves systemically infected by TMV-cg at6 dpi. These experiments were repeated at least twice with similar results.
ET but Not SA Contributes to Virus Susceptibility.
To determine the role of both SA and ET in antiviral resistance, we examined their antiviral effect by infecting wild-type plants that were pretreated with aminocyclopropane-1-carboxylic acid (ACC) or SA. As shown in Fig. 5C, ACC treatment dramatically increased the accumulation of TMV-cg in systemically infected leaves; however, SA had no obvious effect (Fig. S2A). In the upper systemic leaves of infected plants, viral RNAs were detected at 4 dpi in ACC-treated plants but at 5 dpi in control plants (Fig. 5C). The accumulation of viral RNA also was significantly higher in plants pretreated with ACC. Thus, ET, but not SA, contributes to virus susceptibility. Therefore, we further analyzed the role of ET in antiviral defense.
Fig. 5.
The role of ET in defense against TMC-cg infection. (A) Expression of ACS6 in wild-type leaves systemically infected dpi by TMV-cg at 3, 4, 5, 6, and 7. (B) Expression of ERF104 in wild-type leaves systemically infected by TMV-cg at 3, 4, 5, 6, and 7 dpi. (C) ACC promotes the accumulation of TMV-cg in systemically infected leaves. Three leaves of 26-d-old wild-type plants treated or not treated with 100 µM ACC were inoculated with TMV-cg. Leaf collection, RNA isolation, and RNA blot analysis of TMV-cg CP were performed as in Fig. 1. (D) Mutation of acs6 or erf104 inhibits the accumulation of TMV-cg in systemically infected leaves. Inoculation, leaf collection, RNA isolation, and RNA blot analysis of TMV-cg CP were performed as in Fig. 1. (E) Mutation of eight acs genes (acs1/2/4/5/6/7/9/11) or ein2 inhibits the accumulation of TMV-cg in systemically infected leaves. Inoculation, leaf collection, RNA isolation, and RNA blot analysis of TMV-cg CP were performed as in Fig. 1. These experiments were repeated at least twice with similar results.
Based on the microarray data, the expression of ACS6 was one of the strongest among the ET-synthesized or -responsive genes upon TMV-cg infection in wrky8 mutants. Thus, acs6 mutants were used to analyze its possible role in antiviral defense. First, expression analysis showed that ACS6, unlike WRKY8, was strongly induced by TMV-cg infection in systemically infected leaves (Fig. 5A). Second, as shown in Fig. 5D, mutation of acs6 (Fig. S2B) significantly inhibited TMV-cg accumulation in systemic leaves. In the upper systemic leaves of infected plants, accumulation of viral RNAs was delayed for 1 d and also was greatly decreased in acs6-mutant plants. Another ET-responsive gene, ERF104, which previously was shown to be involved in plant basal immunity and stress-related interfascicular cambium initiation (47, 48), was analyzed for its antiviral ability. As shown in Fig. 5 B and D, in addition to its induced expression by TMV-cg infection in systemically infected leaves, which is similar to that in acs6 mutants, mutation of erf104 also inhibited TMV-cg in systemic leaves. To dissect the role of ET in antivirus defense further, two other ET-related mutants, ein2 and the octuple acs mutant (acs1/2/4/5/6/7/9/11), were used. As shown in Fig. 5E, mutation in these genes also greatly inhibited the accumulation of TMV-cg in systemic leaves. Thus, based on these results, we deduced that ET plays a negative role in anti–TMV-cg defense; these results also validated the function of WRKY8 in anti–TMV-cg defense.
In Vivo Interaction of WRKY8 with Target Promoters.
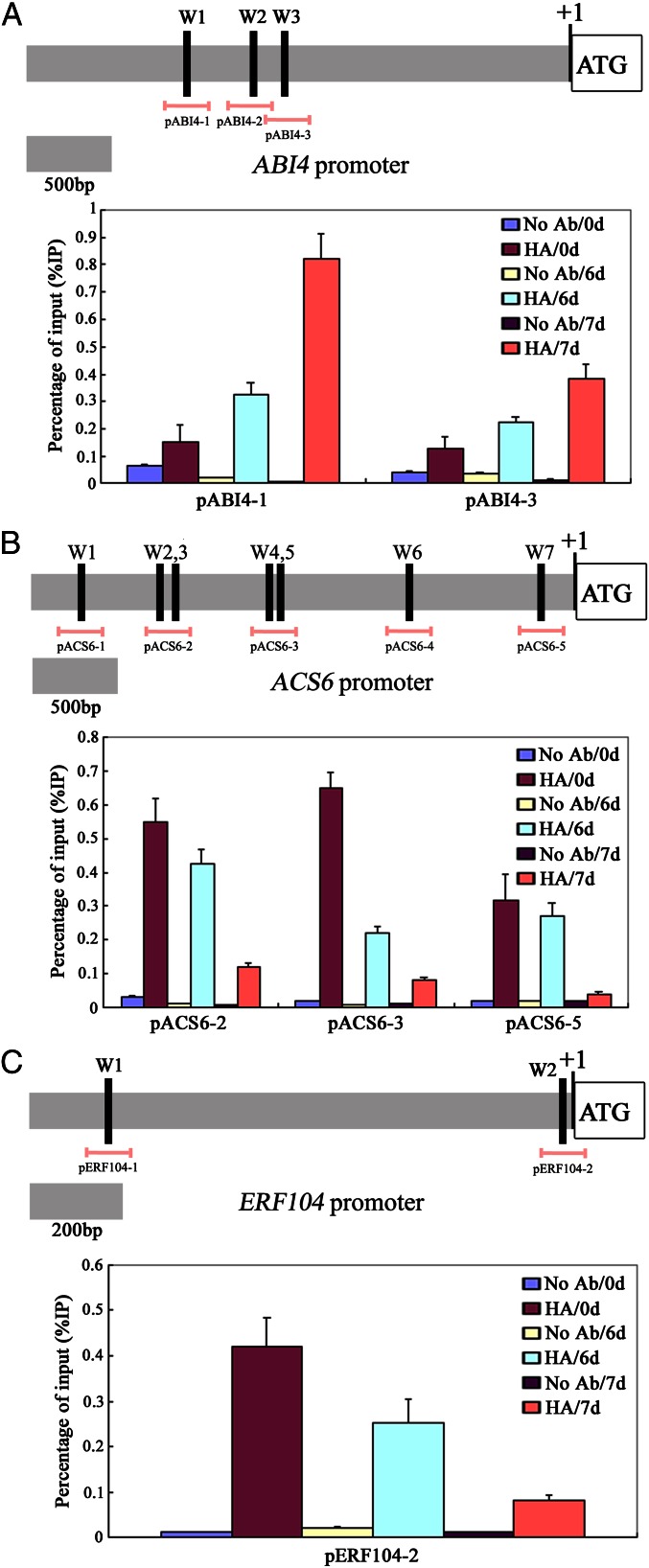
WRKY transcription factors function by binding directly to a putative cis-element, the W-box (T/CTGACC/T), of their target gene promoters (23, 35). Our data suggested that TMV-cg infection inhibits the accumulation of WRKY8, which positively modulates the expression of positive regulators of antivirus defense involved in the ABA signaling and negatively modulates the expression of ET-synthesized or -responsive genes in ET signaling. Interestingly, by searching the genome sequence, several putative W-box elements were found in the promoters of the genes involved in these pathways, indicating that modulation could be caused partly by direct interaction with WRKY factors, including WRKY8. To examine whether several of the genes are direct targets of WRKY8, in vivo ChIP assays were performed using a transgenic line expressing a WRKY8 cDNA construct with an N-terminal HA tag under the control of its native promoter in the wrky8-1 mutant (designated HA-WRKY8). Formaldehyde cross-linking was performed on control, 6-dpi, and 7-dpi plant material. Based on their clear differential expression profiles of mutant versus wild type in microarray and/or qRT-PCR analyses and on the presence of W-box elements within their respective promoters, we chose ABI4, ACS6, and ERF104 as candidate direct target genes of WRKY8. Then we performed detailed ChIP analyses. As shown in Fig. 6A, WRKY8 interacted with the ABI4 promoter when the primer combinations encompassing either W1 or W3 were used under normal conditions, and there was a stronger interaction between WRKY8 and ABI4 promoter upon TMC-cg interaction. WRKY8 also could bind selectively to several putative W-boxes of the ACS6 and ERF104 promoters under normal conditions, whereas the interaction between the WRKY8 protein and W-boxes of the ACS6 or ERF104 promoters was severely impaired by TMV-cg infection (Fig. 6 B and C). These results and the reduction or increase of ABI4, ACS6, and ERF104 transcript levels in wrky8 mutants compared with wild-type plants provide evidence that WRKY8, as a transcription factor, functions as a direct transcriptional activator or repressor of these three genes by binding selectively to the W-box motifs within their promoters.
Fig. 6.
ABI4, ACS6, and ERF104 are direct targets of WRKY8. (A–C) (Upper) Schematic representations of the ABI4, ACS6, and ERF104 promoter regions containing W-box clusters. Only prefect W-boxes (T/CTGACC/T, black bar) are depicted. The diagram indicates the number and relative position of the W-boxes in the respective promoters relative to the ATG start codon. In promoter fragment names, the prefix ”p” indicates promoter. Pink lines indicate the sequences detected by ChIP assays. (Lower) ChIP assays were performed with chromatin prepared from HA-WRKY8 plants infected b TMV-cg at 0, 6, and 7 dpi using an anti-HA antibody (IP) or preimmune serum (no Ab) as a negative control. ChIP results are presented as a percentage of input DNA, SDs were calculated from three technical repeats. One representative experiment is presented. Fig. S3 shows other two ChIP results. The experiments were repeated three times with similar results.
Discussion
Numerous studies have demonstrated that WRKY transcription factors function as important components in complex signaling processes during various stress responses, especially in biotic stresses. However, far less information is available about the functions of WRKY proteins in antiviral defense than about their functions in defense against bacterial or fungal pathogens. Here, we report the role of a WRKY transcription factor in anti–TMV-cg defense.
Mutation of WRKY8 Affects Virus Long-Distance Movement.
We reported previously that WRKY8 is induced by several stressful stimuli and is involved in regulating plant basal defense (43). In this study, we monitored the transcriptional reprogramming induced by TMV-cg infection, explored the role of WRKY8 in anti–TMV-cg defense, and explained how it regulates its downstream target genes to respond to TMV-cg infection.
We observed no obvious differences between the wild type and mutants in TMV-cg accumulation within locally incubated leaves, indicating that replication appears not to be accelerated in wrky8 mutants. However, an obvious difference between the wild type and wrky8 mutants was observed in systemically infected leaves, especially at 6 dpi. Thus, WRKY8 may play a role in inhibiting long-distance movement of the virus. For the virus to move systemically, it first must load into the vascular system from cells within the primary lesions and then translocate through the phloem, finally to be unloaded at distant susceptible tissues. Based on the GUS staining results, the expression of WRKY8 occurs mainly in the vascular bundles when responding to rubbing treatments, and its expression is inhibited by TMV-cg infection. These results suggest that the WRKY8 might function as an inhibitor of virus transport in vascular bundles.
WRKY8 Is a Negative Regulator for ACS6 or ERF104 Transcription but Functions as a Positive Regulator for ABI4 Transcription.
Virus infection causes numerous biochemical and physiological changes in compatible host plants (49). At the molecular level, the expression of numerous host genes is affected in both up-regulation and down-regulation (49). The virus-induced modifications in host gene expression occur transiently and concomitantly with viral replication as they spread from cell to cell away from the site of inoculation. Thus, by determining the expression patterns of the host plant during virus infection, researchers may obtain some clues to understand the complex interactions between the virus and its host plant.
TMV-cg can infect Arabidopsis plants efficiently but does not cause severe symptoms, nor does it induce cell death; thus it is an excellent model for studying virus–plant interactions (45). The qRT-PCR and microarray results showed that ABI4, one possible downstream target gene of WRKY8, was reduced by TMV-cg infection in systemically infected leaves, whereas the other possible target genes, ACS6 or ERF104, were strongly induced. Thus, the expression of ABI4 was similar to that of WRKY8, but ACS6 and ERF104 had the opposite pattern. Therefore, we proposed that WRKY8 might positively modulate the expression of positive regulators of antivirus defense involved in the ABA signaling while negatively modulating the expression of ET-synthesized or -responsive genes in ET signaling. The involvement of WRKY8 in the anti–TMV-cg defense response may be mediated via these two pathways.
Roles of ET and ABA in Anti–TMV-cg Defense.
Several previous studies showed that ET signaling might play an important role in antiviral defense. Overexpression of NtERF5, an ET-response transcription factor, conferred enhanced resistance to tobacco mosaic virus infection, showing reduced size of local hypersensitive-response lesions and impaired systemic spread of the virus (50). Mutations in two ET-signaling components, etr1 and ein2, delayed the formation of systemic symptoms caused by CaMV infection compared with the wild-type components (20). The results presented here also suggest that WRKY8 mediates ET-signaling pathways that are involved in anti–TMV-cg defense. In wrky8 mutants, several ET-synthesized or responsive transcription factors, such as ACS6 and ERF104, were more strongly induced in TMV-cg systemically infected leaves. Functional analysis using mutants showed that acs6, erf104, ein2, or octuple acs mutants had reduced accumulation of TMV-cg RNA in systemically infected leaves compared with the wild type, indicating an important role of ET in anti–TMV-cg defense.
ET (C2H4), an important plant hormone, is involved in both developmental processes and stress responses in plants (51). Various stresses, such as wounding, drought, and pathogen/insect invasion, enhance the production of ET (52–54). ACC synthases function as the rate-limiting enzymes during ET biosynthesis (55). ACS6 is a type I ACS isoform, one of nine ACS members. Interestingly, the expression of each ACS gene is differentially regulated by developmental and environmental signals (52, 55–57), suggesting their unique and overlapping functions during plant growth and stress response. Although at least three ACS genes (ACS2, ACS6, and ACS7) were strongly activated by B. cinerea infection (39), only ACS6 showed relatively higher expression upon TMV-cg infection in systemically infected leaves, suggesting that ACS6 may play a major role in anti–TMV-cg defense. ERF104, an ET response factor that previously was shown to participate in plant resistance to nonadapted bacterial pathogens (47), is also involved in anti–TMV-cg defense. Consistent with our results, exogenous applications of ACC also induce susceptibility to TMV-cg infection. Thus, our results demonstrate that ET plays a negative role in TMV-cg–plant interactions.
The plant hormone ABA is a stress hormone that has essential roles in the initiation of adaptive responses to various environmental conditions. ABA has both negative and positive roles in plant biotic stress responses, and its efficacy is dependent on the host–pathogen combination (46). In plant–virus interactions, several previous studies demonstrated a positive correlation between ABA levels and antiviral resistance. In tobacco, exogenous application of ABA increases resistance to TMV infection, and TMV infection also increases ABA concentrations (58). Recently, Iriti and Faoro (59) showed that exogenous application of ABA induces a significant resistance to tobacco necrosis virus. Furthermore, ABA also can exert its positive effect on virus infection by downregulating the transcriptional level of β-1,3-glucanase genes (19). Our results also suggest that ABA plays an important role in defense against TMV-cg infection. Exogenous application of ABA greatly inhibited the accumulation of TMV-cg RNA in systemically infected leaves; studies using both ABA-deficient and -insensitive mutants showed that the transportation of TMV-cg was faster in these ABA-related mutants than in wild-type plants. Thus, our results also showed that ABA has a positive role in TMV-cg infection.
Mechanism of ABI4, ACS6, and ERF104 Regulated by WRKY8.
Although different WRKY genes may have different functions, they act primarily by binding to cis-acting DNA elements with the minimal consensus T/CTGACC/T W-box sequence in the promoters of specific targets to modulate the temporal and spatial expression of these genes (23, 35). Both the differential expression patterns of WRKY genes under various environmental conditions and the transcriptional-activating or -repressing activity of WRKY proteins may constitute molecular mechanisms by which they perform their specific roles. In addition, WRKY proteins could constitutively occupy the W-box sequences in the promoters of defense genes under normal growth conditions; however, upon infection or elicitor treatment, they may be competitively displaced by other family members (60).
As shown in Fig. S4, WRKY8 shows basal expression in various tissues. Therefore, WRKY8 protein is present in the preformed WRKY factor pool and may mediate transcriptional activation or repression of potential target genes. Based on our ChIP results, when plants were not affected by TMV-cg, WRKY8 repressed ACS6 expression by binding to W-boxes upstream in the ACS6 promoter to reduce ET production. Upon TMV-cg infection, consistent with the inhibited expression of WRKY8, the direct binding of WRKY8 protein to ACS6 promoter was released, and the expression of ACS6 or ERF104 was induced to enhance ET production or ET signaling, ultimately promoting the systemic transport of TMV-cg in infected host plants. In contrast, during infection, WRKY8 binds to the W-box elements upstream in the ABI4 promoter, increasing its expression. Therefore, based on our results, WRKY8 participates in TMV-cg defense through both the ET- and ABA-signaling pathways.
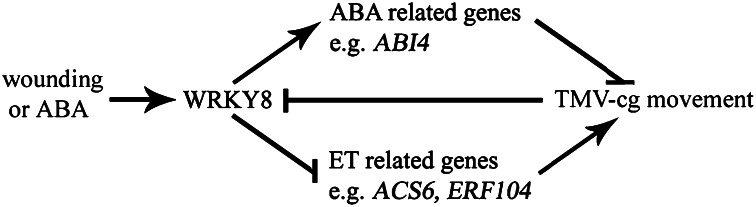
Our results suggested that WRKY8 might mediate crosstalk between ET and ABA signaling when plants are infected by TMV-cg. In the compatible TMV-cg–Arabidopsis interaction, to infect Arabidopsis systemically, TMV-cg may use an unknown mechanism to repress ABA signaling, in addition to enhanced ET synthesis and signaling to facilitate long-distant movement. Thus, WRKY8 may participate in TMV-cg defense by counteracting the effect of TMV-cg host modification. Finally, we propose a simplified model to depict the role of WRKY8 during TMV-cg–Arabidopsis interaction (Fig. 7).
Fig. 7.
A simplified model for the function of WRKY8 during TMV-cg–Arabidopsis interaction. Wounding or ABA treatment induces WRKY8 expression, whereas infection by TMV-cg inhibits WRKY8 expression. WRKY8 participates in the TMV-cg defense response by both activating the expression of ABI4 and repressing ET-related genes such as ACS6 and ERF104 during TMV-cg–Arabidopsis interaction.
Based on our results, we deduced that ET and ABA have opposite roles in the anti–TMV-cg defense response and may interact antagonistically during plant–virus interaction. Indeed, numerous studies have shown that ABA interacts with ET-mediated signaling. High concentrations of ABA inhibit ET production, and the ABA- and ET-signaling pathways interact mostly antagonistically both during plant development and in the stress response (61–64). Elucidating the role of WRKY- mediated ET and ABA signaling in anti–TMV-cg defense adds to our understanding of the complicated phenomenon of plant–virus interaction.
Materials and Methods
Materials.
We obtained [32P]-dATP (>3,000 Ci/mmol) from the Beijing Furui Biotechnology Co., Ltd. ABA, SA, ACC, and X-glucuronide were purchased from Sigma Co. Ltd; Taq DNA polymerase was purchased from TaKaRa Biotechnology (Dalian) Co. Ltd; and agarose and agar were from purchased from Shanghai Sangon Biotechnology Co. Ltd. Arabidopsis thaliana plants were grown in an artificial growth chamber at 22 °C under 180 μE·m2·sec–1 light with a 10-h light/14-h dark photoperiod. Columbia-0 (Col) was used as the wild type. We obtained wrky8, asc6, erf104, abi4, aba1, aba2, aba3, and octuple acs mutant (CS16651, acs1/2/4/5/6/7/9/11) from the Arabidopsis Biological Resource Center. ein2 were kindly gifted by Zhixiang Chen (Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN).
Two homozygous T3 lines of pWRKY8:GUS transgenic plants (carrying the reporter gene driven by a 1.75-kb promoter fragment of WRKY8) were used in GUS staining (43).
Induction Treatments.
SA was dissolved in water as a 100-mM stock solution and was adjusted to pH 6.5 with KOH. Plants were sprayed with a 2-mM SA solution diluted from the stock. ACC was dissolved in water, and a 2-mM solution was sprayed onto plants. First, 14.1 mg of ABA was dissolved in 90 μL ethanol; then water was added to obtain a 10-mM stock solution. The ABA stock solution was diluted to 100 μM with water and sprayed onto plants.
Northern Blotting and qRT-PCR Analysis.
For Northern blotting analyses, total RNA was extracted using TRIzol reagent (Invitrogen). Approximately 3 μg of RNA was separated on an agarose-formaldehyde gel and then was blotted onto nylon membranes following standard procedures. The membranes were hybridized with (α-32P) -dATP–labeled DNA probes. Hybridization was performed in PerfectHyb Plus Hybridization Buffer (Sigma) for 16 h at 68 °C. The membranes were washed once for 10 min with 2× SSC and 0.5% SDS, twice for 20 min with 0.5× SSC and 0.1% SDS, and once for 20 min with 0.1× SSC and 0.1% SDS at 68 °C and then were exposed to X-ray films at −80 °C. DNA probes for PR2 and TMV-cg CP were obtained from PCR amplifications. The gene-specific primers used are listed in Table S2.
For qRT-PCR analysis, total RNA was extracted using TRIzol reagent (Invitrogen) and was treated with RNase-free DNase (Fermentas) according to the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed in a 20-μL reaction mixture using the SuperScript II (Invitrogen). After the reaction, 1-μL aliquots were used as templates for qRT-PCR. Half-reactions (10 μL each) were performed with the Lightcycler FastStart DNA Master SYBR Green I Kit (Roche) on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer's instructions. ACT2 (AT3G18780) was used as a control in qRT-PCR. The primers used for qRT-PCR amplification of different genes are listed in Table S2.
GUS Staining.
Histochemical detection of GUS activity was performed with 5-bromo-4- chloro-3-indolyl b-d-glucuronic acid (X-gluc) as the substrate. Plant tissues first were prefixed in ice-cold 90% (vol/vol) acetone for 20 min and then were washed three times with GUS staining buffer (without X-gluc) before incubation in X-gluc solution [1 mM X-gluc, 50 mM NaPO4 (pH 7), 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6, and 0.05% Triton X-100] under a vacuum for 10 min at room temperature and then were incubated overnight at 37 °C. Chlorophyll was removed using several changes of 70% (vol/vol) ethanol, and the tissues were photographed.
Virus Infection.
Inoculation of Arabidopsis plants with TMV-cg was performed as previously described (15). TMV-cg was mechanically inoculated on the third, fourth, and fifth true leaves of plants by rubbing the leaf with the virus (5 μg/mL solution in 5 mM sodium phosphate, pH 7.5) mixed with carborundum. Mock inoculation was performed with the phosphate buffer only.
Affymetrix Microarray Analysis.
An Affymetrix microarray chip covering ∼22,747 genes was used. Preparation of cDNA from total RNA and hybridization to ATH1 Arabidopsis Genome Arrays (Affymetrix Inc.) was performed by the Shanghai Jintai Biological Technology Co. Ltd. in accordance with the standard manufacturer’s protocol (Affymetrix 2001). The resulting data files were normalized and analyzed with Expression Console (Affymetrix) and Partek Genomics Suite 6.4 (Partek Inc) and were processed with Microsoft Excel and Access (Microsoft). The expression profiles of TMV-cg–infected wrky8-1 and wild-type plants were compared 6 dpi.
ChIP Assays.
ChIP assays were performed essentially in accordance with previously described protocols (65). A transgenic line was created expressing a WRKY8 cDNA construct with an N-terminal HA tag under the control of its native promoter (2,577 bp) in wrky8-1 mutant plants (designated HA-WRKY8). The 26-d-old HA-WRKY8 plants were incubated with TMV-cg for 0, 6, or 7 d, and then these materials were used for ChIP assays. An HA antibody (Thermofisher Pierce) was used to immunoprecipitate the protein–DNA complex, and the precipitated DNA was purified using a PCR purification kit for qRT-PCR analysis. The ChIP experiments were performed three times. Chromatin precipitated without antibody was used as the negative control, and the isolated chromatin before precipitation was used as the input control. ChIP results are presented as a percentage of input DNA. The primers used for qRT-PCR amplification of different promoters are listed in Table S2.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center at Ohio State University for supplying the wrky8, aba1, aba2, aba3, abi4, acs6 (Salk_138142C), erf104 (Salk_057720C), and octuple acs (CS16651, acs1/2/4/5/6/7/9/11) mutants, Dr. Zhixiang Chen for providing ein2 and TMV-cg, and anonymous reviewers for insightful comments on the manuscript drafts. This research was supported by the Science Foundation of the Ministry of Agriculture of the Peoples’ Republic of China Grant 2009ZX08009-066B, Natural Science Foundation of China Grants 90817003 and 31200915, and the Science Foundation of the Chinese Academy of Sciences 135 program XTBG-F04.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221347110/-/DCSupplemental.
References
- 1.Hills GJ, et al. Immunogold localization of the intracellular sites of structural and nonstructural tobacco mosaic virus proteins. Virology. 1987;161(2):488–496. doi: 10.1016/0042-6822(87)90143-7. [DOI] [PubMed] [Google Scholar]
- 2.Wolf S, Deom CM, Beachy RN, Lucas WJ. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science. 1989;246(4928):377–379. doi: 10.1126/science.246.4928.377. [DOI] [PubMed] [Google Scholar]
- 3.Waigmann E, Lucas WJ, Citovsky V, Zambryski PC. Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc Natl Acad Sci USA. 1994;91(4):1433–1437. doi: 10.1073/pnas.91.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgyán J, Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16(5):265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000;19(5):792–799. doi: 10.1093/emboj/19.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai J, et al. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 2009;57(5):905–917. doi: 10.1111/j.1365-313X.2008.03737.x. [DOI] [PubMed] [Google Scholar]
- 7.Cui X, Fan B, Scholz J, Chen Z. Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell. 2007;19(4):1388–1402. doi: 10.1105/tpc.107.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbers K, Meuwly P, Frommer WB, Metraux JP, Sonnewald U. Systemic acquired resistance mediated by the ectopic expression of invertase: Possible hexose sensing in the secretory pathway. Plant Cell. 1996;8(5):793–803. doi: 10.1105/tpc.8.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, et al. Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J. 2010;62(1):12–23. doi: 10.1111/j.1365-313X.2009.04120.x. [DOI] [PubMed] [Google Scholar]
- 10.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130(3):413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 12.Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25(14):3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313(5783):68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 14.Morel JB, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14(3):629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D, Fan B, MacFarlane SA, Chen Z. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant Microbe Interact. 2003;16(3):206–216. doi: 10.1094/MPMI.2003.16.3.206. [DOI] [PubMed] [Google Scholar]
- 16.Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49(4):493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 17.Jakubiec A, Yang SW, Chua NH. Arabidopsis DRB4 protein in antiviral defense against Turnip yellow mosaic virus infection. Plant J. 2012;69(1):14–25. doi: 10.1111/j.1365-313X.2011.04765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE. 2009;4(3):e4971. doi: 10.1371/journal.pone.0004971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezzonico E, Flury N, Meins F, Jr, Beffa R. Transcriptional down-regulation by abscisic acid of pathogenesis-related beta-1,3-glucanase genes in tobacco cell cultures. Plant Physiol. 1998;117(2):585–592. doi: 10.1104/pp.117.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love AJ, et al. Components of Arabidopsis defense- and ethylene-signaling pathways regulate susceptibility to Cauliflower mosaic virus by restricting long-distance movement. Mol Plant Microbe Interact. 2007;20(6):659–670. doi: 10.1094/MPMI-20-6-0659. [DOI] [PubMed] [Google Scholar]
- 21.Love AJ, Yun BW, Laval V, Loake GJ, Milner JJ. Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol. 2005;139(2):935–948. doi: 10.1104/pp.105.066803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinformatics. 2007;8:42. doi: 10.1186/1471-2105-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5(5):199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 24.Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10(2):71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15(5):247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, et al. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta. 2012;1819(2):120–128. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Zheng ZY, Qamar SA, Chen ZX, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48(4):592–605. doi: 10.1111/j.1365-313X.2006.02901.x. [DOI] [PubMed] [Google Scholar]
- 28.Lai Z, Vinod K, Zheng Z, Fan B, Chen Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008;8:68. doi: 10.1186/1471-2229-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Brader G, Palva ET. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16(2):319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray SL, Ingle RA, Petersen LN, Denby KJ. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol Plant Microbe Interact. 2007;20(11):1431–1438. doi: 10.1094/MPMI-20-11-1431. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Dong Q, Yu D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012;185-186:288–297. doi: 10.1016/j.plantsci.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233(6):1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 33.Zou C, Jiang W, Yu D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot. 2010;61(14):3901–3914. doi: 10.1093/jxb/erq204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren X, Huang W, Li W, Yu D. Tobacco transcription factor WRKY4 is a modulator of leaf development and disease resistance. Biol Plant. 2010;54(4):684–690. [Google Scholar]
- 35.Ulker B, Somssich IE. WRKY transcription factors: From DNA binding towards biological function. Curr Opin Plant Biol. 2004;7(5):491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 36.Shang Y, et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22(6):1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey SP, Roccaro M, Schön M, Logemann E, Somssich IE. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J. 2010;64(6):912–923. doi: 10.1111/j.1365-313X.2010.04387.x. [DOI] [PubMed] [Google Scholar]
- 38.Birkenbihl RP, Diezel C, Somssich IE. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012;159(1):266–285. doi: 10.1104/pp.111.192641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, et al. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 2012;8(6):e1002767. doi: 10.1371/journal.pgen.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Y, Liang G, Yu D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant. 2012;5(6):1375–1388. doi: 10.1093/mp/sss080. [DOI] [PubMed] [Google Scholar]
- 41.Aguilar I, Sánchez F, Martin Martin A, Martinez-Herrera D, Ponz F. Nucleotide sequence of Chinese rape mosaic virus (oilseed rape mosaic virus), a crucifer tobamovirus infectious on Arabidopsis thaliana. Plant Mol Biol. 1996;30(1):191–197. doi: 10.1007/BF00017814. [DOI] [PubMed] [Google Scholar]
- 42.Lartey RT, Voss TC, Melcher U. Tobamovirus evolution: Gene overlaps, recombination, and taxonomic implications. Mol Biol Evol. 1996;13(10):1327–1338. doi: 10.1093/oxfordjournals.molbev.a025579. [DOI] [PubMed] [Google Scholar]
- 43.Chen L, Zhang L, Yu D. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol Plant Microbe Interact. 2010;23(5):558–565. doi: 10.1094/MPMI-23-5-0558. [DOI] [PubMed] [Google Scholar]
- 44.Hu Y, et al. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013 doi: 10.1111/tpj.12159. [DOI] [PubMed] [Google Scholar]
- 45.Pereda S, Ehrenfeld N, Medina C, Delgado J, Arce-Johnson P. Comparative analysis of TMV-Cg and TMV-U1 detection methods in infected Arabidopsis thaliana. J Virol Methods. 2000;90(2):135–142. doi: 10.1016/s0166-0934(00)00230-5. [DOI] [PubMed] [Google Scholar]
- 46.Mauch-Mani B, Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8(4):409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Bethke G, et al. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA. 2009;106(19):8067–8072. doi: 10.1073/pnas.0810206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sehr EM, et al. Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 2010;63(5):811–822. doi: 10.1111/j.1365-313X.2010.04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maule A, Leh V, Lederer C. The dialogue between viruses and hosts in compatible interactions. Curr Opin Plant Biol. 2002;5(4):279–284. doi: 10.1016/s1369-5266(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 50.Fischer U, Dröge-Laser W. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol Plant Microbe Interact. 2004;17(10):1162–1171. doi: 10.1094/MPMI.2004.17.10.1162. [DOI] [PubMed] [Google Scholar]
- 51.Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. New York: Academic; 1992. [Google Scholar]
- 52.Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14(Suppl):S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Broekaert WF, Delauré SL, De Bolle MF, Cammue BP. The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol. 2006;44:393–416. doi: 10.1146/annurev.phyto.44.070505.143440. [DOI] [PubMed] [Google Scholar]
- 54.van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11(4):184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- 56.Tsuchisaka A, Theologis A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004;136(2):2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng HP, Lin TY, Wang NN, Shih MC. Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol Biol. 2005;58(1):15–25. doi: 10.1007/s11103-005-3573-4. [DOI] [PubMed] [Google Scholar]
- 58.Whenham RJ, Fraser RSS, Brown LP, Payne JA. Tobacco mosaic virus-induced increase in abscisic acid concentration in tobacco leaves: Intracellular location in light and dark green areas, and relationship to symptom development. Planta. 1986;168(4):592–598. doi: 10.1007/BF00392281. [DOI] [PubMed] [Google Scholar]
- 59.Iriti M, Faoro F. Abscisic acid is involved in chitosan-induced resistance to tobacco necrosis virus (TNV) Plant Physiol Biochem. 2008;46(12):1106–1111. doi: 10.1016/j.plaphy.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Turck F, Zhou A, Somssich IE. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley. Plant Cell. 2004;16(10):2573–2585. doi: 10.1105/tpc.104.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LeNoble ME, Spollen WG, Sharp RE. Maintenance of shoot growth by endogenous ABA: Genetic assessment of the involvement of ethylene suppression. J Exp Bot. 2004;55(395):237–245. doi: 10.1093/jxb/erh031. [DOI] [PubMed] [Google Scholar]
- 62.Beaudoin N, Serizet C, Gosti F, Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12(7):1103–1115. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghassemian M, et al. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;12(7):1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson JP, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16(12):3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008;3(6):1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.