Abstract
Heat shock protein (Hsp) 104 is a ring-forming, protein-remodeling machine that harnesses the energy of ATP binding and hydrolysis to drive protein disaggregation. Although Hsp104 is an active ATPase, the recovery of functional protein requires the species-specific cooperation of the Hsp70 system. However, like Hsp104, Hsp70 is an active ATPase, which recognizes aggregated and aggregation-prone proteins, making it difficult to differentiate the mechanistic roles of Hsp104 and Hsp70 during protein disaggregation. Mapping the Hsp70-binding sites in yeast Hsp104 using peptide array technology and photo–cross-linking revealed a striking conservation of the primary Hsp70-binding motifs on the Hsp104 middle-domain across species, despite lack of sequence identity. Remarkably, inserting a Strep-Tactin binding motif at the spatially conserved Hsp70-binding site elicits the Hsp104 protein disaggregating activity that now depends on Strep-Tactin but no longer requires Hsp70/40. Consistent with a Strep-Tactin–dependent activation step, we found that full-length Hsp70 on its own could activate the Hsp104 hexamer by promoting intersubunit coordination, suggesting that Hsp70 is an activator of the Hsp104 motor.
Keywords: ClpB, DnaK, Hsp100, molecular chaperone
Molecular chaperones assist protein folding by preventing protein misfolding and aggregation. However, most chaperones do not recover functional protein from aggregates. The ring-forming heat shock protein (Hsp) 100 ATPase is the principal protein disaggregase in yeast (Hsp104), bacteria (ClpB), and plants (Hsp101) and is essential for cell survival during severe stress (1–4). Hsp100 consists of an N-terminal domain (NTD), a middle (M-) domain, and two ATP-binding domains (D1 and D2) (Fig. 1A). The M-domain is essential for protein disaggregation and forms an 85-Å-long coiled-coil (5, 6). Although Hsp100 is an active ATPase, the recovery of functional protein from aggregates requires the synergistic interaction with the cognate Hsp70 and Hsp40 molecular chaperones (Hsp70/40 in yeast and DnaK/DnaJ/GrpE in eubacteria) (7–10), which together form a powerful bichaperone system.
Fig. 1.
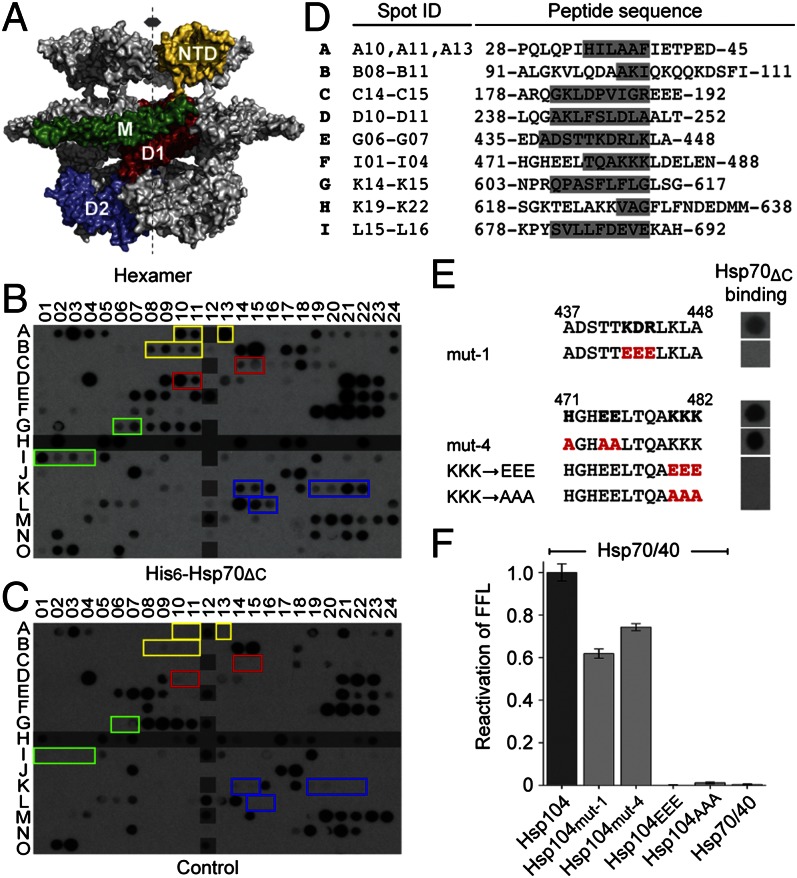
Identifying the Hsp70:Hsp104 binding sites using peptide array technology. (A) 3D structure of Hsp104 based on the cryoEM fit (6). The NTD is colored yellow, the D1-domain red, the M-domain green, and the D2-domain blue. The symbol indicates the sixfold symmetry axis. (B) Hsp104 peptide array probed with His6-Hsp70ΔC and (C) with Hsp70ΔC (without His6-tag), which functions as a negative control. Overlapping peptides containing at least two consecutive spots are boxed and colored according to domain location. Peptides that function as either positive or negative internal binding controls, including His6- and poly-alanine–containing peptides, are highlighted in gray. (D) Sequence of Hsp70ΔC-binding peptides. Consensus motifs are boxed in gray. (E) Binding of His6-Hsp70ΔC to mutated Hsp70-binding motifs in the Hsp104 M-domain. Mutated residues are shown in red. (F) FFL activity recovered by Hsp104 mutants together with Hsp70/40. Averages of three independent measurements ± SD are shown.
In addition to its role in the stress response, Hsp104 also governs the inheritance and maintenance of yeast prions (11, 12). Yeast prions resemble mammalian prions in their non-Mendelian inheritance (13, 14) and amyloid state (15) and may confer evolutionary advantages (16) and, perhaps, long-term memory formation (17). More recently, Hsp104 was implicated in yeast longevity and in the asymmetric distribution of oxidatively damaged, carbonylated proteins (18), and in mediating the sorting of tail-anchored proteins to the endoplasmic reticulum membrane (19), underscoring the multifaceted functional roles of Hsp104.
The Hsp70/40 requirement for protein disaggregation (7) and prion remodeling (20–22) by Hsp104 suggests a shared underlying mechanism. Consistently, ClpB, the bacterial homolog of Hsp104, cooperates with the bacterial Hsp70 system in protein disaggregation (8–10). Although it was presumed that ClpB could not remodel yeast prions (23), it was shown recently that ClpB could replace yeast Hsp104 in vivo when DnaK and GrpE are also present (24).
The prevailing model suggests that the Hsp70 system targets the Hsp104 motor to aggregates (20, 22), from which Hsp104 extracts polypeptides using pore loops in D1 and D2, perhaps with assistance from the NTDs (25), by threading the polypeptide through the central channel of the Hsp104 hexamer ring (20, 21, 26, 27). This threading mechanism is similar to that of the structurally homologous ClpA ATPase, which lacks an M-domain and together with ClpP degrades ssrA-tagged proteins (28–30). Hsp104 neither associates with ClpP nor degrades proteins but instead releases the unfolded polypeptide from the distal end, which then refolds spontaneously or with assistance from Hsp70/40, depending on the substrate.
Although not mutually exclusive, it was demonstrated more recently that Hsp104 can also recognize substrates directly and recover functional protein when its ATPase activities are decelerated asymmetrically (31, 32). Consistent with an innate protein remodeling activity, we recently showed that an engineered Hsp104 variant, which featured a T4 lysozyme insertion in the M-domain (Hsp104T4L), had gained the ability to recover functional protein from aggregates in the absence of Hsp70/40 (6, 33). Because it seems that the Hsp70 system is dispensable for substrate binding, Hsp70 must have other functions in addition to targeting Hsp104 to aggregates.
The species-specific cooperation of the bichaperone system suggested a physical interaction between Hsp104 and Hsp70, Hsp40, or both. In support of a direct interaction, it was shown that replacing the M-domain of yeast Hsp104 with that of bacterial ClpB is sufficient to switch the species-specificity of the bichaperone system so that the yeast Hsp104 chimera now cooperated with the bacterial Hsp70 system in protein disaggregation, and vice versa (33, 34).
Here we used peptide array technology and site-specific photo-cross-linking to map the Hsp70-binding motifs in yeast Hsp104. We found that the principal Hsp70-binding site on the M-domain is spatially conserved across species, despite lack of sequence conservation. Remarkably, inserting an 8-aa Strep-Tactin binding motif at this site elicits an Hsp104 protein disaggregating activity that is now dependent on Strep-Tactin but no longer requires Hsp70/40. Consistent with a Strep-Tactin binding-dependent activation step, we found that Hsp70 alone elicits the protein disaggregating function by promoting intersubunit coordination in Hsp104. Together, our findings provide molecular insight into the mechanism of protein disaggregation by the Hsp104 bichaperone system and suggest that Hsp70 is an activator of Hsp104.
Results
Identifying the Hsp70–Hsp104 Interaction Sites.
The Hsp104 M-domain coiled-coil is composed of four α-helices, which can be subdivided into two smaller coiled-coils, termed motif 1 and motif 2 (5, 6). It was previously shown that mutations in the ClpB M-domain helix 2 had a small but significant effect on substrate recovery (34), whereas mutations in helix 3 abrogated protein disaggregation (35), which was attributed to a potentially impaired interaction between ClpB and DnaK (35).
Because the sequence of the M-domain differs between species, we asked whether the Hsp70-binding motifs are conserved in Eukarya. We therefore mapped the Hsp70 interacting sites in yeast Hsp104 using peptide array technology. Miniaturized libraries of overlapping 12mer peptides were synthesized by walking through the Saccharomyces cerevisiae Hsp104 amino acid sequence, advancing two or three amino acids at a time. The library was probed with a bacterially expressed Hsp70 variant that lacked the 10-kDa C-terminal domain (Hsp70ΔC), a deletion known to eliminate Hsp70 polymerization (36, 37) but that is functional in uncoating clathrin-coated vesicles (36).
Probing our Hsp104 peptide array with His6-Hsp70ΔC (Fig. 1B), and eliminating peptides due to nonspecific binding by the antibody probe (Fig. 1C), resulted in 44 distinct peptides, 25 of which contained at least two consecutive spots on the membrane and were grouped, which resulted in nine consensus motifs (motifs A–I) (Fig. 1D). The remaining 19 peptides, which did not have neighboring spots, are shown in Fig. S1. It is of interest that many of the consensus motifs are enriched in hydrophobic residues (Leu, Ile, Val, Phe, and Tyr), which are often found in Hsp70 substrates (38). Importantly, we found that the location of consensus motifs within M-domain helix 2 (motifs E and F) overlapped with mutation sites in Escherichia coli ClpB, which either impaired ClpB’s innate protein disaggregating activity or affected the functional cooperation between ClpB and DnaK/DnaJ/GrpE in substrate recovery (34). For instance, Hsp70ΔC recognized both Lys442/Asp443/Arg444 (motif E) and His471/Glu474/Glu475 (motif F) (Fig. 1D), which correspond to Lys438/Lys439/Arg440 (Mut-1) and Ala467/Ser470/Gly471 (Mut-4) in ClpB (34). To determine the specificity of the Hsp70–peptide interaction, we replaced Lys442/Asp443/Arg444 with glutamates (mut-1), and His471/Glu474/Glu475 with alanines (mut-4). Although the mut-1 peptide abolished Hsp70ΔC binding, the mut-4 peptide did not (Fig. 1E). However, when introduced into Hsp104, both Hsp104mut-1 and Hsp104mut-4 reduced the recovery of aggregated firefly luciferase (FFL) somewhat (Fig. 1F), indicating that other Hsp104 sites contribute toward Hsp70 binding. Notably, we found that mutating Lys480/Lys481/Lys482 in the M-domain motif 2 downstream of the mut-4 site to glutamate or alanine abolished both Hsp70ΔC interaction and protein disaggregation (Fig. 1 E and F). Together, our findings suggest that the principal Hsp70-binding site on the M-domain is spatially conserved across species, and mutations introduced into the M-domain motif 2, which abrogate Hsp70 binding, interfere with the functional cooperation of the bichaperone system.
Hsp70NBD Recognizes the Hsp104 M-Domain Motif 2.
Hsp70 is a multidomain protein consisting of an N-terminal ATP binding domain (Hsp70NBD) and a substrate binding domain, which can be subdivided into a substrate-binding pocket (Hsp70SBDΔC) and a C-terminal domain (Hsp70CTD) (Fig. 2A). To determine which domain is responsible for Hsp104 interaction, we developed a bead-binding assay using immobilized Hsp104 hexamer as probe to capture purified full-length Hsp70 and truncated variants. We found that Hsp70, Hsp70ΔC, and Hsp70NBD were bound by Hsp104 (Fig. 2B). Binding was strongest in the presence of nucleotide (Fig. 2B). However, neither Hsp70 nor Hsp104 recognized the other protein as pseudosubstrate, because an interaction was observed in the presence of ATP, which weakens Hsp70–substrate interaction (39), as well as in the presence of ADP, which does not support binding of substrates to Hsp104 (40). Moreover, binding was also observed in the absence of nucleotide, albeit more weakly, and with Hsp70NBD that lacked the substrate-binding domain altogether (Fig. 2B). Conversely, Hsp70SBDΔC and yeast Hsp40 did not bind Hsp104 even in the presence of nucleotide (Fig. 2B), suggesting that Hsp70NBD is required for Hsp104 interaction.
Fig. 2.
Hsp70 binds to the M-domain via its nucleotide-binding domain. (A) Domain structure of Hsp70 (52). The ATP-binding domain (NBD) is colored orange, the substrate-binding pocket (SBDΔC) purple, and the C-terminal domain (CTD) gray. The N and C termini are indicated. (B) Bead-binding assay using immobilized Hsp104 hexamer to capture full-length Hsp70, Hsp70ΔC, Hsp70NBD, Hsp70SBDΔC, or Hsp40. Input, final wash, and eluate were analyzed by Western blotting. The eluate from a negative control using beads only (no Hsp104) is also shown. (C) The M-domain consists of four α-helices (H1–H4), which make up motif 1 (cyan) and motif 2 (gold). The locations of the introduced cysteine mutations are shown as purple spheres. (D) Photo-activated cross-linking of BPIA-labeled Hsp104* variants and Hsp70ΔC (ΔC), Hsp70NBD (N), and Hsp70SBDΔC (S) in the presence of ADP. Arrows indicate the cross-linked products with Hsp70ΔC (red) and Hsp70NBD (black).
To confirm the direct interaction between Hsp70 and the Hsp104 M-domain, we probed this interaction by site-specific photo-activated cross-linking using engineered Hsp104 variants that feature a surface-exposed cysteine in the M-domain. To generate these mutants, we replaced Glu435, Asn467, Glu468, His471, or Arg496 with cysteine in a cysteine-less Hsp104 variant (Hsp104*), which assembles into hexamers and cooperates with Hsp70/40 in protein disaggregation (Fig. S2). The introduced cysteines are located at the tip of motif 1 (E435C) or motif 2 (R496C), or in the middle of helix 2 (N467C, E468C, and H471C) (Fig. 2C) and can be labeled with the thiol-specific photo-cross-linker benzophenone-4-iodoacetamide (BPIA) to capture Hsp70 interaction on exposure to UV radiation.
We found that Hsp70ΔC and Hsp70NBD can be cross-linked to Hsp104R496C to yield a ∼160-kDa and ∼150-kDa product, respectively (Fig. 2D). Other Hsp104 variants showed only weak cross-linking, if at all (Fig. 2D). As anticipated, no cross-linked products were observed with Hsp104* or in a mock reaction without Hsp104. Thus, our findings suggest that Hsp70 interacts with motif 2 of the Hsp104 M-domain via Hsp70NBD.
Engineering a Functional Hsp104 Strep-Tag Variant.
Conservation of the principal Hsp70-binding site across species suggested that binding of Hsp70 to the M-domain might have a direct impact on Hsp104. However, capturing an Hsp104:Hsp70 complex is difficult, because the interaction between Hsp104 and Hsp70/40 is transient (7). In light of a direct physical interaction between Hsp70 and the Hsp104 M-domain, we hypothesized that specific binding of a protein to its high-affinity binding site, which is engineered into the M-domain, might be able to elicit the protein disaggregating activity of Hsp104.
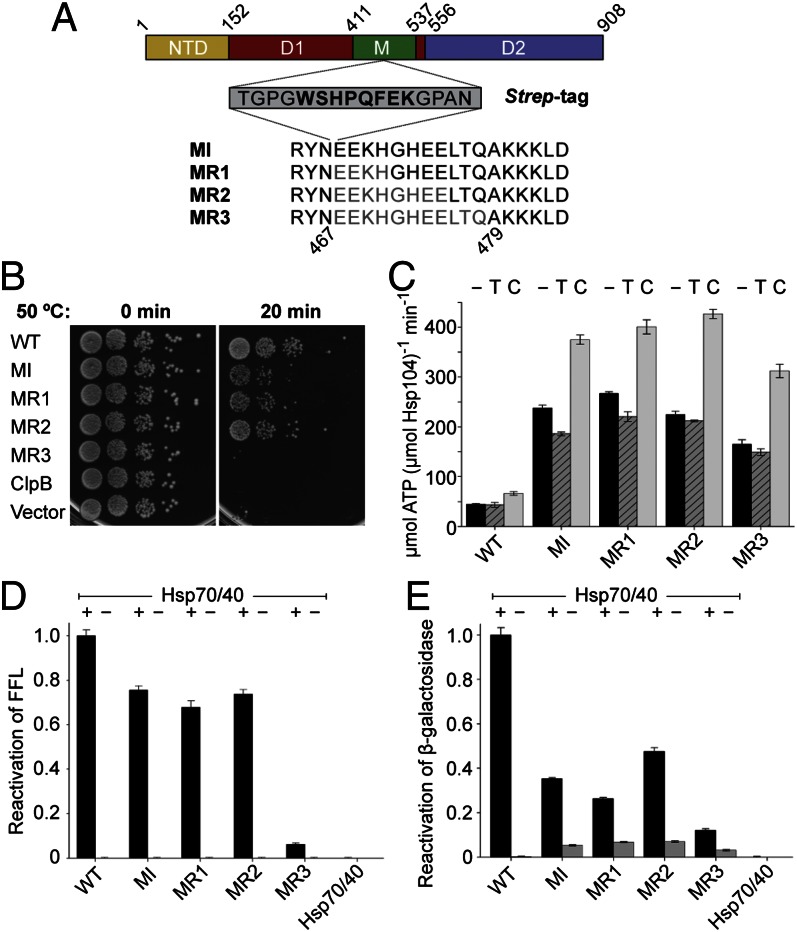
Strep-Tactin is an engineered protein that recognizes the Strep-tag as a homo-tetramer with micromolar affinity (41). To determine whether Strep-Tactin binding to the M-domain can activate the Hsp104 motor, we engineered four Hsp104 variants that featured an eight-residue Strep-tag inserted after Asn467 (Hsp104MI) and replacing 4 (Hsp104MR1), 8 (Hsp104MR2), or 11 residues (Hsp104MR3) in the Hsp70-binding site (Fig. 3A). To ensure that our engineered Hsp104 Strep-tag variants are functional, we examined their ability to provide thermotolerance in Δhsp104 yeast (Fig. 3B and Fig. S3). We found that all Strep-tag variants are viable (Fig. 3B at 0 min) and, except for Hsp104MR3, can provide thermotolerance development in the absence of Hsp104 (Fig. 3B at 20 min). As anticipated, neither ClpB nor an empty vector supported thermotolerance development in Δhsp104 yeast (Fig. 3B at 20 min).
Fig. 3.
Functional analysis of engineered Hsp104 Strep-tag variants. (A) Schematic diagram of Hsp104MI, Hsp104MR1, Hsp104MR2, and Hsp104MR3 showing the location of the inserted Strep-tag (boxed) with the Strep-Tactin binding motif in bold. Residues that were replaced are shown in gray. (B) Induced thermotolerance assay. Δhsp104 yeast, expressing the indicated wild-type or mutant protein, were heat-shocked at 50 °C for 20 min as described in SI Text. Shown are 10-fold serial dilutions. (C) ATPase activities of Hsp104 wild-type and Strep-tag variants without and with 4× Strep-Tactin (T) or κ-casein (C). (D and E) Coupled chaperone activities of Hsp104 wild-type and variants in the presence of Hsp70/40. Averages of three independent measurements ± SD are shown.
Lack of thermotolerance development by Hsp104MR3, however, was not caused by an inability to bind or hydrolyze ATP, because all of our engineered Strep-tag variants are functional ATPases and displayed a three- to fivefold higher basal ATPase activity than Hsp104 wild-type (Fig. 3C). The basal ATPase activity of Hsp104 variants is reduced by Strep-Tactin but further stimulated by κ-casein (Fig. 3C). Despite the elevated basal ATPase activities, none of our Hsp104 Strep-tag variants recovered functional FFL from aggregates in the absence of Hsp70/40 (Fig. 3D). Notably, the Hsp104MR3 protein disaggregating activity with Hsp70/40 was substantially reduced (Fig. 3D). Similar results were also obtained with heat-aggregated β-gal, which showed an overall lower recovery of β-gal activity for all Strep-tag variants in conjunction with Hsp70/40 (Fig. 3E). However, unlike FFL, a small amount of β-gal was also recovered by Hsp104 variants alone, indicating substrate- or aggregate-specific differences.
Hsp104 Strep-Tag Variants Are Activated by Strep-Tactin.
To determine whether Strep-Tactin can elicit the Hsp104 protein remodeling activity, we further examined the functional activity of our Hsp104 Strep-tag variants. Heat-aggregated β-gal was chosen as substrate because it does not require Hsp70/40 for refolding (6). Remarkably, we found that binding of Strep-Tactin to Hsp104MI elicited the Hsp104 protein disaggregating activity in a Strep-Tactin–dependent manner (Fig. 4A and Fig. S4A). Strikingly, at an eightfold excess of Strep-Tactin over Hsp104MI, the recovery of β-gal activity was more than twofold higher than Hsp104MI together with Hsp70/40, even surpassing the level observed with the wild-type bichaperone system (Fig. 4A and Fig. S4A). Protein disaggregation was specific to the interaction between Strep-Tactin and Hsp104MI, because neither Strep-Tactin alone nor Hsp104MI together with BSA, even at an eightfold molar excess, could recover functional protein from aggregates (Fig. 4A). Like Hsp104MI, we found that Hsp104MR1 and Hsp104MR2, and even Hsp104MR3, are activated in a Strep-Tactin–dependent manner (Fig. 4B). However, neither Hsp104 wild-type nor Hsp104 variants that feature a Strep-tag in the D2-domain are responsive to Strep-Tactin (Fig. S5). Similar results were obtained using heat-aggregated α-glucosidase, which is reactivated by Hsp104MI in the presence of Strep-Tactin (Fig. 4C and Fig. S4B). However, the recovered α-glucosidase activity was generally lower than β-gal.
Fig. 4.
Hsp104MI is activated by Strep-Tactin and recovers functional protein in the absence of Hsp70/40. (A–C) Strep-Tactin–dependent protein disaggregation by Hsp104 Strep-tag variants. Strep-Tactin (tetramer; 4 × 15 kDa) and BSA (monomer; 67 kDa) were added in n-fold molar excess of Hsp104 monomer. Recovered enzymatic activities are expressed relative to the wild-type bichaperone system. Averages of three independent measurements ± SD are shown. (D) Cartoon illustrating the proposed model by which Strep-Tactin unlocks the protein disaggregating activity of Hsp104MI. In this model, Hsp104 binds aggregated proteins in an ATP-dependent manner, but requires Strep-Tactin to initiate protein disaggregation. The unfolding and translocation of substrates requires ATP hydrolysis.
Hsp70 Is an Activator of the Hsp104 Motor.
We reasoned that the ability of Strep-Tactin to elicit the protein disaggregating activity of engineered Hsp104 Strep-tag variants may arise from a critical Hsp104 activation step, which was mimicked by Strep-Tactin binding to the modified M-domain (Fig. 4D). Because Hsp70 binds directly to the M-domain of Hsp104 (Figs. 1E and 2D), we examined whether Hsp70 alone could trigger Hsp104 protein disaggregation in the absence of Hsp40. As anticipated, many of our substrates were not reactivated, including chemically denatured FFL, which requires both Hsp70 and Hsp40 for protein refolding (42, 43). Strikingly, we found that both aggregated β-gal and EGFP, which do not require Hsp70/40 for reactivation (6, 31, 44), could be rescued by Hsp104 together with full-length Hsp70 (Fig. 5A and Fig. S6). Neither Hsp70 nor Hsp104 alone or together with Hsp40 could do so (Fig. 5A). However, although the recovery of functionally active protein was dependent on the concentration of Hsp70, it was less effective than the complete Hsp104/Hsp70/40 system (Fig. 5A and Fig. S6). Nevertheless, protein recovery was significant, and up to 70% of β-gal activity was recovered relative to the bichaperone system (Fig. 5A and Fig. S6A). Taken together, our findings demonstrate that Hsp70 alone can elicit the Hsp104 protein disaggregating activity toward aggregated model substrates that do not require Hsp70/40 for refolding.
Fig. 5.
Hsp70 is a potent activator of the Hsp104 motor. (A–D) Hsp70 (monomer) or Strep-Tactin (tetramer) was added in n-fold excess over Hsp104 monomer. Averages of three independent measurements ± SD are shown. (A) Hsp70 activates the Hsp104 protein disaggregating activity in a concentration-dependent manner. Recovered enzymatic activities of heat-aggregated β-gal are expressed relative to the complete Hsp104/Hsp70/40 system. (B) Initial rate of β-gal recovery by mixtures of Hsp104 wild-type and Trap mutant at the indicated ratios without and with Hsp70/40. (Inset) An enlarged view of the innate protein remodeling activity of Hsp104. (C) Relative recovery rate of β-gal by Hsp104/Trap and their respective Hsp104MI variants with Hsp70/40, 10× Hsp70, or 4× Strep-Tactin. The dashed gray line represents the linear decrease expected if the activity of the Hsp104 hexamer is proportional to the number of wild-type subunits present. (D) Relative ATPase activities of Hsp104MI/TrapMI without and with Hsp70/40, 10× Hsp70, or 4× Strep-Tactin.
What is the mechanism for Hsp104 activation? We reasoned that Hsp70 binding to the M-domain regulates the Hsp104 ATPase activity and thereby controls the Hsp104 protein disaggregation function. To test this, we mixed wild-type Hsp104 with an ATP-hydrolysis-deficient Hsp104 mutant (Trap). As previously shown, incorporation of Trap subunits elicits the innate protein remodeling activity of Hsp104 (Fig. 5B) (31, 32), with hetero-hexamers containing four wild-type subunits being most active (Fig. 5B, Inset). Conversely, Hsp70/40-dependent protein disaggregation was substantially reduced by incorporation of one or more Trap proteins (Fig. 5B), indicating a highly cooperative mechanism between subunits when Hsp70/40 is present. Remarkably, increased coordination between Hsp104 subunits was also observed with Hsp70 alone and with our engineered Strep-Tactin system, which were very similar to that with Hsp70/40 (Fig. 5C).
Next, we examined the ATPase activities of our hetero-hexamers in the absence or presence of Hsp70 or Strep-Tactin. We found that both Hsp70 and Strep-Tactin controlled the Hsp104 ATPase activity in a similar manner, which was indistinguishable from mixtures containing Hsp70/40, but distinct from those in the absence of activator (Fig. 5D). Taken together, our findings suggest that Hsp70 and Strep-Tactin activate the Hsp104 motor by promoting intersubunit coordination in the hexamer.
Discussion
Hsp104 harnesses the energy of ATP binding and hydrolysis to rescue stress-damaged proteins from a previously aggregated state. Whereas Hsp70/40 is essential for Hsp104 protein disaggregation, the mechanistic role of Hsp70 remained unclear. It has been reported that Hsp70/40 possesses some protein disaggregating activity (45), which can be stimulated by Hsp110 (an Hsp70-related nucleotide exchange factor of Hsp70), but which is less efficient than the Hsp104 bichaperone system (46).
It has been suggested that the Hsp104 bichaperone system uses distinct mechanisms to remodel different substrates (23). Here we demonstrate that Hsp70 is an activator of the Hsp104 motor. Remarkably, we found that inserting a Strep-Tactin binding motif at the spatially conserved Hsp70-binding site on the M-domain elicits the Hsp104 protein remodeling activity in a Strep-Tactin–dependent manner (Fig. 4 A–C) by promoting intersubunit coordination in the engineered Hsp104 hexamer, as did Hsp70/40 (Fig. 5 C and D). More importantly, we demonstrate that full-length Hsp70 on its own can activate the Hsp104 motor (Fig. 5 C and D). Hsp70 does so in a protein concentration-dependent manner (Fig. 5A). However, protein binding per se is insufficient for Hsp104 activation, because Hsp70NBD, which binds the M-domain, did not activate Hsp104 (Fig. 5A). In light of our findings, we propose that Hsp70 binding to the M-domain controls the Hsp104 ATPase activity, which, in turn, unleashes the protein disaggregating activity of Hsp104.
Although the physical nature of this interaction remains unknown, an ATP-driven M-domain motion has been demonstrated previously for ClpB (35, 47). It is tempting to speculate that Hsp70 binding to the M-domain stabilizes or alters an M-domain conformation from a normally inactive to an active state. Our present data suggest that Hsp70 recognizes the coiled-coil structure of the M-domain motif 2, which is held together by a leucine zipper-like heptad repeat. Consistent with this notion, in Hsp104MR3, where the Strep-tag overlapped and replaced Leu476 in the heptad repeat, the ability to synergize with Hsp70/40 was substantially reduced (Fig. 3 B, D, and E), possibly due to perturbation of the coiled-coil structure.
Fig. 6 summarizes our mechanistic understanding of the Hsp104–Hsp70/40 bichaperone system. Previously proposed models suggested a function for the Hsp70 system in targeting the Hsp104 motor to aggregates (20, 22). Although Hsp104 possesses an innate protein remodeling activity that can be elicited in vitro (31, 32), this activity is not robust (48, 49). Here we propose an additional function for Hsp70 as an activator of the Hsp104 motor, which is on top of Hsp70’s known role in protein folding. Because Hsp70/40 chaperones often bind or colocalize with aggregates (20, 22), our model is in agreement with an upstream role of Hsp70/40 in protein disaggregation to activate Hsp104, and a downstream role in substrate recovery. Finally, our work also suggests that, at least in principle, Hsp104 can be activated by other yeast proteins, which specifically bind to the M-domain to elicit the diverse cellular activities of this machine.
Fig. 6.

Schematic model illustrating the different roles of Hsp70 in Hsp104-mediated protein disaggregation. Hsp70 is required to activate the Hsp104 motor to extract polypeptides from aggregates (Upper) and often binds or colocalizes with aggregates together with Hsp40 (Lower).
Materials and Methods
Cloning.
pHsp70 (50), featuring full-length human Hsp70 (HSPA1A), was used to generate all Hsp70 variants. His6-Hsp70ΔC (residues 1–554) was constructed by inserting stop codons after Glu554. PCR cloning was used to generate all other Hsp70 constructs comprising residues 1–394 (Hsp70NBD), 395–554 (Hsp70SBDΔC), and 555–641 (Hsp70CTD). S. cerevisiae Hsp104 (6) was used to generate all Hsp104 constructs. Hsp104mut-1, Hsp104mut-4, Hsp104EEE, Hsp104AAA, and Hsp104E285Q;E687Q (Trap) were generated by overlap extension PCR. Hsp104 variants, featuring a Strep-tag (WSHPQFEK) flanked by four amino acids on both sides, were generated essentially as described (6). Four constructs were made in which the Strep-tag is inserted after Asn467 by replacing 0 (Hsp104MI), 4 (ΔGlu468-His471; Hsp104MR1), 8 (ΔGlu468-Glu475; Hsp104MR2), or 11 residues of the M-domain (ΔGlu468-Gln478; Hsp104MR3). TrapMI, featuring the Strep-tag–containing cassette of Hsp104MI, was generated from Trap. A cysteine-less Hsp104 variant (Hsp104*) was used to generate Hsp104*E435C, Hsp104*N467C, Hsp104*E468C, Hsp104*H471C, and Hsp104*R496C by site-directed mutagenesis.
Protein Expression and Purification.
His6-tagged Hsp70, Hsp104, and their variants were bacterially overexpressed as described (50). Proteins were purified from cleared lysates by Ni-nitrilotriacetic acid (NTA) affinity and anion exchange chromatography. For Hsp104 and variants, the N-terminal His6-tag was removed before anion exchange chromatography by incubating the protein with His6-Tobacco Etch Virus (TEV) protease during dialysis and passing the cleavage product over a Ni-NTA column. Hsp104 and Strep-tag variants were further purified on Superose 6 10/300 GL in 25 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 5% (vol/vol) glycerol, and 1 mM DTT. For enzymatic assays, Hsp104 and mutants were used after the second Ni-NTA column. His6-Hsp40 (S. cerevisiae Ydj1) and EGFP, which contained a TEV cleavable N-terminal His7-tag and the first 15 amino acids of RepA at the C terminus (EGFP), were purified essentially as described (6, 50).
Peptide Array Synthesis and Analysis.
Miniaturized peptide libraries were prepared on a cellulose membrane using an ASP222 autospot robot (Intavis AG). The membrane was blocked with 1× Superblock in Tris-buffered saline (TBS) and probed at 22 °C for 1 h with 0.5 µM His6-Hsp70ΔC and 1 mM ATP in TBS containing 10% (vol/vol) 1× Superblock and 5% (wt/vol) sucrose. The membrane was washed three times for 10 min in TBS with 50 µM ATP. His6-Hsp70ΔC binding was detected by probing the membrane directly with an anti-His6 monoclonal antibody conjugated to HRP (mAB-HRP; BD Bioscience). Nonspecifically bound peptides were eliminated by subtracting spots that were also detected in a control experiment using nontagged Hsp70ΔC and anti-His6 mAB-HRP.
Bead-Binding Assay.
Hsp104/Hsp104MI hexamers were prepared by mixing Hsp104 (20 µg) with Hsp104MI (4 µg) and incubating on ice for 30 min in reaction buffer [25 mM Hepes (pH 7.5), 50 mM KOAc, 10 mM Mg(OAc)2, and 1 mM DTT] with 2 mM ADP, ATP, or no nucleotide. Hsp104/Hsp104MI hexamers were immobilized on Strep-Tactin Sepharose beads (IBA GmbH) and incubated for 30 min at 4 °C with His6-Hsp70 (14 µg), His6-Hsp70ΔC (12 µg), His6-Hsp70NBD (9 µg), His6-Hsp70SBDΔC (4 µg), or His6-Hsp40 (8 µg). Beads were washed three times with or without the indicated nucleotide, before eluting bound proteins with 10 mM d-desthiobiotin in reaction buffer. Eluted proteins were analyzed by Western blotting using anti-His6 mAB-HRP.
Site-Specific Photo-Cross-Linking.
Hsp104* and cysteine-containing Hsp104* variants were labeled with BPIA (Santa Cruz Biotechnology) by incubating 0.5 mM BPIA and 10–12 µM protein (monomer) for 2 h at 23 °C in 25 mM Hepes (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, and 0.5 mM Tris(2-carboxyethyl) phosphine (TCEP). The reaction was quenched with 5 mM DTT, and free BPIA was removed using a PD-10 column (GE Healthcare). Hsp104* and BPIA-labeled Hsp104* variants were incubated overnight at 4 °C with Hsp70ΔC, Hsp70NBD, or Hsp70SBDΔC in 25 mM Hepes (pH 7.5), 50 mM KOAc, 10 mM Mg(OAc)2, and 0.5 mM TCEP together with 2 mM ADP and cross-linked for 30 min on ice by UV irradiation at 365 nm. Cross-linked products were analyzed on a coomassie-stained 3–8% Tris-acetate SDS/PAGE gel.
ATPase Activity Assay.
ATPase activities of Hsp104 and variants, except for subunit mixing experiments, were measured in buffer [25 mM Hepes (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, and 10 mM DTT] as described (50) but with 5 mM ATP.
Protein Disaggregation Assay.
Protein disaggregation assays with chemically denatured FFL or heat-aggregated β-gal were performed as described (50). α-Glucosidase aggregates were prepared by denaturing 0.5 µM yeast α-glucosidase (Sigma-Aldrich) at 50 °C for 12 min in assay buffer. Heat-aggregated α-glucosidase (0.2 µM) was added to assay buffer containing equimolar concentrations of molecular chaperones (0.7 μM monomer) and a previously described ATP regenerating system (50). Recovered α-glucosidase activity was assayed by adding 1 mM 4-nitrophenyl-α-N-glucopyranoside, incubating for 15 min at 30 °C, and measuring the absorbance at 405 nm. Aggregated EGFP was prepared by denaturing 4.5 µM of EGFP at 80 °C for 10 min in assay buffer. Heat-aggregated EGFP (0.225 µM) was added to 0.5 μM (monomer) of molecular chaperones in the presence of an ATP regenerating system. Real-time recovery of EGFP fluorescence was monitored at 25 °C over 120 min as described (50).
Subunit Mixing Experiments.
Hsp104 or Hsp104MI and their respective Trap mutants were mixed at different ratios to achieve the indicated subunit composition, while keeping the total protein concentration constant. To measure the ATPase activity, 5 µM (monomer) of Hsp104/Trap mixtures were incubated at 4 °C for 15 min, diluted 10-fold, and incubated with 2 mM ATP at 22 °C for 15 min before measuring the amount of released inorganic phosphate using a malachite green assay (51). For protein disaggregation, 10 µM (monomer) of Hsp104/Trap mixtures were incubated at 30 °C for 30 min and diluted 10-fold before measuring protein disaggregation. Recovered β-gal activity was measured at different time points. The linear range of protein recovery was used to calculate the initial rate.
Supplementary Material
Acknowledgments
We thank N. Sung for technical assistance and Dr. T. Wensel for support. This work was funded by National Institutes of Health Grant R01GM104980 and Welch Foundation Grant Q-1530.
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217988110/-/DCSupplemental.
References
- 1.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125(3):443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Bösl B, Grimminger V, Walter S. The molecular chaperone Hsp104—A molecular machine for protein disaggregation. J Struct Biol. 2006;156(1):139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Doyle SM, Wickner S. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci. 2009;34(1):40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Haslberger T, Bukau B, Mogk A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol. 2010;88(1):63–75. doi: 10.1139/o09-118. [DOI] [PubMed] [Google Scholar]
- 5.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115(2):229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, Sielaff B, Lee J, Tsai FTF. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci USA. 2010;107(18):8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 8.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA. 1999;96(13):7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96(24):13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999;274(40):28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268(5212):880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 12.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20(23):8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 14.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5(8):611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuite MF, Serio TR. The prion hypothesis: From biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11(12):823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6(6):435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 18.Erjavec N, Larsson L, Grantham J, Nyström T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21(19):2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40(1):159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32(4):584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68(1):87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 22.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198(3):387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeSantis ME, et al. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151(4):778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reidy M, Miot M, Masison DC. Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics. 2012;192(1):185–193. doi: 10.1534/genetics.112.142307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle SM, Hoskins JR, Wickner S. DnaK chaperone-dependent disaggregation by caseinolytic peptidase B (ClpB) mutants reveals functional overlap in the N-terminal domain and nucleotide-binding domain-1 pore tyrosine. J Biol Chem. 2012;287(34):28470–28479. doi: 10.1074/jbc.M112.383091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279(28):29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 27.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119(5):653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 28.Reid BG, Fenton WA, Horwich AL, Weber-Ban EU. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci USA. 2001;98(7):3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121(7):1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 30.Hinnerwisch J, Reid BG, Fenton WA, Horwich AL. Roles of the N-domains of the ClpA unfoldase in binding substrate proteins and in stable complex formation with the ClpP protease. J Biol Chem. 2005;280(49):40838–40844. doi: 10.1074/jbc.M507879200. [DOI] [PubMed] [Google Scholar]
- 31.Doyle SM, et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14(2):114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaupp AM, Marcinowski M, Grimminger V, Bösl B, Walter S. Processing of proteins by the molecular chaperone Hsp104. J Mol Biol. 2007;370(4):674–686. doi: 10.1016/j.jmb.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 33.Sielaff B, Tsai FTF. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J Mol Biol. 2010;402(1):30–37. doi: 10.1016/j.jmb.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miot M, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci USA. 2011;108(17):6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haslberger T, et al. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell. 2007;25(2):247–260. doi: 10.1016/j.molcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Ungewickell E, Ungewickell H, Holstein SE. Functional interaction of the auxilin J domain with the nucleotide- and substrate-binding modules of Hsc70. J Biol Chem. 1997;272(31):19594–19600. doi: 10.1074/jbc.272.31.19594. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20(4):513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rüdiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol. 1997;4(5):342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 39.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 40.Bösl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J Biol Chem. 2005;280(46):38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt TGM, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2(6):1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 42.Schröder H, Langer T, Hartl FU, Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12(11):4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14(10):2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoskins JR, Doyle SM, Wickner S. Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc Natl Acad Sci USA. 2009;106(52):22233–22238. doi: 10.1073/pnas.0911937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci USA. 2006;103(16):6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE. 2011;6(10):e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, Choi JM, Tsai FTF. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol Cell. 2007;25(2):261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue Y, Taguchi H, Kishimoto A, Yoshida M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J Biol Chem. 2004;279(50):52319–52323. doi: 10.1074/jbc.M408159200. [DOI] [PubMed] [Google Scholar]
- 49.Krzewska J, Melki R. Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. EMBO J. 2006;25(4):822–833. doi: 10.1038/sj.emboj.7600985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biter AB, Lee J, Sung N, Tsai FTF, Lee S. Functional analysis of conserved cis- and trans-elements in the Hsp104 protein disaggregating machine. J Struct Biol. 2012;179(2):172–180. doi: 10.1016/j.jsb.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100(1):95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 52.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106(21):8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.