Significance
Here, we show that FilP, a bacterial cytoskeletal protein related to metazoan intermediate filament (IF) proteins, can self-assemble into a regular network structure. This finding offers a possible explanation for its previously characterized role in cellular rigidity and elasticity and might offer insights into the mechanical role of human IFs. The assembly of FilP cytoskeleton is coupled to the function of the polarisome, a protein complex orchestrating the polar growth characteristic of Streptomyces. These results suggest that apical assembly of a stress-bearing cytoskeleton is a common strategy in tip-growing walled cells, such as filamentous fungi, pollen tubes, and mycelial bacteria.
Abstract
Intermediate filament (IF)-like cytoskeleton emerges as a versatile tool for cellular organization in all kingdoms of life, underscoring the importance of mechanistically understanding its diverse manifestations. We showed previously that, in Streptomyces (a bacterium with a mycelial lifestyle similar to that of filamentous fungi, including extreme cell and growth polarity), the IF protein FilP confers rigidity to the hyphae by an unknown mechanism. Here, we provide a possible explanation for the IF-like function of FilP by demonstrating its ability to self-assemble into a cis-interconnected regular network in vitro and its localization into structures consistent with a cytoskeletal network in vivo. Furthermore, we reveal that a spatially restricted interaction between FilP and DivIVA, the main component of the Streptomyces polarisome complex, leads to formation of apical gradients of FilP in hyphae undergoing active tip extension. We propose that the coupling between the mechanism driving polar growth and the assembly of an IF cytoskeleton provides each new hypha with an additional stress-bearing structure at its tip, where the nascent cell wall is inevitably more flexible and compliant while it is being assembled and matured. Our data suggest that recruitment of cytoskeleton around a cell polarity landmark is a broadly conserved strategy in tip-growing cells.
Coiled coil-rich proteins are emerging as wide-spread and important determinants of cell architecture in bacteria. A subclass of such proteins possesses a segmented coiled-coil architecture, resembling that of metazoan intermediate filament (IF) proteins. Recently, an increasing number of reports have shown that these putatively IF-like proteins assemble into cytoskeletons in evolutionarily diverse bacteria, of which several examples are given below.
Crescentin, which determines the characteristic curved cell morphology in the ubiquitous aquatic bacterium Caulobacter crescentus, was the first and is now the best characterized bacterial IF-like protein (1, 2). Crescentin forms a long cable-like cytoskeletal structure, which is characterized by stability and very low turnover rates (2, 3). Because it is attached to the cell membrane parallel to the long cell axis in a stretched-out state, it applies physical force to the underlying cell envelope, thereby locally reducing the rate of new cell wall incorporation. The opposite sides of the cylindrical cell wall will therefore grow with different rates, resulting in a crescent-shaped curved sacculus (1, 2). Thus, crescentin functions via mechanical interference with the building of the bacterial cell wall. Although less studied, filament-forming coiled coil-rich proteins are important determinants also of the characteristic helical cell shape of Helicobacter species (4, 5). In Corynebacterium glutamicum, an essential coiled-coil protein, rod shape morphology protein (RsmP) forms a cytoskeletal element involved in determination of the rod shape of the cells (6). Another example of a coiled-coil cytoskeleton in bacteria is presented by the conspicuous cytoplasmic filaments of Treponema (7). We have previously shown that IF-like proteins are present and conserved in several species of the Gram-positive phylum Actinobacteria (see below and ref. 8).
Although most of the distantly related bacterial IF-like proteins do not share obvious conserved sequence motifs with each other or with eukaryotic IF proteins, we refer to them as “IF-like,” based on their structural, biochemical, and functional analogy to eukaryotic IF proteins. In addition to their similar coiled-coil domain architecture, most of the above mentioned bacterial proteins also share a fundamental biochemical property with the eukaryotic IF proteins, which is to spontaneously self-assemble into ordered filaments without requirements for nucleotides or specific cofactors (5, 6, 9, 10). It has been shown for crescentin that, similarly to IF proteins, this property is conferred by the main structural element, the central coiled-coil rod domain (11), which is likely to be the case for other bacterial IF-like proteins. Evidently, both bacteria and eukaryotes have developed cytoskeletal systems based on fibrous proteins with a segmented coiled-coil rod as a molecular blueprint and the structural basis for a cytoskeletal function, suggesting that a coiled-coil cytoskeleton emerged early in evolution.
Differently from actin- and tubulin-based cytoskeletons, a hallmark of the eukaryotic IF cytoskeleton is a wide diversity of cellular structures and functions (12, 13). Because specific types of IF proteins are present in various differentiated animal cells, the lack of easily tractable genetic systems has hampered molecular studies of animal IF proteins. Thus, studies of IF-like proteins in genetically and phenotypically tractable organisms would be a valuable source of new information to understand the basic working principles and the functional versatility of the coiled-coil cytoskeleton.
Here, we report data concerning the IF-like protein FilP in Streptomyces coelicolor and provide mechanistic insight into a divergent IF-like function in bacteria. Streptomyces are ubiquitous soil and aquatic organisms of large industrial and medical importance due to secondary metabolite production (14). Due to their intricate lifestyle involving multicellularity, differentiation, and sporulation, they have also become model organisms in cell and developmental biology research (15–17). A feature relevant for the present work is the pronounced apical growth (18). Vegetative hyphae grow by tip extension and branching to form a multicellular mycelium. We have previously reported that FilP and other members of its conserved family, such as Mb1709 from Mycobacterium bovis, spontaneously oligomerize in vitro. Higher-order structures were also formed by FilP in vivo, which contributed to the normal rigidity and elasticity of the S. coelicolor hyphae, as shown by atomic force microscopy (8). However, because the latter properties are usually determined by the peptidoglycan cell wall in bacteria, the role of FilP remained mysterious. Here, we show that lack of FilP does not substantially affect cell wall structure. We also present data showing that FilP forms a densely cross-linked regular network in vitro and that the in vivo localization is consistent with a cellular network of FilP. These data suggest that FilP is likely to provide direct mechanical support to the cells. Another finding presented here is that the formation of the FilP cytoskeleton is directly connected to polar growth. Apical growth is one of the most extreme manifestations of cellular polarity and has been extensively studied in filamentous fungi, yeasts, and the pollen tubes and root hairs of plants. Polarized actin and tubulin cytoskeletons play key roles in these eukaryotic examples of apical growth. In contrast, the actin and tubulin cytoskeletons are not involved in polar growth in Streptomyces, as mutants lacking FtsZ (tubulin) or MreB (actin) proteins are viable and grow via tip extension (19, 20). In Streptomyces species, and in many other members of Actinobacteria, the coiled-coil protein DivIVA is the key factor in generating cell polarity (15, 18). The current model of DivIVA action is that it assembles into large protein complexes at sites of de novo pole formation. These complexes, likely involving other proteins, are termed bacterial polarisomes and serve to recruit factors needed for new cell envelope synthesis (21–24). Recently, another coiled-coil protein, Scy, was found to be part of the polarisome complexes (also referred to as tip-organizing centers), and to have an important, albeit not essential, role in polar growth (25). Here, we show that the DivIVA polarisomes also recruit gradients of the FilP cytoskeleton, which thereby will extend from sites of incipient polar growth into the hyphal interior during tip elongation, ensuring that an additional stress-bearing structure assembles at the most vulnerable zones of the hyphal envelope. Thus, we have revealed that a dynamic interplay between two coiled-coil cytoskeletal elements is an important process during establishment and maintenance of growth polarity in a tip-growing organism.
Results
FilP Forms Cis-Interconnected Networks in Vitro.
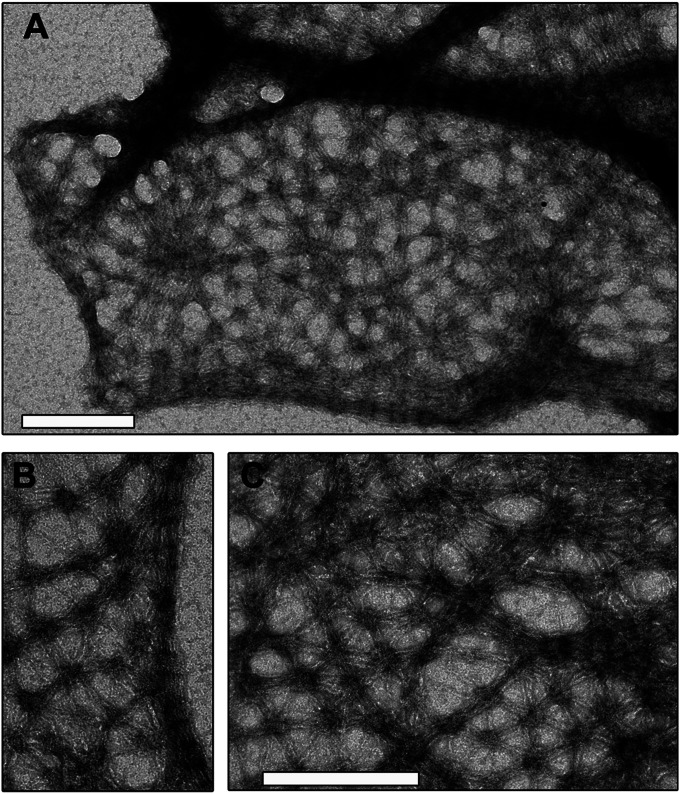
In previous work, we have shown that loss of FilP leads to reduced rigidity and elasticity of the hyphae, suggesting that FilP may either contribute to peptidoglycan cell wall assembly, or provide direct mechanical support, on par with the IF cytoskeleton in eukaryotic cells (8). In an attempt to distinguish between these possibilities, we first examined transmission electron micrographs of thin sections of vegetative hyphae of a wild-type and an filP deletion strain NA883. No obvious differences in thickness and structure of their cell envelopes could be detected. Next, we reasoned that defects in cell wall structure should render the hyphae more susceptible to cell wall damaging agents. Lysozyme and vancomycin were applied on paper discs placed on a thin lawn of young mycelium of wild-type and filP mutant strains, and the inhibition zones were measured. Again, no significant difference was observed between the strains (Table S1). Since these data indicate that the absence of FilP does not result in substantial defects in the peptidoglycan cell wall structure, we sought to explore the potential for a direct structural role for the FilP cytoskeleton. We began by investigating the in vitro filaments formed by purified hexahistidine-tagged FilP (His-FilP) under various conditions. Our previous results had revealed that His-FilP formed compact striated filaments in most buffers tested, that were difficult to relate to the cellular function in rigidity (8). Systematic screening of different assembly conditions revealed that several conditions [slow oligomerization at +4 °C and assembly in polymix buffer (26)] favored formation of FilP networks, rather than compact filaments (Fig. 1 and Fig. S1). Polymix is a complex buffer designed to mimic conditions in the cytoplasm (26, 27). Fig. 1 shows a spectacular network, consisting predominantly of hexagonal elements, which at several places coalesces into compact striated filaments. Because the network is clearly continuous with the previously observed striated filaments (Fig. 1B), it suggests that both structures assemble by the same mechanism and differ by the degree of compaction mainly. There is a large body of experimental data, both in vivo and in vitro, demonstrating that the viscoelasticity and mechanical resilience of cytoskeletal networks are determined by the density of cross-links between individual filaments (28, 29). Thus, our data showing that FilP has the ability to self-assemble into extensive networks with frequent and regularly spaced cross-links, without the involvement of additional cross-linking proteins, offer a possible mechanistic explanation for its observed biological role in cell rigidity and elasticity.
Fig. 1.
FilP forms cis-interconnected networks in vitro. Denatured His-FilP in 8 M urea was slowly dialyzed into 50 mM Tris-buffer pH 7 at +4 °C overnight. Filaments were negatively stained and visualized by transmission electron microscopy. (Scale bars: 200 nm.) (A) View of a larger area covered by networks and compact striated filaments. (B) Shows in higher magnification that a network is a continuous structure with a striated filament. (C) Shows a regular hexagonal pattern and areas with variable compaction in the network.
FilP Cytoskeleton Forms Polar Gradients During Active Growth.
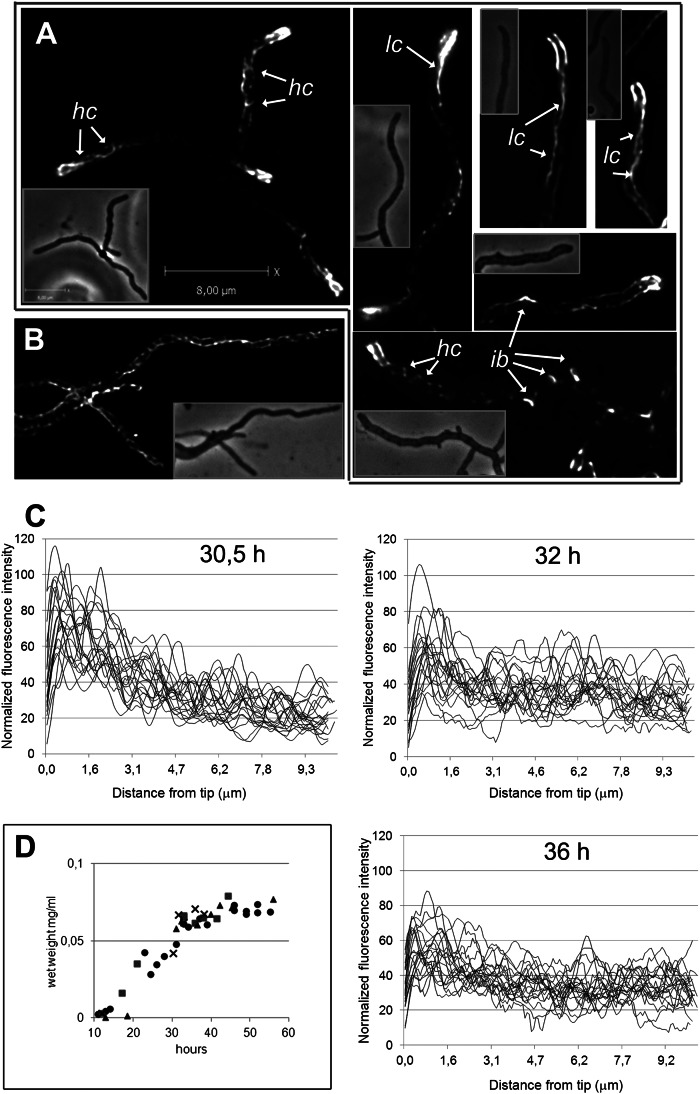
To test whether the in vitro data have relevance for the in vivo situation, we set out to explore the cellular localization of FilP by immunofluorescence microscopy (IFM) using affinity purified anti-FilP antibodies in the wild-type strain M145. The control strain NA883 lacking filP consistently lacked fluorescent signals, showing that the antibodies are highly specific. Interestingly, IFM revealed that, in actively growing cultures, 98% of hyphae exhibited strong staining in the apical region (17 h of growth, n = 200), which appeared as a polar gradient, with the strongest signal being found close to the tip (Figs. 2A and 3C). Deconvolution of several Z-stacks revealed patterns of fluorescence in these apical gradients, reflecting various structures. Some hyphae contained long cables localized asymmetrically along one side of the hypha, whereas others showed shorter helical-looking cables (marked as lc and hc, respectively in Fig. 2A). More faint filamentous structures were present throughout the hyphae but were not readily visible in all hyphae in the figure due to adjustments of the grayscale needed to resolve the brighter apical structures. Interestingly, lateral bulges, suggesting sites of emerging new branches, were often marked by bright FilP-specific fluorescence (ib in Fig. 2A). Notably, the in vitro ability of FilP to form a network, which locally coalesces into cables with variable length and thickness (Fig. 1), provides a straight-forward explanation for the complex localization pattern visualized by IFM (Fig. 2 A and B). The in vivo localization of FilP is also consistent with a cellular meshwork of branching filaments.
Fig. 2.
FilP forms complex cytoskeletal structures in Streptomyces hyphae and is organized into growth-dependent polar gradients. (A and B) Superimposed frames of deconvolved Z-stacks of anti-FilP immunofluorescence microscopy images showing the complex localization pattern of FilP. Corresponding phase contrast images are shown as insets. A and B depict hyphae from actively growing and stationary cultures, respectively. Arrows point to key elements of FilP cellular structures designated as follows: hc, helical cables; lc, lateral cables; ib, incipient branch. All hyphae in A have apical gradients that, due to deconvolution, in some cases appear as thick filaments. (C) Normalized intensity profiles of anti-FilP immunofluorescence along 10 µm starting from the tips in 25 hyphae from cultures with indicated age, corresponding to growth curves in D, showing gradual disappearance of apical gradients during transition from active to stationary growth phase. (D) Four independent growth curves of strain M145 in liquid YEME medium, showing the increase of biomass per sample volume.
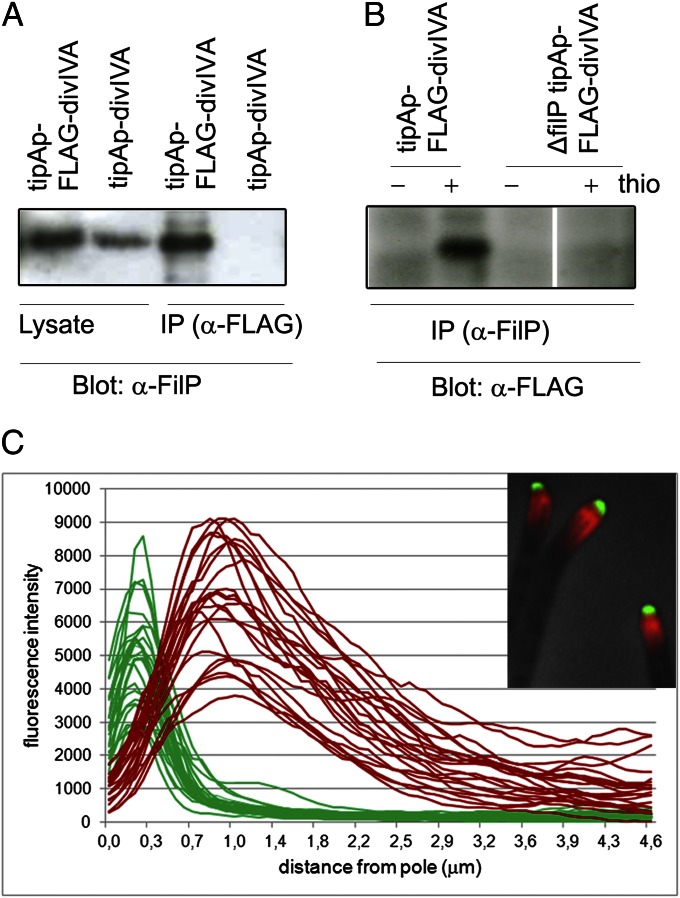
Fig. 3.
FilP and DivIVA interact in coimmunoprecipitation assay but do not colocalize in vivo. (A) FLAG-DivIVA was immunoprecipitated from cell extracts of strains K114 (tipAp-divIVA) and K120 (tipAp-FLAG-divIVA) grown in the presence of thiostrepton, using a monoclonal antibody to the FLAG peptide (α-FLAG). Precipitated samples (IP) and cell lysates (lysate) were analyzed by immunoblotting (Blot) using an anti-FilP polyclonal antiserum (α-FilP). (B) FilP was immunoprecipitated using affinity purified anti-FilP polyclonal antibodies (α-FilP) from cell extracts of strains K120 (contains filP and tipAp-FLAG-divIVA) and NA956 (lacks filP and contains ΔfilP tipAp-FLAG-divIVA), grown with and without thiostrepton (+ and − thio). Precipitated samples (IP) were analyzed by immunoblotting with α-FLAG antibodies. (C) Intensity profiles of DivIVA-EGFP (green lines) and anti-FilP (red lines) fluorescence along the first 4.6 µm of 24 individual hyphae, illustrating the consistent adjacent localization of DivIVA and FilP signals. Position 0.0 indicates the hyphal tip. (Inset) An overlay of phase contrast, anti-FilP immunofluorescence (red) and DivIVA-EGFP fluorescence (green) images showing apical DivIVA-EGFP clusters adjacent to subapical FilP gradients.
Hyphae from stationary cultures were devoid of polar FilP gradients and instead exhibited filamentous structures of relatively uniform intensity throughout the hyphal length (Fig. 2B). To verify that the polar FilP gradients were associated with active growth, we analyzed the localization pattern of FilP in the wild-type strain M145 during growth in a batch culture. Fig. 2D shows growth curves of four independent time-course experiments, based on wet weight of withdrawn samples. IFM showed that, during the onset (30,5 h) and further progression of the stationary phase (32 h, 36 h), apical gradients were present in 73% (n = 300), 47% (n = 150), and 27% (n = 300) of the hyphae, respectively (Fig. 2C). The remaining hyphae contained extensive filamentous structures throughout their length, similar to those in Fig. 2B. Thus, our data indicate that polar accumulation of FilP and the formation of apical cytoskeletal gradients are associated with active growth.
Previously, we have reported a somewhat different localization of FilP as prominent lateral cables and small apical clouds in a strain expressing both FilP and FilP-EGFP (8). However, a dominant negative effect of the nonfunctional FilP-EGFP is likely to distort the normal localization pattern of FilP in that strain. Reasoning that a flexible linker might improve the functionality of the fusion protein and overexpression of wild-type FilP might counteract the dominant negative effect of FilP-YPet, we constructed a strain that has filP-ypet (encoding FilP fused to the yellow fluorescent protein YPet via a flexible linker) at the native chromosomal locus, and tipAp-filP enabling thiostrepton-inducible synthesis of wild-type FilP at an ectopic locus (NA1062). Without thiostrepton, NA1062 exhibited a filP mutant phenotype, and FilP-YPet localized as described for the nonfunctional FilP-EGFP (8) (Fig. S2B). However, overproduction of untagged FilP restored the wild-type morphology and caused relocalization of FilP-YPet into apical gradients in 70% of the hyphae (Fig. S2A). Other characteristic features of FilP localization shown in Fig. 2A, such as network-like structures and accumulation at sites of incipient branches, were also frequent (Fig. S2A). Similar overproduction of FilP in the wild-type background resulted in increased signals of FilP immunofluorescence but did not cause any observable change in cell morphology or in FilP localization. Thus, the localization pattern of FilP, which we have shown here by different methods, IFM and live cell imaging, is likely to reliably reflect the in vivo situation.
FilP and DivIVA Interact but Do Not Colocalize.
Because our data (Fig. 2) suggest that only hyphae undergoing active tip extension contain apical gradients of FilP, the next obvious step was to investigate the role of DivIVA, the essential determinant of polar growth, in FilP localization. First, we asked whether there is an interaction between these two proteins. Anti-FLAG antibodies were used to pull down FLAG-DivIVA with interacting proteins from cell lysates of strains K120 (tipAp-FLAG-divIVA) and K114 (tipAp-divIVA), in which FLAG-DivIVA and untagged DivIVA, respectively, were under the control of the thiostrepton-inducible tipAp promoter (22, 24). Western blot analysis clearly demonstrated that FilP was pulled down by DivIVA from lysates containing FLAG-DivIVA, but not from the control lysates containing untagged DivIVA (Fig. 3A). To further verify these results, we conducted the reciprocal experiment, using anti-FilP antibodies to coimmunoprecipitate FLAG-DivIVA from strains K120 (tipAp-FLAG-divIVA) and NA956, an isogenic strain containing a deletion of filP. Consistent with our previous observations, FLAG-DivIVA was detected in the coimmunoprecipitate from strain K120, but not from the control strain NA956 (Fig. 3B). These data strongly suggest that FilP and DivIVA interact although it is not possible to distinguish between a direct interaction, or an indirect one requiring other interaction partners present in the total cell lysate of S. coelicolor.
To differentiate between a direct versus an indirect interaction between FilP and DivIVA, we used a bacterial two-hybrid assay, based on the interaction-dependent reconstitution of adenylate cyclase activity from the T25 and T18 fragments of the CyaA protein (30). Bait (T25-FilP, T25-DivIVA) and prey (FilP-T18, T18-FilP, DivIVA-T18, and T18-DivIVA) fusion proteins were coexpressed in the BTH101 strain of Eschericia coli in all possible combinations, and interactions were scored as development of blue color on indicator plates containing X-Gal (Fig. S3). As expected, a strong interaction signal was observed for T25-FilP when paired with FilP-T18 or T18-FilP, supporting our previous observations that FilP interacts with itself (8). Similarly, the T25-DivIVA/T18-DivIVA pair also showed a strong interaction signal, consistent with DivIVA oligomerization (24). Interestingly, strong signals were also obtained for three different pairs involving both FilP and DivIVA (T25-FilP/T18-DivIVA, T25-DivIVA/T18-FilP, and T25-DivIVA/FilP-T18). The DivIVA-T18 fusion failed to produce a signal with either bait construct. To rule out a possibility of unspecific interactions between coiled-coil proteins in overexpression conditions, T25-DivIVA was coexpressed with prey constructs encoding T18 fusions to crescentin (10) and SCO3114 (a putative coiled-coil protein sharing conserved motifs with FilP). No interaction signal was observed for any of these combinations (Fig. S3). These data collectively suggest that FilP and DivIVA interact directly.
Next, we visualized the localization of both proteins simultaneously by studying strain K112, containing divIVA-egfp, by fluorescence microscopy and found to our surprise that the FilP gradients initiated just behind the tip focus of DivIVA-EGFP and there was no colocalization of FilP and DivIVA. Fig. 3C shows apical intensity profiles of both DivIVA-EGFP fluorescence and FilP immunostaining in 24 hyphae and effectively illustrates the consistent adjacent localization of the cellular zones occupied by DivIVA and FilP and the formation of apical gradients by FilP.
In Situ Proximity Ligation Assay Confirms Close Cellular Proximity of FilP and DivIVA Molecules.
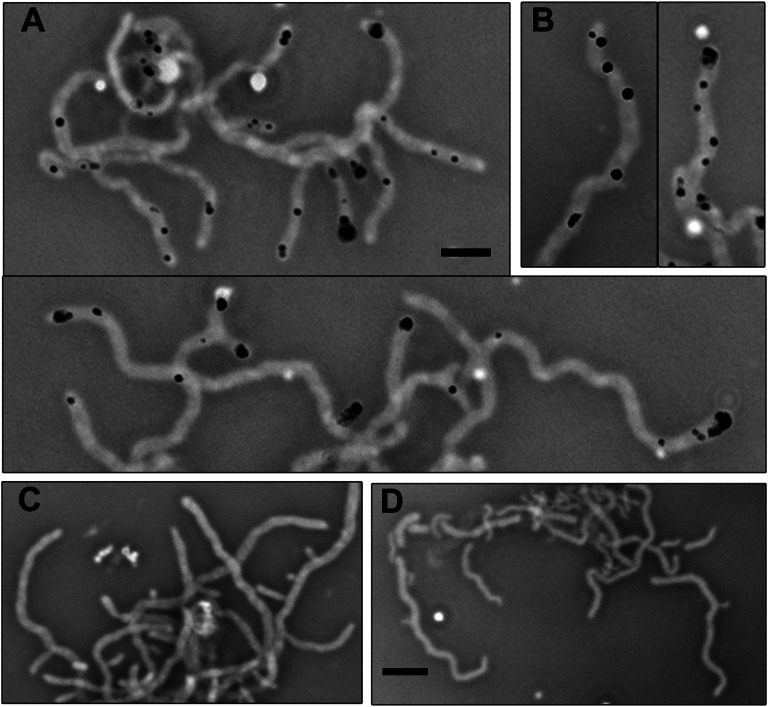
To understand the seemingly contradicting data showing interaction, but no colocalization, between FilP and DivIVA, we asked whether the interaction occurs at the interface of the FilP and DivIVA cellular zones. To visualize the interaction between FilP and DivIVA in S. coelicolor hyphae, we used in situ proximity ligation assay (PLA) (31–33). Affinity purified rabbit anti-FilP antibodies and monoclonal mouse anti-FLAG antibodies were first bound to FilP and FLAG-DivIVA, respectively, in fixed hyphae of strain K120 (tipAp-FLAG-divIVA). Proximity was detected by addition of PLA probes—secondary antibodies coupled to DNA oligonucleotides—that template hybridization and ligation of two subsequently added connector oligonucleotides, which enable rolling circle amplification. Thus, signals can be expected only in subcellular locations where FilP and DivIVA molecules are in close proximity and not further apart than 40 nm. We detected distinct fluorescent signals in cells of strain K120 (Fig. 4 A and B) whereas no signals were observed in the control strains NA956 (containing FLAG-DivIVA and lacking FilP) and M145 (containing FilP and lacking FLAG-DivIVA) (Fig. 4 C and D). The number of signals correlated to the level of induction of FLAG-divIVA. The tipAp promoter gives low level of expression even without inducer, and a few hyphae exhibited signals when grown without thiostrepton. However, the vast majority of hyphae contained one or multiple signals after a pulse of strong induction of FLAG-divIVA with 10 µg/mL thiostrepton for 1 h, followed by growth without inducer for 1 h (Fig. 4 A and B). Signals were localized close to the tips and along the lateral sides of the hyphae. The apical signals were often a short distance away from the very tip, consistent with the convening edges of the DivIVA and FilP zones (Figs. 3C and 4 A and B). Several hyphae exhibited multiple lateral signals (panels with individual hyphae in Fig. 4A), which is a localization pattern characteristic of DivIVA overproduction (see below and ref. 23). Because no signals were observed in the controls, and the lateral signals appeared only after induction of FLAG-divIVA, it is likely that they represent locations where newly formed clusters of DivIVA interact with FilP. These results suggest that interaction between these two proteins occurs very early during hyphal branch formation because little to no lateral outgrowth was associated with the lateral PLA signals. In summary, the PLA assay confirmed that FilP and DivIVA are likely to interact with each other at the interface of their convening cellular zones.
Fig. 4.
PLA analysis visualizes cellular locations where FilP and DivIVA are situated in close proximity. (A–D) Negative images of overlaid phase contrast and PLA fluorescence micrographs. A and B show strain K120 (tipAp-FLAG-divIVA) grown to late log-phase in liquid YEME medium, after which production of FLAG-DivIVA was induced by thiostrepton for 1 h. Two representative images of young branching mycelia are shown in A. (B) Individual hyphae exhibiting multiple lateral signals, which were also frequent. C and D show wild-type strain M145 and strain NA956, respectively. NA965 was grown and treated with thiostrepton similarly as K120 in A and B. (Scale bars: A and B, 3 µm; C and D, 4 µm.)
It is possible that other protein partners modulate the interaction between DivIVA and FilP in vivo during the complex process of tip elongation in Streptomyces. A potential candidate is the large coiled-coil protein Scy, which has been shown to interact with both FilP and DivIVA (25). We studied the localization of FilP in a scy null mutant strain (NA336). Although the scy mutation caused a severe morphological defect, FilP was still seen to form apical gradients in the actively growing hyphae of strain NA336 (Fig. S4), indicating that the recruitment of FilP gradients by DivIVA in vivo does not require Scy.
DivIVA Clusters Recruit the FilP Cytoskeleton.
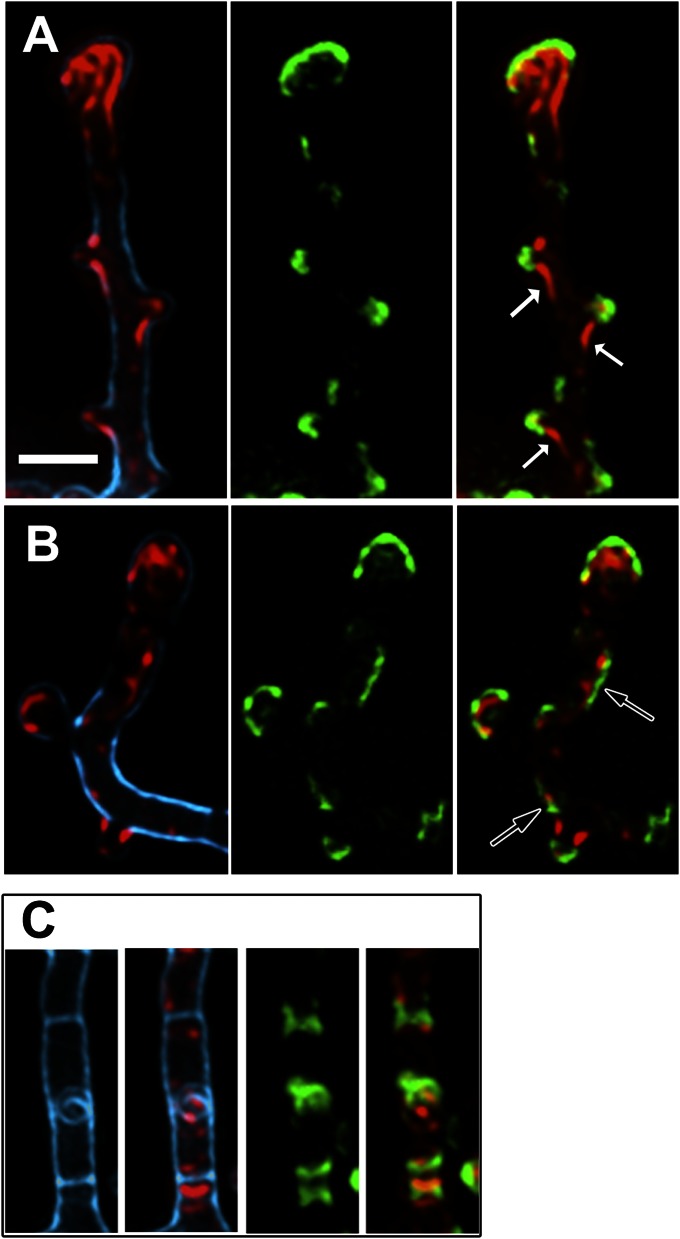
Our data show that FilP readily and spontaneously assembles into a network structure without cofactors in vitro, but in vivo the most prominent FilP structures are found at the sites of polar growth. Therefore, we postulate that there are factors that regulate the assembly of FilP in vivo, and one such candidate is DivIVA. The demonstrated interaction between DivIVA and FilP might promote oligomerization of FilP into cytoskeletal structures, resulting in the formation of apical gradients as the tip elongates and the DivIVA cluster moves ahead. If this assumption is true, recruitment of FilP should be visible immediately following the formation of DivIVA foci. To study this putative recruitment process, we used strain K121, containing divIVA-egfp under the native divIVA promoter and a thiostrepton-inducible tipAp-divIVA, in which we can induce formation of multiple ectopic foci along the lateral sides of the hyphae by overproduction of DivIVA (made visible due to incorporation of DivIVA-EGFP). Some of these foci go on to establish new cell poles by recruiting peptidoglycan synthesis complexes and ultimately cause outgrowth of new hyphal branches (23). Clear correlation between the localization of FilP and DivIVA clusters was observed in this strain after 1 and 2 h of DivIVA induction (Fig. 5). Significantly, lateral foci of DivIVA-EGFP that were not associated with bulges and thus might represent the earliest stage of formation of DivIVA clusters were in most cases flanked by increased FilP signal (Fig. 5B, arrows): of 304 lateral DivIVA foci counted, 289 (95%) were associated with an enhanced FilP signal. Thus, the data indicate that recruitment of FilP is an early event following formation of a DivIVA cluster. Also, in the cases where new branch outgrowth had been initiated, giving rise to lateral bulges in the hyphal wall, enhanced FilP fluorescence was consistently associated with DivIVA-EGFP foci. FilP was often seen as thicker cables emanating from the weaker FilP cytoskeleton in the main hypha and reaching out into the nascent branch, suggesting a dynamic remodeling of FilP polymers in response to the redistribution of cellular DivIVA (Fig. 5A, solid arrows). Consistent with our previous observations, also the swollen hyphal tips (such swelling is a hallmark of DivIVA overexpression) contained DivIVA and a FilP apical gradient (Fig. 5 A and B). We observed that, in overexpression conditions, DivIVA also localized to the cross-walls dividing the vegetative hyphae into compartments. These bands of DivIVA were flanked by enhanced zones of FilP (Fig. 5C). Remarkably, there is little overlap ever seen between the DivIVA and FilP signals, even in overexpression conditions. These results support the proposal that FilP gradients are recruited by cellular clusters of DivIVA soon after their assembly, thus making sure that each new branch will acquire the FilP cytoskeleton.
Fig. 5.
DivIVA recruits FilP cytoskeleton. (A–C) Superimposed deconvolved Z-stacks of fluorescence microscopy images of strain K121 after induction of divIVA for 2 h. Blue, cell wall stain (wheat germ agglutinin); green, DivIVA-EGFP fluorescence; red, anti-FilP immunofluorescence. Arrows in A point to filamentous structures of FilP trailing from main hyphae into newly forming branches; arrows in B point to examples of lateral DivIVA signals, not associated with bulges, but associated with enhanced signals of FilP. (C) A hyphal segment with two clearly visible crosswalls (blue), both flanked by DivIVA-GFP and FilP signals. (Scale bar: A, 2 µm; applies to all images.)
To test whether DivIVA is the sole factor needed for formation of FilP apical gradients, we expressed FilP and DivIVA-EGFP from inducible promoters in E. coli as a heterologous host (Fig. S5). After a short (30 min) induction of FilP in E. coli cells, which already contained polarly localized DivIVA-EGFP, small clusters of FilP had formed at random positions in the cells (Fig. S5A), showing that DivIVA alone is not able to recruit FilP to polar locations in E. coli. Upon prolonged expression, FilP filled the E. coli cells with structures visually similar to those observed in stationary phase S. coelicolor hyphae (Fig. 2B and Fig. S5B). These data suggest that FilP can independently form cytoskeletal structures in vivo, but the formation of apical gradients during active growth is a complex process and is controlled by other Streptomyces-specific factors in addition to DivIVA.
Discussion
The Achilles' heel of apically growing walled cells, such as filamentous fungi, pollen tubes, yeasts, and others, is the inevitable structural weakness of the growing tip. The nascent cell wall is being assembled and matured at the tip, which is by necessity more flexible and compliant than the more highly cross-linked pole-distal parts (34, 35). This tip plasticity is also a prerequisite for polar growth. Current models suggest that the localized compliance of the apical cell wall enables extension of the cell in response to physical forces, provided by turgor pressure, cytoskeleton, or another mechanism (36). There are experimental observations showing that the tip regions of Streptomyces hyphae are more susceptible to treatments with cell wall damaging agents than the tip-distal parts, suggesting that plasticity of the tip is also a feature of Streptomyces hyphae (37). It is still not clear how cells deal with this inherent problem, but cytoskeletal elements may provide support to the growing tips. Here, we would like to propose and discuss a model of polar growth in Streptomyces, whereby recruitment of FilP is directly coupled to the process of tip extension, to build up an additional stress-bearing structure to compensate for the inherent weakness of the apical cell walls.
The first aspect of this model concerns the ability of FilP to provide structural support. It has been established by numerous in vitro and in vivo studies that cytoskeletal fibers need to be cross-linked into a network to obtain the mechanical properties needed for maintenance of cell shape and integrity and protection against physical stress (28). We show here that FilP is able to self-assemble into a cis-linked, hexagonal network in the absence of any accessory proteins (Fig. 1 and Fig. S1). Cis-acting factors have also been proposed to cause formation of IF cellular networks because, in contrast to actin and tubulin, very few IF cross-linking proteins are known (38, 39). For example, divalent cations and sites at the extreme C termini of the subunits have been shown to mediate interactions between individual 10-nm-wide filaments and cause bundling (40–44). However, network formation has not been visualized. Modeling of the hypothetical vimentin and neurofilament networks using arbitrary values ranging from 0.3 to 0.7 µm for the average distance between cross-links, has yielded elastic properties consistent with those of the cells (43, 44). In comparison, the cross-links in a FilP network occur approximately at every 60 nm, an order of magnitude more frequent than estimated for animal IF networks. Intuitively, this high density of cross-links should render the FilP network a mechanically highly resilient material and would explain how FilP can make a significant contribution to the mechanical properties of the cells as measured by atomic force microscopy (8) without overtly affecting the peptidoglycan structure (Table S1). Furthermore, the complex localization pattern of FilP in the cells (Fig. 2), featuring areas of almost uniform localization interspaced with filamentous cables of varying shape and size, is consistent with the notion of a cytoskeletal network with various extent of compaction (Fig. 1 and Fig. S1). Cellular structures of FilP might also consist of an irregular meshwork of branching filaments with different diameters, corresponding to those formed by FilP in standard buffers, as reported previously (8). One can envision that different cellular cues, such as membrane domains of varying composition or local differences in the availability of FilP subunits, might influence the density of FilP polymers in the living cells.
Secondly, there is the question of how the FilP cytoskeleton is recruited to the extending tips, as strong apical accumulation of FilP was observed only in actively growing cultures and disappeared upon cessation of growth (Figs. 2 and 3). Based on in vitro and in situ data revealing an interaction between FilP and DivIVA (Figs. 3 and 4 and Fig. S3), and showing that de novo formation of DivIVA foci was immediately followed by increased accumulation of FilP cytoskeleton in the flanking areas (Fig. 5), we propose that DivIVA (or a component of the DivIVA-containing polarisome) stimulates FilP oligomerization. This process would ensure that FilP gradients are always associated with DivIVA foci and move forward with the elongating tip. Moreover, in order for polar gradients to form, the FilP structures must exhibit significant turnover rates in vivo. In analogy with animal IF proteins and crescentin, we can assume that FilP polymers are not intrinsically dynamic; thus the turnover of FilP cellular structures is likely to be facilitated by additional mechanisms, such as phosphorylation or targeted degradation (45). Interestingly, DivIVA itself is subject to serine/threonine phosphorylation that seems to affect the assembly and disassembly of the polarisome complexes (46). In this respect, it might be significant that a recent report also identified FilP as a potential phosphoprotein in Streptomyces (47). Furthermore, numerous cases of phosphorylation of IF proteins in animal cells have been reported. As one example, regulated phosphorylation of vimentin subunits in motile fibroblasts leads to a spatially restricted disassembly and retraction of the otherwise stable vimentin cytoskeleton at the cell surface where lamellipodia form (48). There must also be mechanisms in place to control the assembly of FilP. We observed that, under optimal in vitro assembly conditions, there is very low background of free subunits, suggesting that FilP has high self-affinity. The defined localization patterns we observed for FilP in vivo argue against a spontaneous and immediate posttranslational incorporation into existing cytoskeletal structures, suggesting instead that there may exist some sort of chaperone mechanism to control the assembly of subunits into filaments. Interaction with DivIVA could trigger the release of such inhibition, thereby providing spatial control over the assembly of the FilP cytoskeleton. This idea is consistent with our data showing that, in E. coli cells possessing polarly localized DivIVA but lacking the mechanisms discussed above, FilP does not form polar gradients and fills the cells with structures visually similar to those observed in stationary phase S. coelicolor hyphae (Fig. S5).
Regardless of the exact mechanism, recruitment of the FilP cytoskeleton by DivIVA, directly or indirectly, is highly reminiscent of processes occurring in polarized eukaryotic cells, where a cellular landmark complex orchestrates the assembly of actin- and tubulin-based cytoskeletal structures to help the rest of the cell to orient itself toward the landmark (49). It is likely that polarity needs to be communicated also in tip-growing bacteria, but the mechanisms have remained a mystery. Because there is no evidence of motor proteins in Streptomyces, there seems to be little need for intrinsically polar cytoskeletal tracks, as is the case in eukaryotic organisms, and instead a gradient might be used as a measure of the distance from the tip. In this regard, it is possible that the polarly oriented FilP gradient also has a role in propagation of polarity, in addition to being a structural support. Because several distantly related actinobacterial genera, such as Mycobacterium and Frankia, also possess homologs with high sequence similarity to FilP, it is likely that the cellular role of FilP is broadly conserved in this group of bacteria.
Although there remain many fascinating questions to address, we present here a mechanism to couple polar growth and the formation of an IF cytoskeleton in a bacterium, suggesting that dynamic recruitment of cytoskeleton by a polar landmark complex is a general strategy adopted by evolutionarily distant groups of organisms exhibiting cellular polarity.
Materials and Methods
Bacterial Strains and Media.
The S. coelicolor and E.coli strains used in this work are listed in Table S2. Cultivation of E.coli strains was performed as described in ref. 50. For bacterial two-hybrid (BTH) assays, strains were grown in Luria Agar plates containing ampicillin (100 µg/mL), kanamycin (50 µg/mL), isopropyl β-D-1-thiogalactopyranoside (IPTG), and X-Gal, as specified in ref. 30. S. coelicolor strains were grown on mannitol soy flour agar plates or in liquid yeast extract-malt extract medium (YEME) as described in ref. 51.
Construction of Plasmids and Recombinant S. coelicolor Strains.
The plasmids and strains used in this paper are listed in Table S2. DNA manipulation and cloning were carried out according to standard protocols. Constructs for BTH screening were generated by PCR amplification of filP, divIVA, creS, and SCO3114 and cloning into the vectors pKT25, pUT18, and pUT18c (30). All constructs were verified by nucleic acid sequencing. Strain NA883 is a marker-free derivative of strain NA335 (8), obtained by excision of the apramycin-resistance cassette as described in ref. 52. Strain NA336 was constructed by replacing SCO5367 by an apramycin-resistance cassette according to ref. 52. Plasmid pKF67 was conjugated into strain NA883 (24) to obtain strain NA956 as described in ref. 51. Sequences of primers used for cloning and construction of mutants are specified in Table S3.
Protein Techniques.
For coimmunoprecipitation experiments, the cell lysates were prepared as described in ref. 53. Forty microliters of Anti-FLAG M2 Affinity Gel (Sigma-Aldrich) was used to pull down FLAG-DivIVA complexes from 1 mL of cell lysate according to the manufacturer's guide. Protein A Sepharose (Sigma-Aldrich) was used to pull down FilP complexes from 1 mL of cell lysate incubated with 75 μL of anti-FilP antiserum. Samples were eluted by boiling with loading dye for 15 min. Supernatants were run on 10% SDS/PAGE gels, and FilP and FLAG-DivIVA were detected by Western blotting using a polyclonal rabbit anti-FilP antiserum and monoclonal mouse anti-FLAG antibodies (Sigma) as primary antibodies, respectively. Secondary horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies (Sigma) were used for detection. Blots were visualized using ECL reagents (GE Healthcare). Recombinant His-FilP was purified as in ref. 8. Anti-FilP antibodies were affinity purified from a polyclonal rabbit antiserum (Innovagen) as described in ref. 54.
Light Microscopy.
The samples for phase contrast and fluorescence microscopy were obtained from cultures grown in liquid YEME medium containing 17% (wt/vol) sucrose and 0.5% glycine. For immunofluorescence microscopy (IFM), cells were treated as described in ref. 55. Affinity purified rabbit anti-FilP antibodies and secondary goat anti-rabbit antibodies conjugated to Alexa Fluor568 (red) and Alexa Fluor488 (green) were used to visualize FilP. Direct EGFP fluorescence was used to visualize DivIVA-EGFP, and cell walls were visualized by staining with Alexa Fluor350-conjugated wheat germ agglutinin. All micrographs were generated using a Zeiss Axio Imager.Z1 microscope equipped with X-Cite 120 Illumination (EXFO Photonic Solutions) and a 9100–02 EM-CCD camera (Hamamatsu Photonics), and images were processed with Volocity5 software (Improvision).
Electron Microscopy.
Oligomerization assays of FilP were performed essentially as described in ref. 8. Dialysis was performed for 4.5 h on ice or 2 h at room temperature with two bath changes. The polymix buffer is composed of 5 mM magnesium acetate, 0.5 mM calcium chloride, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate, 95 mM potassium chloride, 5 mM ammonium chloride, and 1 mM DTT titrated to pH 7.5. The dialyzed samples were adsorbed for 2 min onto glow-discharged carbon-coated copper grids, washed in H2O, and immediately negatively stained in 50 μL of 1.5% uranyl acetate solution for 30 s. Negative stained samples were examined on a Philips CM120 transmission electron microscope operating at 100 kV. Digital images were recorded using a MegaView III CCD camera (1376 × 1032 pixels) and analySIS software (Olympus Soft Imaging Solutions). At 110,000× magnification (Fig. 1 B and C), the pixel size used for visualization is 0.63 nm.
Proximity Ligation Assay.
Cells were grown in YEME medium as described for IFM. To induce FLAG-DivIVA, an actively growing culture was incubated with10 µg/mL thiostrepton for 1 h, whereafter the cells were washed and grown in YEME medium for another hour. Cells were fixed, blocked, and treated with primary antibodies as described for IFM. After washing twice in PBS, the PLA probes of anti-Mouse PLUS and anti-Rabbit MINUS (Olink) diluted (1:5) in Antibody Diluent (Olink) were applied and incubated for 1 h at 37 °C. After washing twice in PBS or 1× Wash buffer A (Olink), the ligation and subsequent rolling circle amplification were performed using the Duolink II Detection Reagents Orange (Olink) according to the Duolink II User Manual for Fluorescence (Olink). After washing in 1× Wash Buffer B twice and then 0.01× Wash Buffer once (Olink), the slide was dried, mounted with Duolink II Mounting Medium, and analyzed by fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank Sergej Masich and Prof. Bertil Daneholt for technical expertise. N.A. and K. Flärdh. were supported by grants from the Swedish Research Council and the Carl Trygger Foundation. L.S. was supported by a European Molecular Biology Organization Long-Term Fellowship and by the Royal Swedish Academy of Sciences for electron microscope operation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305358110/-/DCSupplemental.
References
- 1.Cabeen MT, et al. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28(9):1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: In vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23(9):1131–1144. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esue O, Rupprecht L, Sun SX, Wirtz D. Dynamics of the bacterial intermediate filament crescentin in vitro and in vivo. PLoS ONE. 2010;5(1):e8855. doi: 10.1371/journal.pone.0008855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Specht M, Schätzle S, Graumann PL, Waidner B. Helicobacter pylori possesses four coiled-coil-rich proteins that form extended filamentous structures and control cell shape and motility. J Bacteriol. 2011;193(17):4523–4530. doi: 10.1128/JB.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waidner B, et al. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 2009;5(11):e1000669. doi: 10.1371/journal.ppat.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiuza M, et al. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum. J Biol Chem. 2010;285(38):29387–29397. doi: 10.1074/jbc.M110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izard J. Cytoskeletal cytoplasmic filament ribbon of Treponema: A member of an intermediate-like filament protein family. J Mol Microbiol Biotechnol. 2006;11(3-5):159–166. doi: 10.1159/000094052. [DOI] [PubMed] [Google Scholar]
- 8.Bagchi S, Tomenius H, Belova LM, Ausmees N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol. 2008;70(4):1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann H, Aebi U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 10.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell. 2003;115(6):705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 11.Cabeen MT, Herrmann H, Jacobs-Wagner C. The domain organization of the bacterial intermediate filament-like protein crescentin is important for assembly and function. Cytoskeleton (Hoboken) 2011;68(4):205–219. doi: 10.1002/cm.20505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrmann H, Hesse M, Reichenzeller M, Aebi U, Magin TM. Functional complexity of intermediate filament cytoskeletons: From structure to assembly to gene ablation. Int Rev Cytol. 2003;223:83–175. doi: 10.1016/s0074-7696(05)23003-6. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann H, Strelkov SV. History and phylogeny of intermediate filaments: Now in insects. BMC Biol. 2011;9:16. doi: 10.1186/1741-7007-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat Prod Rep. 2011;28(7):1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 15.Flärdh K, Buttner MJ. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7(1):36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 16.Jakimowicz D, van Wezel GP. Cell division and DNA segregation in Streptomyces: How to build a septum in the middle of nowhere? Mol Microbiol. 2012;85(3):393–404. doi: 10.1111/j.1365-2958.2012.08107.x. [DOI] [PubMed] [Google Scholar]
- 17.McCormick JR, Flärdh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36(1):206–231. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flärdh K. Cell polarity and the control of apical growth in Streptomyces. Curr Opin Microbiol. 2010;13(6):758–765. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Mazza P, et al. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol Microbiol. 2006;60(4):838–852. doi: 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- 20.McCormick JR, Su EP, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14(2):243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 21.Richards DM, Hempel AM, Flärdh K, Buttner MJ, Howard M. Mechanistic basis of branch-site selection in filamentous bacteria. PLOS Comput Biol. 2012;8(3):e1002423. doi: 10.1371/journal.pcbi.1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flärdh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol Microbiol. 2003;49(6):1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 23.Hempel AM, Wang SB, Letek M, Gil JA, Flärdh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol. 2008;190(22):7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SB, et al. Domains involved in the in vivo function and oligomerization of apical growth determinant DivIVA in Streptomyces coelicolor. FEMS Microbiol Lett. 2009;297(1):101–109. doi: 10.1111/j.1574-6968.2009.01678.x. [DOI] [PubMed] [Google Scholar]
- 25.Holmes NA, et al. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci USA. 2013;110(5):E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jelenc PC, Kurland CG. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci USA. 1979;76(7):3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner EG, Jelenc PC, Ehrenberg M, Kurland CG. Rate of elongation of polyphenylalanine in vitro. Eur J Biochem. 1982;122(1):193–197. doi: 10.1111/j.1432-1033.1982.tb05866.x. [DOI] [PubMed] [Google Scholar]
- 28.Gardel ML, Kasza KE, Brangwynne CP, Liu J, Weitz DA. Chapter 19: Mechanical response of cytoskeletal networks. Methods Cell Biol. 2008;89:487–519. doi: 10.1016/S0091-679X(08)00619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardel ML, et al. Stress-dependent elasticity of composite actin networks as a model for cell behavior. Phys Rev Lett. 2006;96(8):088102. doi: 10.1103/PhysRevLett.96.088102. [DOI] [PubMed] [Google Scholar]
- 30.Karimova G, Ullmann A, Ladant D. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 2000;328:59–73. doi: 10.1016/s0076-6879(00)28390-0. [DOI] [PubMed] [Google Scholar]
- 31.Weibrecht I, et al. Proximity ligation assays: A recent addition to the proteomics toolbox. Expert Rev Proteomics. 2010;7(3):401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 32.Banér J, Nilsson M, Mendel-Hartvig M, Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26(22):5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 34.Koch AL. Bacterial Growth and Form. New York: Chapman & Hall; 1995. Apical growth of streptomycetes and fungi; pp. 311–325. [Google Scholar]
- 35.Goriely A, Tabor M. Biomechanical models of hyphal growth in actinomycetes. J Theor Biol. 2003;222(2):211–218. doi: 10.1016/s0022-5193(03)00029-8. [DOI] [PubMed] [Google Scholar]
- 36.Harold FM. Force and compliance: Rethinking morphogenesis in walled cells. Fungal Genet Biol. 2002;37(3):271–282. doi: 10.1016/s1087-1845(02)00528-5. [DOI] [PubMed] [Google Scholar]
- 37.Gray DI, Gooday GW, Prosser JI. Apical hyphal extension in Streptomyces coelicolor A3(2) J Gen Microbiol. 1990;136(6):1077–1084. doi: 10.1099/00221287-136-6-1077. [DOI] [PubMed] [Google Scholar]
- 38.Beil M, et al. Sphingosylphosphorylcholine regulates keratin network architecture and visco-elastic properties of human cancer cells. Nat Cell Biol. 2003;5(9):803–811. doi: 10.1038/ncb1037. [DOI] [PubMed] [Google Scholar]
- 39.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol. 2011;12(11):695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 40.Ma L, Yamada S, Wirtz D, Coulombe PA. A ‘hot-spot’ mutation alters the mechanical properties of keratin filament networks. Nat Cell Biol. 2001;3(5):503–506. doi: 10.1038/35074576. [DOI] [PubMed] [Google Scholar]
- 41.Kim JS, Lee CH, Coulombe PA. Modeling the self-organization property of keratin intermediate filaments. Biophys J. 2010;99(9):2748–2756. doi: 10.1016/j.bpj.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C-H, Coulombe PA. Self-organization of keratin intermediate filaments into cross-linked networks. J Cell Biol. 2009;186(3):409–421. doi: 10.1083/jcb.200810196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y-C, et al. Divalent cations crosslink vimentin intermediate filament tail domains to regulate network mechanics. J Mol Biol. 2010;399(4):637–644. doi: 10.1016/j.jmb.2010.04.054. [DOI] [PubMed] [Google Scholar]
- 44.Yao NY, et al. Elasticity in ionically cross-linked neurofilament networks. Biophys J. 2010;98(10):2147–2153. doi: 10.1016/j.bpj.2010.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon KH, et al. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J Cell Biol. 2001;153(3):503–516. doi: 10.1083/jcb.153.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hempel AM, et al. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci USA. 2012;109(35):E2371–E2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manteca A, Ye J, Sánchez J, Jensen ON. Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J Proteome Res. 2011;10(12):5481–5492. doi: 10.1021/pr200762y. [DOI] [PubMed] [Google Scholar]
- 48.Helfand BT, et al. Vimentin organization modulates the formation of lamellipodia. Mol Biol Cell. 2011;22(8):1274–1289. doi: 10.1091/mbc.E10-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422(6933):766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2 Ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood D. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 52.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100(4):1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: A possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34(29):9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 54.Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. In: Guthrie C, Fink GR, editors. Methods in Enzymology. Vol 194. San Diego: Academic; 1991. pp. 565–602. [DOI] [PubMed] [Google Scholar]
- 55.Schwedock J, McCormick JR, Angert ER, Nodwell JR, Losick R. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol Microbiol. 1997;25(5):847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.