Abstract
Procaspase-3 (P3) and procaspase-7 (P7) are activated through proteolytic maturation to form caspase-3 (C3) and caspase-7 (C7), respectively, which serve overlapping but nonredundant roles as the executioners of apoptosis in humans. However, it is unclear if differences in P3 and P7 maturation mechanisms underlie their unique biological functions, as the structure of P3 remains unknown. Here, we report structures of P3 in a catalytically inactive conformation, structures of P3 and P7 bound to covalent peptide inhibitors that reveal the active conformation of the zymogens, and the structure of a partially matured C7:P7 heterodimer. Along with a biochemical analysis, we show that P3 is catalytically inactive and matures through a symmetric all-or-nothing process. In contrast, P7 contains latent catalytic activity and matures through an asymmetric and tiered mechanism, suggesting a lower threshold for activation. Finally, we use our structures to design a selection strategy for conformation specific antibody fragments that stimulate procaspase activity, showing that executioner procaspase conformational equilibrium can be rationally modulated. Our studies provide a structural framework that may help to explain the unique roles of these important proapoptotic enzymes, and suggest general strategies for the discovery of proenzyme activators.
Keywords: cell death, protease, allostery, Fab, phage display
Proteases play a primary role in catalyzing some of the most dramatic physiological transformations in multicellular organisms, including apoptosis, innate immune system activation, blood clotting, and tissue remodeling (1, 2). The vast majority of proteases are stored as inactive zymogens requiring proteolysis for activation, thus ensuring that the magnitude of proteolytic events is appropriate for a given physiological context (1, 3). The apoptotic caspases represent an excellent model system for studying protease mechanism and zymogen activation because of their critical role in mediating the proteolytic events controlling programmed cell death (2). A variety of studies have suggested that the close homologues caspase-3 (C3) and caspase-7 (C7) (52% identical in humans) play primary but nonredundant roles as the executioners of apoptosis, with caspase-6 (C6) having more specialized roles in specific physiological contexts (4–7).
Executioner procaspases are homodimeric proteins composed of two ∼32-kDa protomers. Enzymatic cleavage by upstream proteases at an extended loop known as the intersubunit linker or loop-2 (L2) generates a large and small subunit within each protomer and leads to enzyme activation (8, 9) (Fig. S1A). A variety of studies have revealed that mature caspases exist in a dynamic equilibrium between an inactive and active conformation (10–12), and it is likewise thought that executioner caspase proenzymes may be able to adopt a catalytically active conformation in the absence of cleavage. Escape from apoptosis is a defining feature of many cancers, and significant effort has thus been placed in identifying small molecules that are capable of directly activating executioner procaspases in cancer cells (13, 14). However, subsequent studies have revealed that such small-molecule proenzyme activators function via nontraditional mechanisms, and it is not clear if or how such molecular entities could be further developed into viable therapeutic agents (15–17). Most of these studies have focused on procaspase-3 (P3), the primary executioner in humans; however, a central problem in developing P3 activators is the lack of any structural information for this proenzyme. Furthermore, for executioner P3 and procaspase-7 (P7), it is not clear if the catalytically active conformation of the enzyme is accessible in the absence of L2 cleavage, and thus the conformational changes required for allosteric proenzyme activation are unknown. Finally, although P7 has served as a model for understanding P3 activation, these two enzymes are functionally distinct in vitro and in vivo, and it is unclear if mechanistic differences governing proenzyme activation may play a role in these unique phenotypes (5, 7, 18).
To answer these questions, we determined a series of structures outlining a detailed molecular framework for the activation of P3 and P7. These structures, as well as a biochemical analysis, reveal distinct mechanisms of activation for P3 and P7 that may underlie the unique biological roles of these critical mediators of cell death. The structures then allow us to rationally generate conformation-selective antibody fragments [i.e., fragments antigen-binding (Fabs)] that specifically bind a covalently trapped conformation of P3 bound to substrate mimetics. Strikingly, these Fabs are capable of significantly stimulating P3 activity against peptidic substrates without inducing zymogen cleavage, suggesting a pathway for the discovery of allosteric proenzyme activators.
Results
Structure of Human P3.
We cloned a P3 expression construct (ΔN-P3-C163A; Fig. S1B), allowing us to crystallize and solve an experimentally phased structure of P3 to 2.5 Å (P3-1), as well as a second structure by molecular replacement to 2.9 Å (P3-2; Table S1 and Fig. S2A). The P3-1 structure contains a dimer in the asymmetric unit and resolves ∼80% of the amino acids due to a number of disordered loops. The structure is globally similar to available structures of P7, aligning with Cα rmsd values of 1.9 Å and 1.8 Å among commonly resolved residues to structures 1K88 and 1GQF [Protein Data Bank (PDB) ID codes], respectively (Fig. 1A). Portions of loop-2 (L2), which forms the intersubunit linker, occupy the central cavity of the caspase dimer, a common feature of other caspase proenzymes (8, 9). Loop-3 (L3), or the “activation loop,” which forms the primary substrate binding groove in the active enzyme, is pulled out of the active site in both P3 monomers, although one is partially disordered whereas the other is fully resolved (Fig. 1 A and B). This could be caused in part by asymmetric crystal packing contacts that appear to stabilize the single L3 resolved in our structure, although the existing structures of P7 (PDB ID codes 1K88 and 1GQF) also reveal a variety of ordered and disordered L3s, suggesting that this loop samples a large number of conformations in P3 and P7 (8, 9).
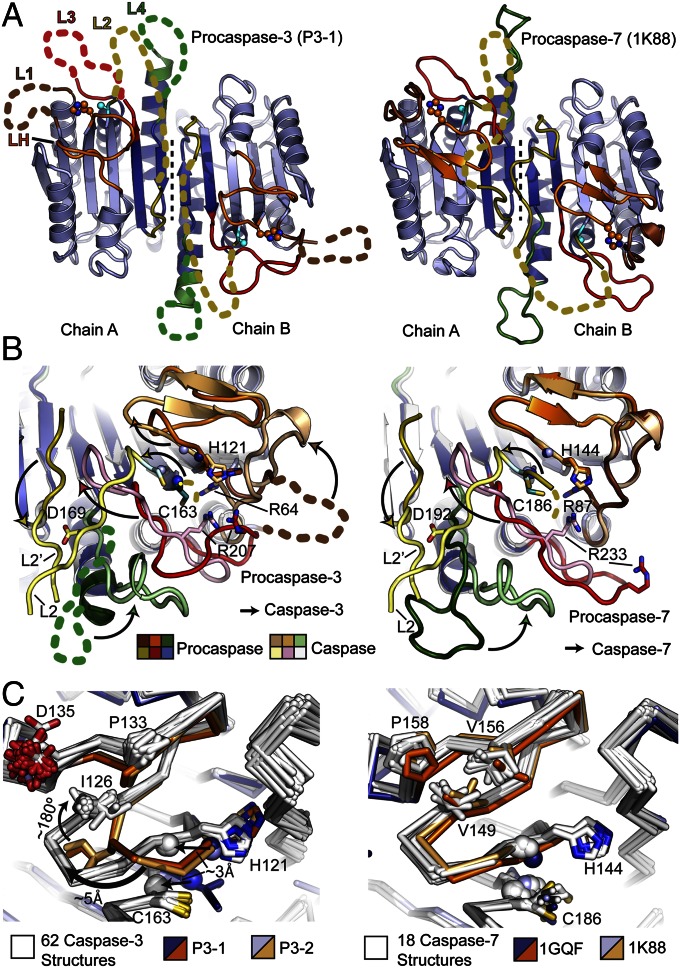
Fig. 1.
Structural comparison of P3 and P7. (A) Comparison of P3-1 (Left) and P7 (Right; PDB ID code 1K88) with disordered loops (colored dashed lines), catalytic cysteine (cyan spheres) and histidine (orange spheres) side chains, dimer interface (black dashed line), and small (dark blue) and large (light blue) subunits shown (8). (B) Conformational changes (arrows) required for maturation in P3 (Left) and P7 (Right) colored as in A. Caspase structures used are PDB ID code 2DKO (Left) and PDB ID code 1F1J (Right) (24, 42). Side chains involved in substrate binding and catalysis (sticks) and backbone amides forming the oxyanion hole (blue spheres) are shown. (C) An alignment of P3-1 and P3-2 with 62 C3 structures shows that the LH structure is unique to the proenzyme (Left). An alignment of available P7 structures with 18 C7 structures shows that LH, the catalytic histidine, and elements of the oxyanion hole (spheres) are in nearly identical conformations (Right). Catalytic side chains and LH residues unique to C3 or C7 (Fig. S4B) are shown as sticks.
In addition to these similarities, the structural superposition also reveals three distinct differences between P3 and P7 (Fig. 1 A and B). First, loop-1 (L1), which to our knowledge is visible in every C3 and C7 structure solved to date, is disordered in the structure of P3. This includes disorder of Arg64, one of the primary residues responsible for binding the P1 aspartate in caspase substrates. Second, the two- to three-strand β-sheet [hereafter referred to as loop-H (LH)] containing residues contributing to the oxyanion hole as well as the catalytic histidine (His121) has been disrupted, and moves ∼5 Å away from its conformation in structures of the mature enzyme (Fig. 1 B and C). Third, three or four residues typically forming part of loop-4 (L4) have instead been rewound into the adjacent α-helix, forming a complete additional turn (Fig. 1B). These structural differences are most clearly visualized in energy-minimized linear interpolations comparing P3 and P7 maturation (Movie S1). Importantly, we can rule out crystal packing as a cause of these changes in P3, as the same three features are also present in the P3-2 crystal structure, which demonstrates a unique packing arrangement and aligns with an rmsd of 0.7 Å across commonly resolved Cα atoms (Fig. S3).
Compared with all known structures of C3, the change in LH results in a ∼2- to 3-Å shift in the Cα atoms of His121, and of the adjacent backbone amide from Gly122 that forms part of the oxyanion hole (Fig. 1C). In contrast to this, LH, the catalytic histidine and the adjacent backbone amide are in nearly identical conformations in all C7 and P7 structures (Fig. 1C). This conformation of LH in P3 appears to be stabilized in part by a ∼180° flip of Ile126 from a solvent-exposed position in C3 to a buried position in P3 that is itself stabilized by structures changes in Tyr197 and Leu136 (Fig. S4A). The dramatic flip of Ile126 may be caused by destabilization of the LH β-sheet influenced by Pro133 and Gly125 respectively, which are conserved among C3, but not C7, homologues (Fig. S4B). Interestingly, this same region has been observed to undergo a strand to helix conformational change in C6, suggesting that LH metastability may be a common feature regulating catalytic activity in executioner caspases (19, 20).
In Vitro Biochemical Analysis of P3 and P7.
To determine if the structural differences in P3 and P7 result in functional differences, we conducted a kinetic analysis of noncleavable P3 (P3-D3A) and P7 (P7-D2A) in optimized assay buffers at pH 7.4 (Fig. S1B). Trace levels of mature or partially mature enzyme in these preparations were inactivated by titrating the covalent inhibitor acetyl-Asp-Glu-Val-Asp-chloromethylketone (Ac-DEVD-CMK) up to 10% of the total protein as described previously (16). Although P3-D3A was catalytically inactive as reported previously (16), P7-D2A displayed weak but measureable catalytic activity, with a catalytic efficiency of 0.23 M−1⋅s−1 (Table S2 and Fig. S4C). The P7 Km (976 ± 96 μM) is ∼35 fold higher, and kcat (2.2 × 10−4 ± 9.3 × 10−6⋅s−1) is ∼3.3 × 104 fold lower, than those of C7, leading to a zymogenicity (i.e., kcat/Km enzyme divided by kcat/Km proenzyme) of ∼1.13 × 106 for P7 (Table S2). Overall, P7 is at least 10-fold more active than the estimated rate for P3 (16).
A variety of studies have suggested that procaspase activation during apoptosis is stimulated by decreases in intracellular pH (21, 22). We tested this concept in vitro by performing kinetic assays on P3 and P7 at pH 6.0 to 7.5 in a variety of common caspase buffers (Fig. 2A). The results show that decreases in pH stimulate P7 activity as much as 10-fold, whereas only trace levels of activity are observed for P3 at lower pH. We next used this assay to determine if conserved substitutions in LH could affect P3 and P7 catalytic activity. Although P7 residues were not able to restore activity when inserted into P3, proline substitutions in the LH β-sheet of P3 reduced catalytic activity to nearly undetectable levels when inserted into P7 (Fig. 2A and Fig. S4 D and E). Interestingly, a previous study in which LH residues (including V156 and P158) in C7 were mutated to the corresponding residues in C3 showed relatively little change in the kinetic constants of the mature enzymes (23). Overall, the combined structural and biochemical observations suggest that LH β-sheet stability plays an important role in regulating procaspase activity, although our inability to restore catalytic activity to P3 suggests that additional substitutions underlie the full range of mechanistic differences between P3 and P7.
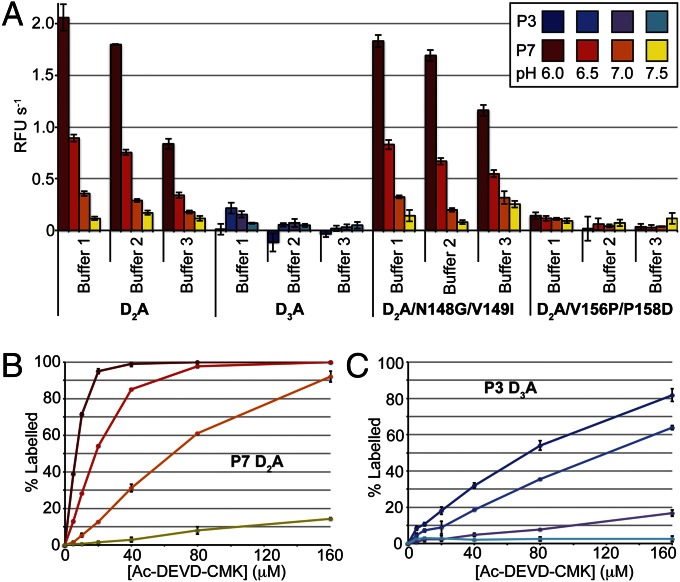
Fig. 2.
Biochemical analysis of P3 and P7. (A) Maximum catalytic rates for P7, P3, and P7 mutants measured at 500 μM Ac-DEVD-AFC are shown under a variety of buffer conditions and pH. Error bars are ± SD (n = 3). (B) MS-based quantification of P7 labeling by the ABP Ac-DEVD-CMK in buffer 2 at a variety of pH values (colored according to A). Error bars are ± SD (n = 2). (C) As in B for P3.
Given the low activities measured in our biochemical assay, we next used the activity-based probe (ABP) Ac-DEVD-CMK to more sensitively compare the activities of P3 and P7. Although this covalent inhibitor does not serve as a direct measure of catalytic activity, it serves as highly sensitive measures of catalytic site accessibility and cysteine nucleophile reactivity. We thus mixed P3 or P7 with increasing concentrations of Ac-DEVD-CMK in assay buffer, and followed percent labeling by MS. At near-physiological pH (7.5), the results show no increase in P3 labeling with increasing concentrations of Ac-DEVD-CMK (Fig. 2 B and C). However, P7 shows a linear increase in labeling with increasing substrate concentrations, again supporting the structural and biochemical results showing that P7 is more active than P3. However, at lower pH, we observed a dramatic increase in labeling for P3 and P7, suggesting that P3 catalytic activity may also be stimulated by slightly acidic pH despite the fact that we could not measure its activity in our substrate cleavage assays (Fig. 2 B and C). Overall, these observations are somewhat paradoxical, given that the catalytic histidine-cysteine dyad would be expected to be protonated and thus less reactive at low pH. However, changes in pH may also affect the conformation or dynamics of the active site in the proenzyme, and thus increase binding affinity or nucleophilic attack on the ABP through an unknown mechanism.
Structures of P3 and P7 Bound to Ac-DEVD-CMK.
We hypothesized that the population of Ac-DEVD-CMK–labeled P3 and P7 identified in our labeling assays might be trapped in the otherwise elusive “active-proenzyme” conformation. By using these assays as a guide, we labeled and purified crystallographic constructs lacking the N-terminal prodomain, and containing a single mutation in one of the primary caspase processing sites (ΔN-P3-D175A and ΔN-P7-D198A; Fig. S1B). The Ac-DEVD-CMK–labeled ΔN-Pro3-D175A (P3-DEVD) and ΔN-Pro7-D198A (P7-DEVD) proteins readily gave crystals diffracting to 1.8 Å and 2.15 Å, respectively, and were solved by using molecular replacement (Table S1). An analysis of the crystals by SDS/PAGE confirmed that the intact proenzyme was present in both cases, and showed no evidence of active enzyme by Coomassie staining (Fig. S2 B and C).
The P3-DEVD structure contains a full procaspase dimer in the asymmetric unit and resolves all loops except for portions of the intersubunit linker (Fig. 3A). The dimer is highly symmetrical (Cα rmsd of 0.3 Å between protomers), and the overall structure aligns much more closely to the mature C3 enzyme than to P3 (Fig. S5A). In both active sites, L1, LH, L3, and L4 are in the active conformation, forming contacts with the bound Ac-DEVD-CMK inhibitor, which shows strong density in simulated annealing omit electron density maps (Fig. 3B and Fig. S5B). The catalytic histidine and cysteine are thus in their active conformations, with the cysteine covalently bound to the CMK group of the inhibitor. Interestingly, the 1.8-Å electron density maps clearly show a tetrahedral transition state intermediate in which the cysteine sulfur atom is covalently linked to the ketone carbonyl C atom rather than to the ketomethylene of the CMK group, the chlorine atom is absent, and the P1 carbonyl group is perfectly positioned in the oxyanion hole (Fig. S5C). The same arrangement has been observed in the 1.06-Å structure of C3 bound to Ac-DEVD-CMK, where it was shown to be induced by synchrotron radiation (24). Overall, the primary feature distinguishing the structure from the proenzyme and the active enzyme is the position of the intersubunit linker (i.e., L2). In the active proenzyme presented here, movement of L3 into the active conformation requires that L2′ is ejected from the central cavity. This allows L2′ to extend into the active conformation, forming a partial L2 bundle (Fig. 3A). However, as the intersubunit linker is still intact in the proenzyme, the remainder of L2 is physically prevented from forming the complete bundle and is instead disordered in solution. This prevents Asp169, a residue important for caspase activity, from reaching its active conformation and fully stabilizing the loop bundle (Fig. 3B) (25). The conformational changes required for P3 to adopt the active substrate-bound conformation are most clearly visualized in energy minimized linear interpolations (Movie S2).
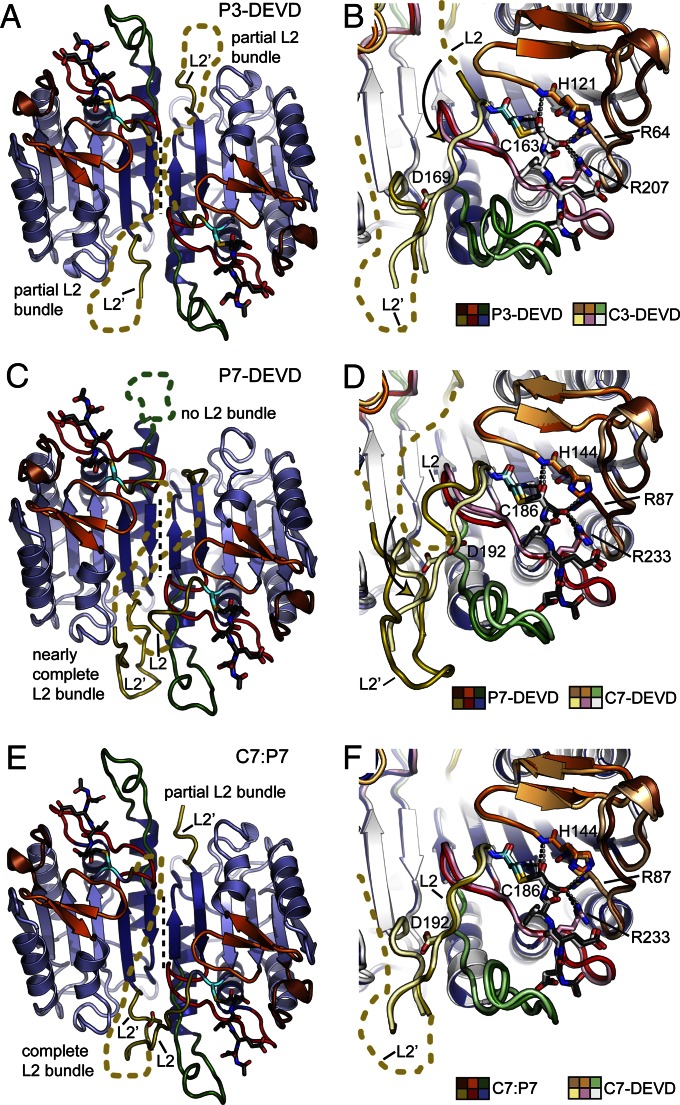
Fig. 3.
Structures of P3 and P7 and a C7:P7 heterodimer bound to Ac-DEVD-CMK. (A) Representation of P3-DEVD as in Fig. 1A. The tetrapeptide substrate is shown as dark gray sticks with oxygen atoms colored red and nitrogen atoms colored blue. (B) Overlay of P3-DEVD and C3-DEVD (PDB ID code 2DKO) represented as in Fig. 1B. Dotted lines indicate hydrogen bonds or salt bridges between bound inhibitor and substrate binding residues. (C) Representation as in A for P7-DEVD. (D) Representation as in B for P7-DEVD and C7-DEVD (PDB ID code 1F1J). (E) Representation as in A for the C7:P7 heterodimer. (F) Representation as in B for C7:P7 and C7-DEVD (PDB ID code 1F1J).
The P7-DEVD structure contains a single procaspase dimer within the asymmetric unit, resolving all of the protein with the exception of one L4 and portions of the intersubunit linker (i.e., L2; Fig. 3C). Again, the overall structure is a striking match for the mature C7 enzyme, whereas it is a poorer match for the proenzyme (Fig. S5D). Like the P3-DEVD structure, L1, LH, and L3 are in the active conformation in both active sites, forming contacts with the covalently bound Ac-DEVD-CMK inhibitor, which shows strong electron density (Fig. 3D and Fig. S5 E and F). Interestingly, the P7-DEVD dimer appears significantly more asymmetric than P3-DEVD (rmsd of 0.3 Å between protomers), with an rmsd of 2.1 Å between protomers. This is because neither the L2′, L2, or L4 loops are in the active conformation in one catalytic site, whereas the L4, L2′, and a portion of the L2 loop have folded into an almost fully matured structure in the other catalytic site (Fig. 3C). Indeed, Asp192 in P7 (structurally corresponding to Asp169 in P3), which is critical for catalytic activity, is present in one active site and poised to adopt the conformation observed in structures of fully mature C7 (26) (Fig. 3D). These differences between P3 and P7 catalytic activation in the absence of proteolytic maturation are clearly visualized in energy minimized linear interpolations (Movie S2).
Structure of a C7:P7 Heterodimer.
We were fortuitous enough to identify a single crystal in our P7-DEVD crystallization trials in which an unknown source of limited proteolysis cleaved ∼50% of P7-DEVD, thus generating a crystal containing one half procaspase and one half caspase (Fig. S5G). We were able to obtain a complete dataset to 1.65 Å and solve the structure by using molecular replacement (Table S1). The resulting structure appears to contain a C7:P7 heterodimer. As a result, one loop bundle is fully formed whereas the other loop bundle is only partially formed (Fig. 3E), an observation supported by simulated annealing omit maps (Fig. S5H). A single catalytic site in the heterodimer, which contains contributions from the P7 and C7 subunits, overlays almost perfectly with the active site in mature C7 (Fig. 3F). A comparison of relative B-factors between the fully formed and partially formed catalytic sites in this structure suggests that complete L2 bundle formation structurally stabilizes the active site loops, and in particular L4 (Fig. S5I). This observation is also consistent with the P3-DEVD and P7-DEVD structures, in which increased loop bundle formation appears to correlate with proper positioning of L4 (Fig. 3 B, D, and F). Thus, a single cleavage event in P7 appears sufficient for formation of a single loop bundle, which stabilizes a single catalytic site in the dimer. Strikingly, this observation confirms previous biochemical evidence showing that a single cleavage event in P7 produces an enzyme with full activity at a single catalytic site (26). These results also suggest that the individual subunits of P7 can behave completely independently with respect to catalytic activation and/or allosteric communication, in contrast to the cooperative activation observed for caspase-1 (C1) (10, 27).
Conformation-Selective Antibody Fragments Stimulate P3 Activity.
We hypothesized that molecules binding specifically to the unique L2 docking surface, or to the substrate-binding loops observed in our P3-DEVD or P7-DEVD structures might act as L2 mimics to allosterically stimulate catalysis in the caspase proenzymes (Fig. S6A). As an initial test of this idea, we used phage display to generate conformation-selective antibody fragments (i.e., Fabs) that bound preferentially to P3-DEVD or P7-DEVD (Fig. S6B). We isolated 25 unique P3-DEVD binding Fabs and 16 unique P7-DEVD binding Fabs; however, only the P3 selections produced Fabs displaying strong binding selectivity for the Ac-DEVD-CMK–labeled proenzyme (Fig. S7A). We identified a clone (F1-8) that appeared to weakly stimulate the activity of P3-D3A (Fig. S7B), and generated an affinity matured variant (NT5-14) with improved binding and P3-D3A stimulating characteristics (Fig. S7 C–E). Size-exclusion chromatography revealed that the NT5-14:P3-DEVD complex is ∼160 kDa, consistent with a binding stoichiometry of 1:1 (Fab:procaspase monomer; Fig. S7F). Competition ELISAs were used to estimate affinity using the methods of Martineau (28), and revealed strong conformational selectivity, with NT5-14 binding P3, C3, C3-DEVD, and P3-DEVD with estimated Kd values of 7.0 (±0.8) μM, 440 (±70) nM, 19 (±0.7) nM, and 6.3 (±0.6) nM, respectively (Fig. 4A).
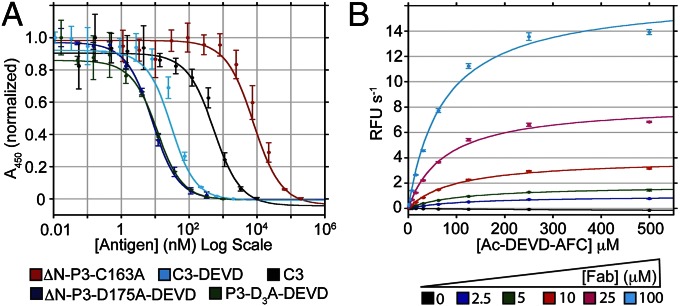
Fig. 4.
Conformation-selective antibody fragments (i.e., Fabs) stimulate P3 catalytic activity. (A) Competition ELISA results showing the strong conformational selectivity of Fab NT5-14 for P3-DEVD over other forms of the enzyme. Error bars are ± SD (n = 3). See main text for calculated dissociation constants. (B) Michaelis–Menten kinetic analysis of 5 μM P3-D3A in the presence of increasing Fab NT5-14 concentrations (values summarized in Table S2). Error bars are ± SD (n = 3).
Dose–response analysis reveals that conformation-selective Fab NT5-14 stimulates P3 activity against small fluorogenic substrates, and that the Fab preparation contains no contaminating Ac-DEVD-AFC hydrolase activity (Fig. S7G). Although we were not able to fully saturate the dose–response as a result of the high concentrations of Fab required, our estimated EC50 value of ∼26 μM is similar in magnitude to the Kd against P3, but ∼3,000-fold higher than the Kd against P3-DEVD. This suggests that the Fab is capable of stabilizing the active conformation of P3 at the energetic price of reduced affinity. This observation may be explained by a high energetic barrier for proenzyme activation—which is supported by the remarkable high zymogenicity of P3. A full kinetic analysis shows that increasing concentrations of Fab NT5-14 restore measurable catalytic activity by increasing the kcat and decreasing the Km of P3, suggesting that Fab NT5-14 alters the conformation of the active site to improve substrate binding and catalysis (Table S2 and Fig. 4B). At saturating levels of Fab, the catalytic efficiency of P3 is 164 M−1⋅s−1, revealing that Fab NT5-14 can stimulate P3 activity by >1,000 fold. However, assays conducted at physiological concentrations (∼100 nM) of WT P3 (29), or mixtures of WT P3 and P7 (in the presence of 10% Ac-DEVD-CMK, with all proteins containing WT caspase processing sites) produced no autoactivation at saturating levels of Fab NT5-14. This could be a result of the low activity of the Fab–procaspase complex (remaining ∼9,000-fold less active than mature C3), or to steric exclusion created by the bound Fab that prevents binding and cleavage of large protein substrates.
Discussion
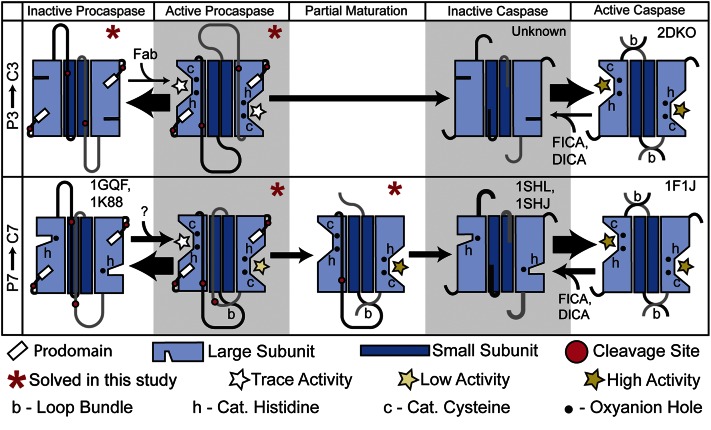
The structural and biochemical data presented here highlight significant differences in the intrinsic catalytic activity and maturation mechanism of P3 and P7 (Fig. 5). Our structure of P3, the primary human executioner of apoptosis, reveals that this enzyme is trapped in a highly inactive conformation (Fig. 1). As a result, in each active site of both the P3-1 and P3-2 structures, all substrate binding loops, and all components of the catalytic machinery are displaced from their positions in the mature enzyme. In contrast, half of the catalytic center and key substrate binding loops appear preactivated in all structures of P7. Our biochemical studies provide strong support for the structural observations (Fig. 2 and Table S2). Interestingly, our studies show that, even though P7 is significantly more active than P3, C3 is significantly more active than C7 (Table S2). This observation fits with the biological need to tightly control the activity of the primary executioner caspase (caspase-3), but also presents a unique evolutionary conundrum. For P3 and P7, nature has driven the simultaneous evolution of two opposing functions into different conformational states of the same protein.
Fig. 5.
Distinct mechanisms of P3 vs. P7 activation. A model of proposed conformational equilibrium and maturation in P3 and P7 based on the structural and biochemical results reported here and elsewhere. PDB ID codes are indicated for structural examples known before this work. Allosteric modulators that affect caspase or procaspase conformational equilibrium [5-fluoro-1H-indole-2-carboxylic acid (2-mercapto-ethyl)-amide (FICA), 2-(2,4-dichloro-phenoxy)-N-(2-mercapto-ethyl)-acetamide (DICA), and Fab NT5-14] are shown. Arrow thickness indicates proposed conformational equilibrium based on activity measurements and structural data. The intersubunit linkers from each protomer are colored light gray and black. Stars indicate catalytically competent active sites (see key).
Despite the low catalytic activities of P3 and P7, our structures of ΔN-P3-D175A and ΔN-P7-D198A bound to Ac-DEVD-CMK are strikingly similar to the mature enzymes (Fig. 3 B and D). The only region that cannot adopt the catalytically active conformation is L2, which is physically prevented from folding into the active structure in the proenzyme. Importantly, this shows that intersubunit linker cleavage is not explicitly required for the proenzyme to adopt a catalytically active structure. Rather, this suggests that the primary purpose of cleavage is to generate the loop bundle, which functions as an allosteric activator of the mature enzyme. This observation is supported by studies showing that mutations within the loop bundle can alter catalytic activity without altering the structure of the active enzyme, and by the observation that phosphorylation of the loop bundle can regulate activity in C6 (25, 30, 31).
The structure of the C7:P7 heterodimer provides a molecular basis for understanding previous biochemical studies that used ABPs showing that P7 (but not P3) may pass through a partially cleaved intermediate (26, 32). Most importantly, it confirms that such an intermediate contains all the structural elements required for full catalytic activity at a single site (Fig. 3F). Taken together, the collection of structures presented here may provide insight into why P7, but not P3, has been observed to mature through such a partially cleaved intermediate. Although our structures of P3 shows symmetric intersubunit linkers, existing structures of P7 as well as our structure of the Ac-DEVD-CMK–bound proenzymes reveals more asymmetric linkers (Figs. 1A and 3 A and C). This asymmetry may favor preferential cleavage and activation of a single P7 subunit in biological systems and via limited proteolysis in our crystallization experiments. When viewed along with the biochemical data, our structural results support an asymmetric-stepwise P7 activation mechanism, but suggest that P3 activation proceeds in an all-or-none fashion, as its intersubunit linkers appear equally accessible to proteolysis in all known structures (Fig. 5). A similar phenomenon is observed in the structure of a caspase-9 (C9) dimer revealing one active and one inactive catalytic site, suggesting that such stepwise activation may be a common feature regulating caspase function (33).
In light of the small but detectable level of catalytic activity present in P7, as well as the recent discovery of a C7 exosite that can stimulate cleavage against specific substrates (23), it is an intriguing possibility that P7 may cleave specific physiological substrates in the absence of complete maturation. The partially cleaved C7:P7 intermediate would then represent an intermediate level of activity between the proenzyme and mature enzyme. Interestingly, P7 is labeled by ABPs early in apoptosis, suggesting that it could control the threshold for apoptotic activation in cells (32). Further consistent with this idea is the dramatic stimulation of P7 activity we observe at reduced pH, which has been shown to correlate with executioner procaspase activation in cellular models of apoptosis (Fig. 2A) (22). This could also explain the somewhat puzzling observation that C3(−/−) mouse embryonic fibroblasts are actually more sensitive to apoptotic inducers than C7(−/−) mouse embryonic fibroblasts, despite the otherwise dominant role of C3 in apoptosis (5). Thus, the timing and magnitude of early P7 activation could regulate entry into apoptosis, with P3 activation serving as a point of no return for committing to apoptosis. Consistent with a need to tightly regulate P7 activation early in apoptosis, C7 contains an inhibitor of apoptosis binding motif (IBM) not present in C3, appears to be more efficiently ubiquitinated by inhibitors of apoptosis (IAPs) and is more readily targeted for proteasomal degradation than C3 (34, 35). A similar concept has recently been proposed for procaspase-6 (P6), which is also observed to activate via a series of partially cleaved intermediates when examined with ABPs in apoptotic cell lysates (36). In such contexts, proteolytic maturation could serve to regulate a tiered activation mechanism (Fig. 5), allowing for limited proteolysis in specific physiological contexts, and possibly explaining some of the nonapoptotic functions more recently attributed to the apoptotic caspases (37).
The dramatic stimulation of P3 induced by Fab NT5-14 shows that conformation-selective binding entities can be used to overcome large energetic barriers and allosterically stimulate otherwise catalytically undetectable enzymes (Fig. 4 and Table S2). However, the 3,000-fold reduction in estimated EC50 relative to the Kd against P3-DEVD highlights the challenges associated with inducing the active conformation of such an inactive zymogen, suggesting that picomolar binding to P3-DEVD might be required to fully activate P3 at an EC50 in the nanomolar range (Fig. 4A and Fig. S7G). Given the unusually high zymogenicity of P3 (16), it is likely that most other proteases, including many extracellular enzymes, may be more robustly stimulated and possibly even fully autoactivated by using similar methods. Conformation-selective binders generated in such a fashion could inform the design of novel therapeutic agents, and would serve as powerful gain-of-function probes to study proteases in vitro and in vivo.
Experimental Procedures
Protein Cloning, Expression, and Purification.
Plasmids encoding full-length C3 and C7 with C-terminal His6 tags were used to express and purify WT C3, C7, P3, and P7 as described previously (13). P3-D3A and P7-D2A proenzymes were expressed for only 2 h at 30 °C to limit adventitious activation by Escherichia coli proteases. All purifications were conducted at 4 °C.
Crystallization and Data Collection.
Crystals were grown in hanging drop format by using a Mosquito nanoliter pipetting system (TTP LabTech). Data were collected at Advanced Light Source Beamline 8.3.1 at 100 K. Datasets were processed by using HKL2000, solved and refined using PHENIX, and built by using Coot (38–40).
Biochemical Assays.
Protease activity assays were conducted at room temperature on a SpectraMax M5 plate reader (Molecular Devices). C3, C7, P3, and P7 biochemical assays were conducted in optimized C3 (buffer 2) or C7 (buffer 1) assay buffers at pH 7.4 as described previously (13, 16).
Phage Display, Affinity Maturation, and Fab Characterization.
The first round of phage display and Fab characterization was conducted essentially as described previously (41).
Additional Methods.
Additional methods are described in SI Experimental Procedures.
Supplementary Material
Acknowledgments
Thanks to Prof. S. Sidhu (Banting and Best Department of Medical Research, University of Toronto) for phage display libraries; S. Pfaff for assistance with surface plasmon resonance; C. Waddling and the University of California, San Francisco Macromolecular Structure Group for access to protein crystallization facilities; J. Tanamachi, J. Holton, and G. Meigs at Advanced Light Source Beamline 8.3.1; Scott Gradia (California Institute for Quantitative Biosciences MacroLab) for expression plasmids; and Patrick Weinkam and J.A.W. laboratory members for helpful discussions. Research was supported by Damon Runyon Cancer Research Foundation Grant 2082-11 (to N.D.T.) and National Institutes of Health Grant R01 CA136779 (to J.A.W.). N.D.T. is the Suzanne and Bob Wright Fellow of the Damon Runyon Cancer Research Foundation. J.T.K. is a Fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4JQY (P3-1), 4JQZ (P3-2), 4JR0 (P3-DEVD), 4JR1 (P7-DEVD), and 4JR2 (C7:P7)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306759110/-/DCSupplemental.
References
- 1.Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation (a review) Proc Natl Acad Sci USA. 1976;73(11):3825–3832. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RC, Cullen SP, Martin SJ. Apoptosis: Controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9(3):231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 3.Stroud RM, Kossiakoff AA, Chambers JL. Mechanisms of zymogen activation. Annu Rev Biophys Bioeng. 1977;6:177–193. doi: 10.1146/annurev.bb.06.060177.001141. [DOI] [PubMed] [Google Scholar]
- 4.Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276(10):7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 5.Lakhani SA, et al. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science. 2006;311(5762):847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142(4):637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh JG, et al. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci USA. 2008;105(35):12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai J, et al. Crystal structure of a procaspase-7 zymogen: Mechanisms of activation and substrate binding. Cell. 2001;107(3):399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 9.Riedl SJ, et al. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci USA. 2001;98(26):14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci USA. 2006;103(20):7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci USA. 2004;101(34):12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proc Natl Acad Sci USA. 2009;106(9):3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326(5954):853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putt KS, et al. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nat Chem Biol. 2006;2(10):543–550. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- 15.Zorn JA, Wille H, Wolan DW, Wells JA. Self-assembling small molecules form nanofibrils that bind procaspase-3 to promote activation. J Am Chem Soc. 2011;133(49):19630–19633. doi: 10.1021/ja208350u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorn JA, Wolan DW, Agard NJ, Wells JA. Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. J Biol Chem. 2012;287(40):33781–33795. doi: 10.1074/jbc.M112.386128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson QP, et al. PAC-1 activates procaspase-3 in vitro through relief of zinc-mediated inhibition. J Mol Biol. 2009;388(1):144–158. doi: 10.1016/j.jmb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agard NJ, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA. 2012;109(6):1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya S, Velázquez-Delgado EM, Abbruzzese G, Hardy JA. Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J Mol Biol. 2011;406(1):75–91. doi: 10.1016/j.jmb.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumgartner R, et al. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J. 2009;423(3):429–439. doi: 10.1042/BJ20090540. [DOI] [PubMed] [Google Scholar]
- 21.Roy S, et al. Maintenance of caspase-3 proenzyme dormancy by an intrinsic “safety catch” regulatory tripeptide. Proc Natl Acad Sci USA. 2001;98(11):6132–6137. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: Early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2(6):318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 23.Boucher D, Blais V, Denault JB. Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc Natl Acad Sci USA. 2012;109(15):5669–5674. doi: 10.1073/pnas.1200934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganesan R, Mittl PR, Jelakovic S, Grütter MG. Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis. J Mol Biol. 2006;359(5):1378–1388. doi: 10.1016/j.jmb.2006.04.051. [DOI] [PubMed] [Google Scholar]
- 25.Feeney B, Pop C, Swartz P, Mattos C, Clark AC. Role of loop bundle hydrogen bonds in the maturation and activity of (Pro)caspase-3. Biochemistry. 2006;45(44):13249–13263. doi: 10.1021/bi0611964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denault JB, et al. Engineered hybrid dimers: Tracking the activation pathway of caspase-7. Mol Cell. 2006;23(4):523–533. doi: 10.1016/j.molcel.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Datta D, Scheer JM, Romanowski MJ, Wells JA. An allosteric circuit in caspase-1. J Mol Biol. 2008;381(5):1157–1167. doi: 10.1016/j.jmb.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martineau P. Affinity measurements by competition ELISA. In: Kontermann R, Dübel S, editors. Antibody Engineering. Vol 1. Berlin: Springer; 2010. pp. 657–665. [Google Scholar]
- 29.Svingen PA, et al. Components of the cell death machine and drug sensitivity of the National Cancer Institute Cell Line Panel. Clin Cancer Res. 2004;10(20):6807–6820. doi: 10.1158/1078-0432.CCR-0778-02. [DOI] [PubMed] [Google Scholar]
- 30.Velázquez-Delgado EM, Hardy JA. Phosphorylation regulates assembly of the caspase-6 substrate-binding groove. Structure. 2012;20(4):742–751. doi: 10.1016/j.str.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Q, et al. Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays. J Biol Chem. 2012;287(19):15371–15379. doi: 10.1074/jbc.M112.351213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger AB, et al. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23(4):509–521. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci USA. 2001;98(25):14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YE, et al. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. J Biol Chem. 2009;284(19):12772–12782. doi: 10.1074/jbc.M807550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhuang M, Guan S, Wang H, Burlingame AL, Wells JA. Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol Cell. 2013;49(2):273–282. doi: 10.1016/j.molcel.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgington LE, et al. An optimized activity-based probe for the study of caspase-6 activation. Chem Biol. 2012;19(3):340–352. doi: 10.1016/j.chembiol.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuranaga E, Miura M. Nonapoptotic functions of caspases: Caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007;17(3):135–144. doi: 10.1016/j.tcb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat Protoc. 2007;2(6):1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- 42.Wei Y, et al. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7(6):423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.