Abstract
Congenital defects, trauma, and disease can compromise the integrity and functionality of the skeletal system to the extent requiring implantation of bone grafts. Engineering of viable bone substitutes that can be personalized to meet specific clinical needs represents a promising therapeutic alternative. The aim of our study was to evaluate the utility of human-induced pluripotent stem cells (hiPSCs) for bone tissue engineering. We first induced three hiPSC lines with different tissue and reprogramming backgrounds into the mesenchymal lineages and used a combination of differentiation assays, surface antigen profiling, and global gene expression analysis to identify the lines exhibiting strong osteogenic differentiation potential. We then engineered functional bone substitutes by culturing hiPSC-derived mesenchymal progenitors on osteoconductive scaffolds in perfusion bioreactors and confirmed their phenotype stability in a subcutaneous implantation model for 12 wk. Molecular analysis confirmed that the maturation of bone substitutes in perfusion bioreactors results in global repression of cell proliferation and an increased expression of lineage-specific genes. These results pave the way for growing patient-specific bone substitutes for reconstructive treatments of the skeletal system and for constructing qualified experimental models of development and disease.
Keywords: embryonic stem cells, mesodermal progenitors, dynamic culture, bone regeneration, microarray analysis
Current treatments of large bone defects, which rely on the use of alloplastic materials or transplantation of bone grafts, have limited clinical potential, and new approaches are required to develop effective therapies for complex bone reconstructions (1). Biomimetic tissue-engineering strategies have recently been explored for the ex vivo cultivation of functional, anatomically shaped bone substitutes, by culturing human mesenchymal stem cells on 3D scaffolds resembling the matrix of native bone in bioreactors providing interstitial flow of culture medium (2). This method has recently been extended to the engineering of mechanically functional human cartilage interfaced with bone that was also derived from mesenchymal stem cells (3). However, the creating of vascular and nerve compartments in engineered bone requires other cell sources.
Pluripotent stem cells are promising candidates for the construction of fully functional bone substitutes, for they can give rise to all specialized cell types constituting the human bone (4, 5). Previous studies have demonstrated that human embryonic stem cells (hESCs) differentiate into osteogenic cells (6–8) and form compact bone tissue matrix when cultured on osteoconductive scaffolds in perfusion bioreactors (9). However, ethical controversy and concerns regarding immune properties of hESCs in allogeneic conditions hinder their clinical translation (10). Human-induced pluripotent stem cells (hiPSCs) reprogrammed using nonintegrating vectors (11, 12) have the potential to overcome both limitations and be used to give rise to the cells of bone, vasculature, and adjacent tissues and to engineer autologous bone substitutes.
Culture protocols developed for hESCs have been successfully adapted to hiPSCs, which resemble hESCs with respect to their morphology, molecular signature, and differentiation potential (13). However, variable efficiencies to form specific lineages have been reported (14), and it is not clear whether such variability results from different genetic backgrounds, the source tissue for cell line derivation by reprogramming, or the reprogramming method itself. Functional mesenchymal progenitors with good osteogenic properties in vitro and in vivo have been derived from hiPSCs (15, 16). However, little is known about the osteogenic potential of mesenchymal progenitors derived from different hiPSC lines, and the molecular changes associated with osteogenic differentiation. Also, engineering of 3D functional bone substitutes using hiPSC lines has not been attempted.
Based on prior studies (2, 9, 15) and known biological similarities between hESCs and hiPSCs (13), we hypothesized that hiPSC lines can be induced into mesenchymal lineages and then used to engineer large bone grafts of defined geometries (9). To test this hypothesis, we induced three hiPSC lines derived from different source tissues and using different reprogramming methods into the mesenchymal lineage, and explored bone tissue formation in the osteoconductive scaffold–perfusion bioreactor culture model. Throughout the process of cell differentiation and bone formation, we studied the molecular changes in the cells, and assessed their phenotypic stability in a subcutaneous implantation model.
We demonstrate that mature, compact, phenotypically stable bone substitutes can be engineered using hiPSCs derived from different tissues using nonintegrating reprogramming vectors. A combination of differentiation assays, surface antigen profiling, and microarray expression signatures suggested that differences in hiPSC–mesenchymal progenitor differentiation potential could be identified before bone cultivation. Our protocols could be adapted to the large-scale construction of patient-specific bone grafts for personalized applications, and used as a controlled biomimetic model to study bone biology and test drugs using select pools of iPSC lines from patients and healthy individuals.
Results and Discussion
Mesenchymal Differentiation Patterns of hiPSCs.
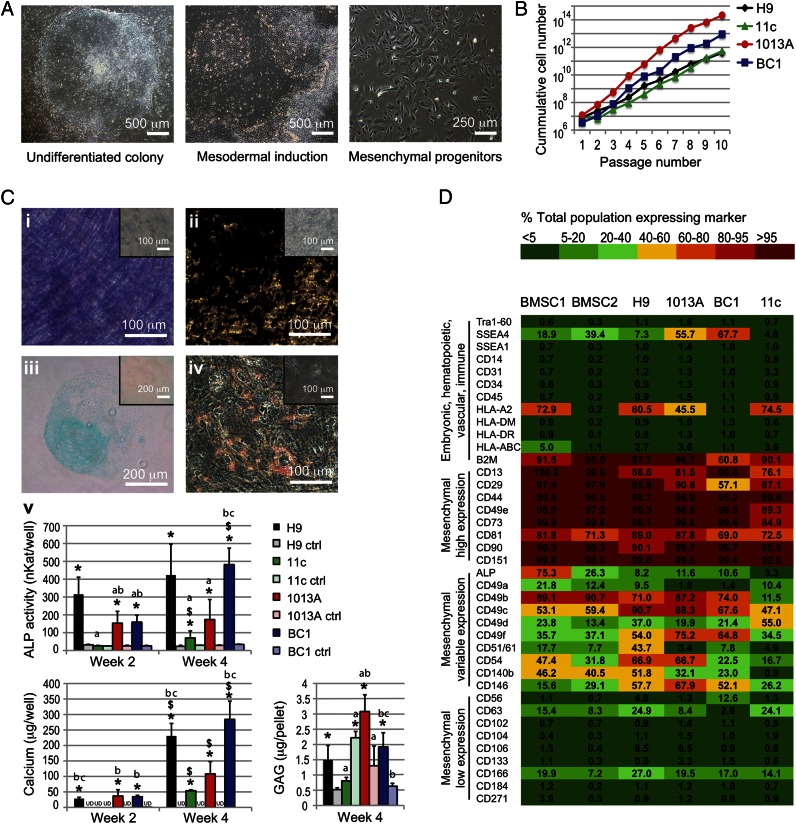
Recent reports have demonstrated feasibility of inducing hiPSCs into the mesenchymal lineage in vitro and in vivo (15, 16). We used our previously developed protocol (9) to derive mesenchymal progenitors from three hiPSC lines that were generated from dermal fibroblasts (11c and 1013A) and bone marrow cells (BC1) using retroviral vectors (11c) (17), Sendai virus (1013A) (18), and episomal vectors (BC1) (12, 19) (Fig. S1A). Mesenchymal progenitors derived from the hESC line H9 were used as controls (9). Cell pluripotency and normal karyotype were confirmed before mesenchymal induction (Fig. S1 B and C). Both hESC- and hiPSC-derived progenitors exhibited a typical fibroblastic-like morphology (Fig. 1A), high proliferation rate (Fig. 1B), and potential to differentiate toward the osteogenic (Fig. 1C; Fig. S2 A, i and ii and B, i), chondrogenic (Fig. 1C; Fig. S2 A, iii), and adipogenic lineages (Fig. 1C; Fig. S2 A, iv and B, ii). Under differentiation conditions, H9-, 1013A-, and BC1-derived progenitors displayed significantly higher alkaline phosphatase (ALP) gene expression, ALP activity, and calcium deposition, as well as glycosaminoglycans synthesis and lipid accumulation than controls at the same time points (Fig. 1 C, v; Fig. S2 A and B). In contrast, 11c-derived progenitors displayed poor osteogenic and adipogenic differentiation, indicating the low potential of this line to differentiate toward the mesenchymal lineages (Fig. 1C; Fig. S2 A and B). Differences in mesenchymal differentiation potential could result from different source tissue used for reprogramming (20), as well as different genetic background of the donor cells (13). Cytogenetic analysis confirmed normal karyotypes for hESC- and hiPSC-derived progenitors following extended in vitro expansion (Fig. S2C), an important consideration for potential clinical application of pluripotent stem cell-derived progenitors.
Fig. 1.
Derivation and characterization of mesenchymal progenitors from hiPSC and hESC lines. (A) Undifferentiated cell lines (Left) were exposed to mesoderm-inducing medium for 1 wk (Center), and the adherent cells were expanded in monolayer culture until they became homogenous for fibroblastic-like morphology (Right). Representative examples shown for line BC1. (B) The derived mesenchymal progenitors exhibited continuous growth over 10 subsequent passages, and (C) variable potential for differentiation into the osteogenic (i, ii, and v), chondrogenic (iii and v), and adipogenic lineages (iv). Examples shown for line BC1; additional results are presented in Fig. S2. (C, i) ALP staining (purple); (C, ii) von Kossa staining of deposited calcium (black); (C, iii) Alcian blue staining of glycosaminoglycans (GAG); (C, iv) Oil Red O staining of lipid vacuoles. (Insets) Cultures in control medium; (C, v) Biochemical determination of ALP activity and calcium deposited in monolayer cultures, and GAG deposited in pellet cultures during the 4-wk culture. Ctrl, control medium; UD, undetected. Data represent averages ± SD (n = 3–5; P < 0.05). Asterisks denote significant difference between differentiation and control media at the same time point; $, significant difference between week 2 and week 4; a, difference to H9; b, difference to 11c; c, difference to 1013A. (D) Flow cytometry characterization of surface antigen expression profiles of hESC- and hiPSC-derived progenitors and BMSCs. Expression patterns are presented as heat maps of the percentage of cells in the total population expressing the marker (see color legend).
Surface Antigens and Molecular Profiling of hiPSC-Derived Progenitors.
We next investigated the expression of a comprehensive list of surface antigens expressed on mesenchymal and nonmesenchymal lineages to evaluate whether specific expression profiles were associated with the observed differentiation potential. Our results suggest that hiPSC-derived progenitors displayed a similar surface antigen profile to that of bone marrow-derived mesenchymal stem cells (BMSCs) and H9-derived progenitors (Fig. 1D). We did not find markers of pluripotency (TRA-1-60), early differentiation (SSEA1), hematopoietic (CD34), endothelial (CD31), immune (CD14, CD45), and neuroectodermal (CD271) lineages, or the HLAs class I (HLA-ABC) and II (HLA-DR and HLA-DM; Fig. 1D). A group of markers commonly associated with mesenchymal stem cells was highly expressed in all lines. However, differences in the mesenchymal differentiation potential of the three hiPSC lines corresponded to subtle differences in the expression of some antigens, including CD13, CD29, CD49e, CD73, ALP, and CD49b (Fig. 1D), with the 11c line exhibiting minimal differentiation (Fig. 1C; Fig. S2 A and B) and lower antigen expression (Fig. 1D).
Interestingly, progenitors from other lines expressed a variable level of SSEA-4, found in pluripotent stem cells, as well as multipotent subpopulations of human MSCs isolated from bone marrow and other tissues (21). We therefore analyzed the coexpression of SSEA-4 with CD73, a marker commonly found in mesenchymal progenitors from pluripotent cell lines (4, 9, 15) and with other expressed antigens (Fig. S3), to identify distinct subsets of cells coexpressing specific markers. These results indicated a high expression of mesenchymal markers in SSEA4+/CD73+ and SSEA4−/CD73+ cell subpopulations, indicating the existence of SSEA4+ subpopulation in the mesenchymal population distinct from the SSEA4+/CD73− subpopulation that was negligible and displayed a reduced coexpression of most investigated markers. It remains to be determined whether the SSEA4+/CD73+ and SSEA4−/CD73+ subpopulations exhibit different functional differentiation. Phenotypic differences in surface markers and gene profiling are recognized to influence the regenerative properties of different BMSC subpopulations (22–24), and could explain the differences observed with the hiPSC-derived progenitors used in this study.
hiPSC-Derived Mesenchymal Progenitors Form Dense Bone-Like Tissue Matrix in Perfusion Culture on Osteoconductive Scaffolds.
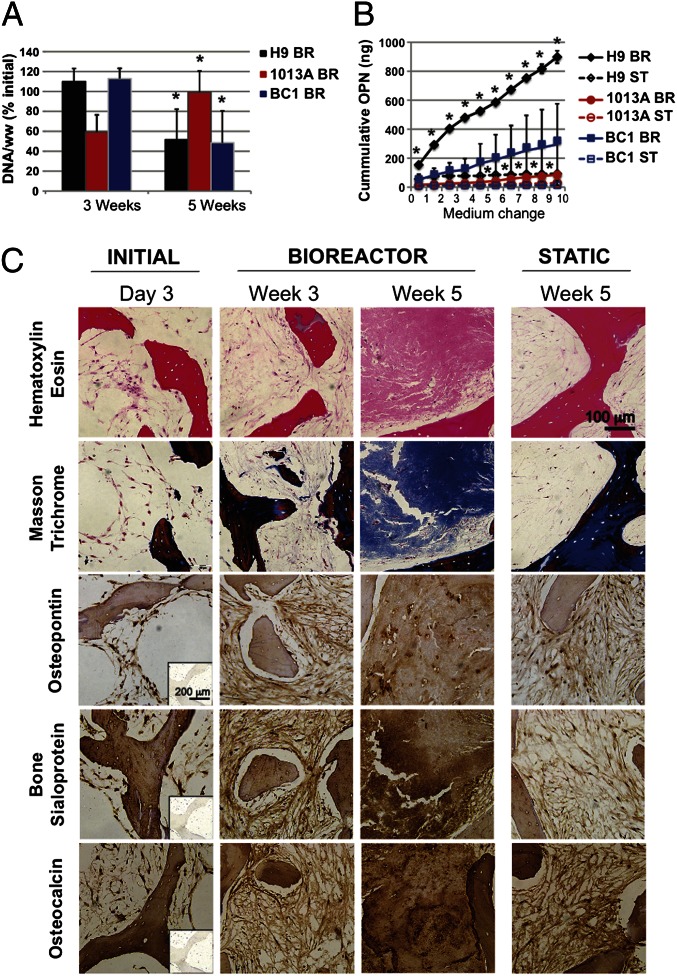
We previously reported that perfusion culture is critical for engineering large compact bone grafts from adult and hESC-derived mesenchymal progenitors (2, 9, 25). In the present study we extended our osteoconductive scaffold–perfusion bioreactor culture model to test the bone-forming potential of the mesenchymal progenitors derived from the hiPSC lines BC1 and 1013A, and hESC line H9, which exhibited excellent osteogenic potential in monolayer assays (Fig. 1; Fig. S2). We followed the same construct assembly and perfusion culture protocol as previously (2, 9), and found increased cellularity and higher cell viability in perfusion bioreactors compared with static controls (Fig. S4). These data supported the vital role of interstitial flow for cell survival. DNA analysis suggested different dynamics of cell growth in perfused culture between the three tested cell lines (Fig. 2A), with H9- and BC1-derived progenitors exhibiting a decrease in DNA content between weeks 3 and 5 of culture, and 1013A-derived progenitors exhibiting an increase in DNA content (similar to our previous study) (9). Differences in the internal architecture of the native bone scaffolds and subtle differences in cell seeding efficiency could affect the cell growth patterns, as observed previously (26). Alternatively, differences in cell growth could reflect dissimilar developmental dynamics of osteogenic progenitors derived from different hiPSC lines, in relation to their progression through stages of proliferation, differentiation, and apoptosis during tissue formation (27).
Fig. 2.
Perfusion bioreactor culture supported bone matrix deposition by hiPSC–mesenchymal progenitors. (A) DNA content per wet weight (ww) of tissue constructs was expressed as percent initial value and found to significantly increase from 3 to 5 wk in the 1013A constructs, in contrast to a significant decrease found in constructs of H9 and BC1 (P < 0.05; asterisk denotes significant difference from week 3). (B) Cumulative osteopontin (OPN) release into culture medium was significantly higher when H9 and 1013A where cultured in perfusion bioreactors compared with static conditions. No significant differences were found for line BC1 (P < 0.05; asterisk denotes significant difference from static culture). (C) Histological analyses of seeded (day 3) and cultured (weeks 3 and 5) constructs (examples shown for line BC1, additional results are presented in Figs. S5 and S6) showed an increase in tissue formation over time, and a denser tissue deposition in bioreactors compared with the static cultures after 5 wk of culture (H&E; Top). Dense bone matrix protein deposition was observed in perfusion bioreactors: collagen (Masson trichrome, blue; upper portion of Upper Middle), osteopontin (brown; Middle), bone sialoprotein (brown; Lower Middle), and osteocalcin (brown; Bottom). Minimal staining was observed in constructs from static cultures. (Insets) Negative staining controls.
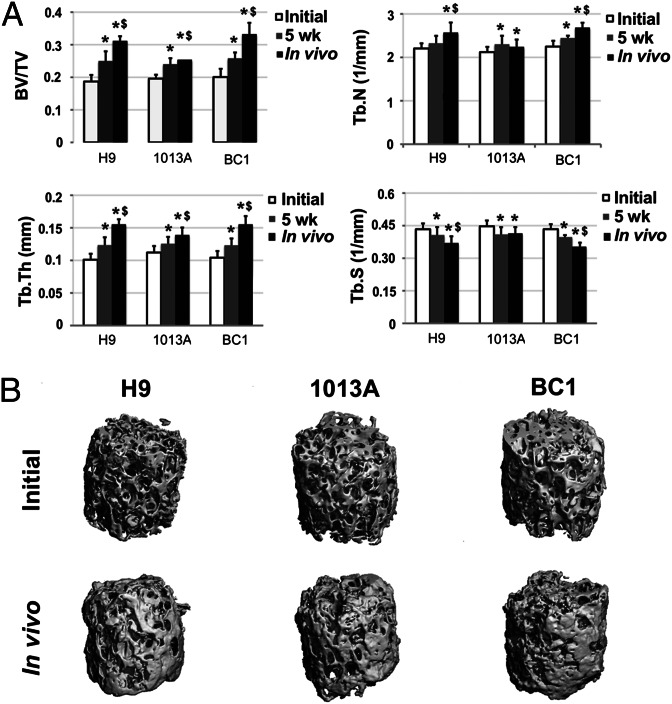
Perfusion culture resulted in increased osteopontin release compared with static culture (Fig. 2B) and a more homogenous deposition of bone matrix, characterized by the presence of collagen, osteopontin, bone sialoprotein, and osteocalcin (Fig. 2C; Figs. S5 and S6) in all three cell lines. BC1-engineered bone constructs displayed regions with the highest density of cells and new bone-like matrix, approaching the density of native tissue (Fig. 2C). Microcomputed tomography (µCT) analysis confirmed maturation of the newly formed tissue during bioreactor culture (Fig. 3), exhibiting significant increases in mineralized tissue portion, trabecular number, and trabecular thickness, along with significant decreases in trabecular spacing. Tissue development, mineral content, and bone structural parameters were similar for constructs engineered using hiPSCs, hESCs, and BMSCs in the current and previous study (9).
Fig. 3.
Mineralization of engineered bone constructs. (A) μCT analyses of cultured bone constructs showed an increase in mineralized tissue deposition during the 5-wk bioreactor culture and 12-wk s.c. implantation-investigated cell lines. Bone volume fraction (BV/TV), trabecular number (Tb.N), and trabecular thickness (Tb.Th) increased significantly in all investigated lines during bioreactor culture, in contrast to trabecular spacing (Tb.Sp), which decreased significantly, indicating bone maturation. Further tissue mineralization was noted in the lines BC1 and H9 during 12-wk s.c. implantation. Data represent averages ± SD (n = 6, P < 0.05; asterisks denote significant difference to initial values; $, significant difference between 5 wk and in vivo). (B) Reconstructed 3D μCT images of the tissue engineered bone constructs from H9-, 1013A-, and BC1-derived progenitors before culture in bioreactors and following 12-wk s.c. implantation in immunocompromised mice showed formation of mineralized tissue.
Maturation of hiPSC-Derived Mesenchymal Progenitors During Bone Tissue Development in Vitro.
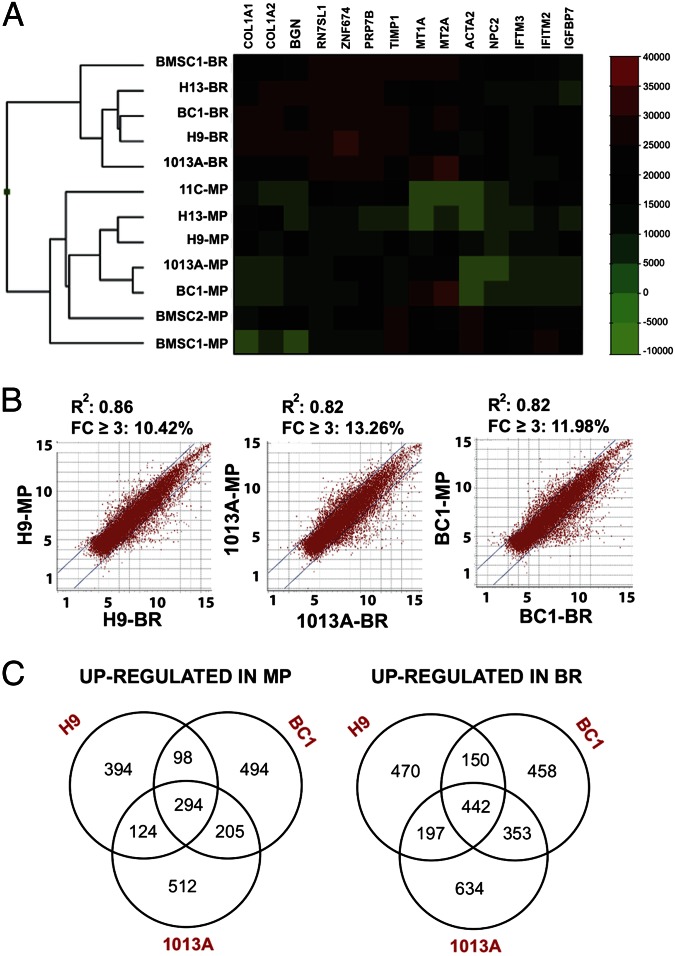
Our data demonstrate the positive effects of perfusion bioreactor culture on bone tissue development from hiPSC- and hESC-derived progenitors. However, the molecular changes occurring in the hiPSC- and hESC-derived progenitors during osteogenic differentiation in perfusion bioreactors have not been explored. We therefore assessed the relationship between the hiPSC- and hESC-derived progenitors and BMSCs from the current and previous study (9), and analyzed the differences in global gene expression profiles before and after bioreactor culture. Hierarchical clustering of hiPSC- and hESC-derived progenitors and BMSCs resulted in two main clusters (Fig. 4A), where hiPSC- and hESC-derived progenitors clustered together with BMSCs before and after culture in bioreactors, indicating that the mesenchymal progenitors display biological similarities with BMSCs, and undergo similar molecular changes during perfusion culture. Among the hiPSC-derived progenitors, 11c-derived progenitors clustered away from the other cell lines, whereas the two hiPSC- and the two hESC-derived progenitors clustered together, and the two BMSC lines clustered away from this group, corroborating the differences observed in proliferation ability, surface antigens profiling, and differentiation potential (Figs. 1 and 4A; Fig. S2) (9).
Fig. 4.
Global gene expression profiles of mesenchymal progenitors and BMSCs before and after bioreactor culture. (A) Hierarchical clustering of BMSCs and hESC- and hiPSC-derived progenitors before and after culture in bioreactors. The dendrogram shows two main clusters of BMSCs, hESC-derived mesenchymal progenitors (H13 and H9) and hiPSC-derived mesenchymal progenitors (11c, 1013A, and BC1) before and after culture in bioreactors. (B) Scatter plot analysis of microarray data exhibited similar transcriptional differences for H9-, 1013A-, and BC1-derived mesenchymal progenitors before and after culture in perfusion bioreactors. Genes within the lines (10–14% of the entire gene set) exhibited expression fold change ≤±3. (C) Venn diagrams showing the relationships between the genes up-regulated and down-regulated with a fold change ≥±3 during bioreactor culture of H9, 1013A, and BC1 mesenchymal progenitors. BR, bioreactor; MP, mesenchymal progenitor.
Scatter plot analysis of microarray data revealed a similar extent of transcriptional changes in H9-, 1013A-, and BC1-derived progenitors during bioreactor culture (Fig. 4B), with a comparable proportion of the genes being down-regulated or up-regulated with a fold change (FC) of ≥3. Many differentially expressed genes were shared between the three lines, indicating a similar molecular response to culture conditions in bioreactors (Fig. 4C). Genes commonly down-regulated in all three lines during bioreactor culture encoded for proteins involved in cell cycle, DNA replication, spindle assembly, and mitotic division, as well as proteins associated with carcinogenesis, including CDC, POL, GINS, MCM, and CENP genes and associated factors (Fig. S7A). The high degree of interconnectivity among these genes suggested a strong repression of proliferation, and could indicate the occurrence of a proliferation/differentiation switch associated with the progression of osteogenesis and tissue maturation. It will be important to elucidate the regulatory machinery controlling bone development in bioreactor culture—e.g., the composition of specific activator protein-1 dimers, known regulators of bone development and homeostasis (28).
Functional classification of genes up-regulated during bioreactor culture revealed increased expression of genes involved in extracellular matrix synthesis and remodeling; osteoblast differentiation; and bone formation, including the hub genes MMP2, STAT3, and TGFB3 (Tables S1–S3 and Fig. S7B). Strong induction toward the osteoblastic phenotype was confirmed by PCR verification of microarray data (Fig. S7C). Taken together, our work provides comprehensive analysis of the hiPSC-derived progenitor transcriptome after bioreactor cultivation, and suggests a functional maturation of the osteogenic lineage with common molecular switches in different cell lines. It will be critical to evaluate a larger number of mesenchymal progenitors derived from different hiPSC lines to confirm the common molecular and functional properties before and upon bioreactor cultivation.
Stability of hiPSC-Engineered Bone Tissue After 12 wk in Vivo.
Transplantation of undifferentiated pluripotent stem cells can lead to the formation of teratomas, limiting their use for clinical applications (29). In a recent report, differentiation of pluripotent stem cells toward bone-forming cells was achieved without formation of teratomas in orthotopic defects, by implantation on bone morphogenetic protein-2–releasing scaffolds, underlining the importance of osteoinductive environment (16). We previously demonstrated that mesenchymal induction and osteogenic differentiation in perfusion bioreactors were sufficient to prevent teratoma formation in engineered bone during 8 wk of s.c. implantation (9). In the current study, we tested whether the same culture protocol yields stable bone-like grafts using hiPSC lines reprogrammed with nonintegrating vectors.
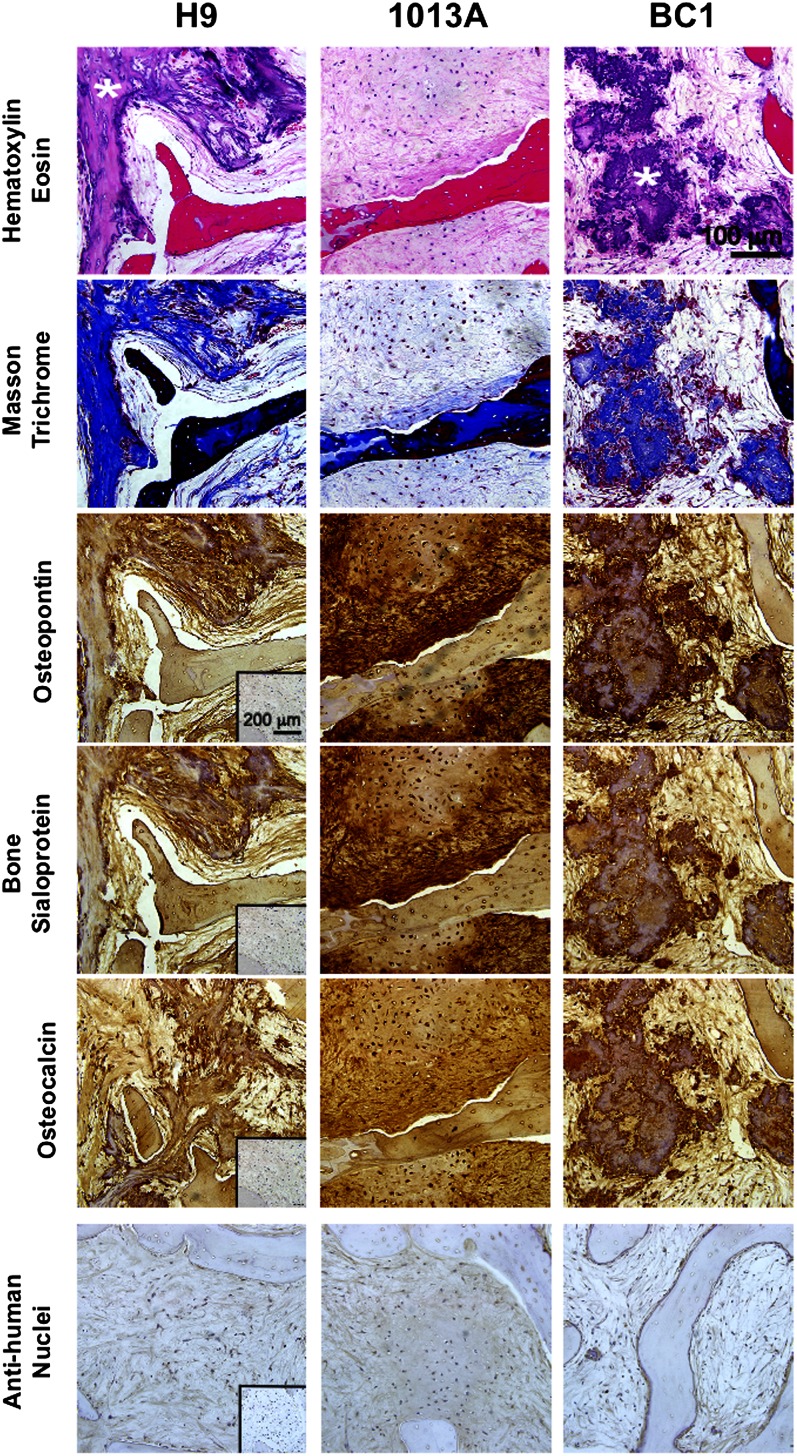
Lack of teratoma formation of hESC-derived progenitors is presumably associated with down-regulation of genes involved in pluripotency, stemness, and cell proliferation, and increased expression of lineage-specific genes (30). Our microarray analysis revealed the occurrence of a proliferation/differentiation switch during bioreactor culture, providing the molecular evidence for strong lineage commitment and suppression of the teratoma-forming ability. Indeed, stable phenotype was confirmed upon 12 wk of s.c. implantation into immunodeficient mice. Explanted hESC- and hiPSC-engineered bone constructs revealed a mature, dense, bone-like tissue that did not undergo differentiation into other lineages but was characterized by the presence of the bone matrix protein collagen, osteopontin, bone sialoprotein and osteocalcin, and cells of human origin (Fig. 5; Fig. S8A). Engineered constructs were surrounded by a loose connective tissue capsule (Fig. S8A) and displayed microvasculature ingrowth across the entire construct thickness (Fig. S8 B and C). We found recruitment of osteoclastic cells at the construct edges as previously (Fig. S8B) (9), suggesting an initiation of scaffold resorption and tissue remodeling, events recognized to promote new matrix deposition and bone formation (31). In addition, bone constructs engineered using H9- and BC1-derived progenitors displayed areas of dystrophic calcification, opposite to constructs of 1013A-derived progenitors that displayed uniform bone-like tissue with matrix-embedded cells. The different patterns of bone tissue maturation in vivo possibly reflect different regenerative properties of the investigated lines, in accord with the observed differences in cell growth (Fig. 2) discussed previously.
Fig. 5.
Stability of engineered bone constructs in vivo. Histological analysis indicated stability of mature bone matrix in H9-, 1013A-, and BC1-engineered bone constructs after 12 wk of s.c. implantation in immunodeficient mice. Thick connective tissue-like matrix devoid of other tissue types was observed in all cell lines investigated (H&E; Top), staining positively for collagen (Masson trichrome, blue; Upper Middle), osteopontin (brown; Middle), bone sialoprotein (brown; Lower Middle), and osteocalcin (brown; Bottom). Anti-human nuclei staining (purple) confirming the human origin of engineered bone tissue for all lines investigated. (Insets) Negative staining controls. In addition, H9- and BC1-engineered bone constructs displayed dispersed regions of dystrophic ossification (asterisks; Top Row).
The µCT examination of explanted constructs demonstrated significant increase in mineralized tissue portion and structural parameters compared with the constructs at the time of implantation (Fig. 3), evidencing continued maturation of hiPSC-engineered bone and indicating a potential for bone defect regeneration. Bone constructs engineered using H9- and BC1-derived progenitors displayed higher mineral content compared with progenitors derived from 1013A, consistent with the presence of regions of dystrophic calcification (Fig. 5; Fig. S8A). It remains to be evaluated how the differences observed in bone tissue development from mesenchymal progenitors derived from different hiPSC lines affect their potential for repairing skeletal defects.
In conclusion, our results demonstrate a potential for osteogenic commitment of hiPSC-derived progenitors and phenotypic stability of engineered bone constructs in vivo, indicating that the osteoconductive scaffold–perfusion bioreactor culture model can be used to engineer customized patient-specific substitutes for reconstructive therapies of bone defects. Further investigation in orthotopic implantation models is required to validate the functionality and safety of bone grafts engineered with hiPSC lines reprogrammed using nonintegrating vectors.
Methods
Detailed experimental methods are provided in SI Methods.
Cell Culture.
Three hiPSC lines (11c, 1013A, and BC1) (12, 17–19) were expanded and induced into mesenchymal lineage as in our previous studies (9). The hESC lines H9 and H13 (WiCell Research Institute) served as controls in the experiments. Expression of surface antigens was determined by flow cytometry and compared with BMSCs of the same batches as used in previous studies (9). In vitro differentiation potential was evaluated in monolayers and pellet cultures as previously (9).
Engineering Bone in Perfusion Bioreactors.
Decellularized bone cylinders were seeded with hiPSC- and hESC-derived progenitors at passage 5, cultured in perfusion bioreactors, and analyzed as in our previous studies (9).
Microarray Analyses of Maturing Mesenchymal Progenitors.
Global gene expression profiles were evaluated for hiPSC- and hESC-derived progenitors before and after bioreactor cultivation. For comparison, BMSCs and progenitors derived from the hESC line H13 used in our previous work were included (9). Samples were processed for RNA extraction, amplified, labeled, and analyzed using the Illumina HumanHT-12 v4 BeadChip (for details, see SI Methods).
Implantation.
Phenotype stability of perfused bone constructs was assessed after 12 wk of s.c. implantation in immunodeficient (SCID-beige) mice. All animal experiments were approved by the Columbia University Institutional Animal Care and Use Committee animal protocol.
Statistical Analyses.
Significant differences were evaluated using Student’s paired and unpaired t tests, ANOVA, and one-way repeated-measures ANOVA using SigmaPlot 12.3 software. Pairwise multiple comparisons were performed using the Holm–Sidak test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Haiqing Hua and Dieter Egli for hiPSC lines and for help with animal studies; Kevin Eggan and Linzhao Cheng for hiPSC lines; and Edward X. Guo for the use of μCT. Funding was provided by the New York Stem Cell Foundation-Helmsley Investigator Award (to D.M.); the Leona M. and Harry B. Helmsley Charitable Trust; Robin Chemers Neustein; Goldman Sachs Gives, at the recommendation of Alan and Deborah Cohen; New York State Stem Cell Science Shared Facility Grant C024179; National Institutes of Health Grants DE016525 and EB002520 (to G.V.-N.); and the New York Stem Cell Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE46315).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301190110/-/DCSupplemental.
References
- 1.Meijer GJ, de Bruijn JD, Koole R, van Blitterswijk CA. Cell-based bone tissue engineering. PLoS Med. 2007;4(2):e9. doi: 10.1371/journal.pmed.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayson WL, et al. Engineering anatomically shaped human bone grafts. Proc Natl Acad Sci USA. 2010;107(8):3299–3304. doi: 10.1073/pnas.0905439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhumiratana S, Eton RE, Wan LQ, Vunjak-Novakovic G. Physiologically stiff cartilage with integration to subchondral bone engineered in vitro from hBMSCs. ORS Transactions. 2012;36:0172. [Google Scholar]
- 4.Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med. 2005;2(6):e161. doi: 10.1371/journal.pmed.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James D, et al. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat Biotechnol. 2010;28(2):161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sottile V, Thomson A, McWhir J. In vitro osteogenic differentiation of human ES cells. Cloning Stem Cells. 2003;5(2):149–155. doi: 10.1089/153623003322234759. [DOI] [PubMed] [Google Scholar]
- 7.de Peppo GM, et al. Osteogenic potential of human mesenchymal stem cells and human embryonic stem cell-derived mesodermal progenitors: A tissue engineering perspective. Tissue Eng Part A. 2010;16(11):3413–3426. doi: 10.1089/ten.TEA.2010.0052. [DOI] [PubMed] [Google Scholar]
- 8.Kuznetsov SA, Cherman N, Robey PG. In vivo bone formation by progeny of human embryonic stem cells. Stem Cells Dev. 2011;20(2):269–287. doi: 10.1089/scd.2009.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marolt D, et al. Engineering bone tissue from human embryonic stem cells. Proc Natl Acad Sci USA. 2012;109(22):8705–8709. doi: 10.1073/pnas.1201830109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swijnenburg RJ, et al. Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc Natl Acad Sci USA. 2008;105(35):12991–12996. doi: 10.1073/pnas.0805802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou BK, et al. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21(3):518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bock C, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26(3):313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 15.Villa-Diaz LG, et al. Derivation of mesenchymal stem cells from human induced pluripotent stem cells cultured on synthetic substrates. Stem Cells. 2012;30(6):1174–1181. doi: 10.1002/stem.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levi B, et al. In vivo directed differentiation of pluripotent stem cells for skeletal regeneration. Proc Natl Acad Sci USA. 2012;109(50):20379–20384. doi: 10.1073/pnas.1218052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulting GL, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(3):279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua H, et al. iPS derived beta cells model diabetes due to glucokinase deficiency. J Clin Invest. 2013 doi: 10.1172/JCI67638. 10.1172/JCI67638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Dowey SN, Huang X, Chou BK, Ye Z, Cheng L. Generation of integration-free human induced pluripotent stem cells from postnatal blood mononuclear cells by plasmid vector expression. Nat Protoc. 2012;7(11):2013–2021. doi: 10.1038/nprot.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(12):1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riekstina U, et al. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009;5(4):378–386. doi: 10.1007/s12015-009-9094-9. [DOI] [PubMed] [Google Scholar]
- 22.Tormin A, et al. Characterization of bone marrow-derived mesenchymal stromal cells (MSC) based on gene expression profiling of functionally defined MSC subsets. Cytotherapy. 2009;11(2):114–128. doi: 10.1080/14653240802716590. [DOI] [PubMed] [Google Scholar]
- 23.Rada T, et al. Osteogenic differentiation of two distinct subpopulations of human adipose-derived stem cells: an in vitro and in vivo study. J Tissue Eng Regen Med. 2012;6(1):1–11. doi: 10.1002/term.388. [DOI] [PubMed] [Google Scholar]
- 24.Leyva M, et al. Characterization of mesenchymal stem cell subpopulations from human amniotic membrane with dissimilar osteoblastic potential. Stem Cells Dev. 2013;22(8):1275–1287. doi: 10.1089/scd.2012.0359. [DOI] [PubMed] [Google Scholar]
- 25.Grayson WL, et al. Optimizing the medium perfusion rate in bone tissue engineering bioreactors. Biotechnol Bioeng. 2011;108(5):1159–1170. doi: 10.1002/bit.23024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcos-Campos I, et al. Bone scaffold architecture modulates the development of mineralized bone matrix by human embryonic stem cells. Biomaterials. 2012;33(33):8329–8342. doi: 10.1016/j.biomaterials.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch MP, Capparelli C, Stein JL, Stein GS, Lian JB. Apoptosis during bone-like tissue development in vitro. J Cell Biochem. 1998;68(1):31–49. [PubMed] [Google Scholar]
- 28.Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis. 2002;61(2, Suppl 2):ii40–ii42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong CY, Gauthaman K, Bongso A. Teratomas from pluripotent stem cells: A clinical hurdle. J Cell Biochem. 2010;111(4):769–781. doi: 10.1002/jcb.22775. [DOI] [PubMed] [Google Scholar]
- 30.de Peppo GM, et al. Human embryonic mesodermal progenitors highly resemble human mesenchymal stem cells and display high potential for tissue engineering applications. Tissue Eng Part A. 2010;16(7):2161–2182. doi: 10.1089/ten.TEA.2009.0629. [DOI] [PubMed] [Google Scholar]
- 31.Pederson L, Ruan M, Westendorf JJ, Khosla S, Oursler MJ. Regulation of bone formation by osteoclasts involves Wnt/BMP signaling and the chemokine sphingosine-1-phosphate. Proc Natl Acad Sci USA. 2008;105(52):20764–20769. doi: 10.1073/pnas.0805133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.