Abstract
The role of the mitotic phosphorylation of the amino (NH2) terminus of Centromere Protein A (CENP-A), the histone variant epigenetic centromeric marker, remains elusive. Here, we show that the NH2 terminus of human CENP-A is essential for mitotic progression and that localization of CENP-C, another key centromeric protein, requires only phosphorylation of the CENP-A NH2 terminus, and is independent of the CENP-A NH2 terminus length and amino acid sequence. Mitotic CENP-A nucleosomal complexes contain CENP-C and phosphobinding 14-3-3 proteins. In contrast, mitotic nucleosomal complexes carrying nonphosphorylatable CENP-A–S7A contained only low levels of CENP-C and no detectable 14-3-3 proteins. Direct interactions between the phosphorylated form of CENP-A and 14-3-3 proteins as well as between 14-3-3 proteins and CENP-C were demonstrated. Taken together, our results reveal that 14-3-3 proteins could act as specific mitotic “bridges,” linking phosphorylated CENP-A and CENP-C, which are necessary for the platform function of CENP-A centromeric chromatin in the assembly and maintenance of active kinetochores.
Histone variants are nonallelic isoforms of conventional histones. It is widely accepted that the incorporation of histone variants generally confers novel structural and functional properties to the nucleosome (1). Centromere Protein A (CENP-A) is a histone variant, which replaces the canonical histone H3 at the centromere (2) and marks epigenetically the centromeres and the kinetochores (for reviews see refs. 3 and 4). The presence of CENP-A is required for the assembly of active kinetochores and its depletion results in numerous mitotic problems, such as chromosome misalignments and segregation defects, generation of chromosome bridges, aneuploidy, etc. (5). The resulting mitotic defects, following CENP-A depletion, were associated also with notable alterations in the composition and organization of the kinetochore, including the delocalization of the inner kinetochore proteins CENP-C, CENP-I, and CENP-H as well as the outer kinetochore components Highly Expressed in Cancer protein 1 (HEC1), Mitotic Arrest Deficient 2-like protein (Mad2), and CENP-E (5).
During recent years, the studies of CENP-A were focused mainly on its histone-fold domain. The NH2 terminus of CENP-A, which is not required for centromeric targeting (6, 7) appeared, however, to play an important role in both mitosis and meiosis. In yeast, the NH2 tail of Chromosome Segregation Protein 4 (Cse4p) (the homolog of mammalian CENP-A) has an essential function distinct from that of the histone-fold domain in chromosome assembly and segregation (8). The reported data in Arabidopsis thaliana suggested the existence of a meiosis-specific loading pathway for CENP-A, requiring its NH2 terminus (9). In addition, human CENP-A is phosphorylated in its NH2 terminus at serine 7 in mitosis but the role of this phorphorylation is far from being clear (10, 11).
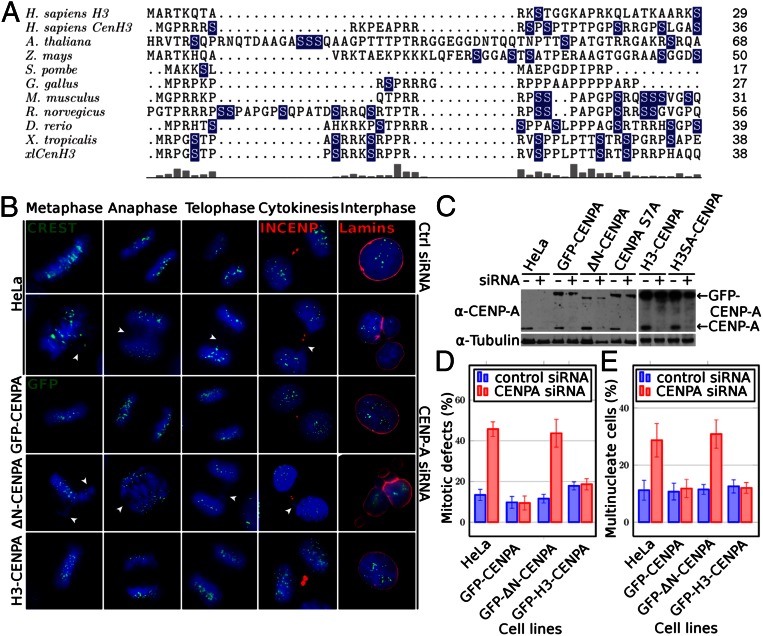
Sequence alignments of the NH2 termini of CENP-A from different species show very low sequence conservation in terms of amino acid composition, sequence, and length (Fig. 1A). However, expression of CENP-A with its NH2 terminus deleted is lethal in yeast (12) and plants (13). These data create a paradox because on one hand, the NH2 terminus of CENP-A, in certain organisms, is required for their survival and on the other hand, it appears to be nonessential because there is no evolutionary pressure to conserve at least some specific sequence elements.
Fig. 1.
The NH2 tail of CENP-A is required for mitosis and the H3 swapped NH2 tail CENP-A chimera rescues the CENP-A null cell phenotype. (A) Sequence alignment of CENP-A NH2 termini from different species. Names of species are indicated at Left. Serine residues are indicated in blue. Conservation between different species is shown (Lower). (B) Cell cycle visualization, after CENP-A suppression by siRNA treatment of naïve HeLa cells (second row) or HeLa cells stably expressing siRNA-resistant full-length GFP–CENP-A (third row) or tailless GFP–ΔN–CENP-A (fourth row) or GFP–H3–CENP-A swapped tail mutant (fifth row). First row shows naïve cells not treated with siRNA. A CREST antibody was used to visualize the centromeres in naïve cells; GFP fluorescence was used to visualize CENP-A in GFP fusion-expressing cells. An antibody against inner centromere protein (INCENP) and antilamina antibody were used to detect the midbody during cytokinesis and the nuclear envelope in interphase cells (shown in red). Blue, DNA; white arrowheads point to misaligned or lagging chromosomes and to chromatin bridges. (Scale bar, 5 µm.) (C) Detection of the GFP–CENP-A fusions and endogenous CENP-A in control (−) and siRNA treated (+) cells at 72 h posttransfection by Western blot. Cells used are indicated (Upper). Arrows indicate the positions of the GFP–CENP-A fusions or endogenous CENP-A. (D) Histograms showing the percentage of mitotic defects at 72 h posttransfection with siRNA against CENP-A in the indicated cell lines. HeLa, naïve cell line. (E) Same as D, but showing the percentage of multinucleate cells. For each experiment, at least 400 cells were counted. Data are means and SEM of five independent experiments.
Here, we have analyzed the mitotic function of the NH2 terminus of human CENP-A and its phosphorylation. We show the mere phosphorylation of CENP-A, but not its length and amino acid sequence, is required for the localization of CENP-C, a key mediator between centromeric chromatin and the outer kinetochore components. Our data reveal that in mitosis, the phosphorylated CENP-A nucleosomes are “bridged” to CENP-C via the phosphobinding 14-3-3 proteins. These 14-3-3 mitotic bridges are essential for the assembly of active kinetochores.
Results
CENP-A NH2 Terminus per Se, but Not Its Amino Acid Sequence Composition and Length, Is Essential for Mitotic Progression.
To address the paradox regarding the essential role of the NH2 tail of CENP-A in human cells, we asked first whether its deletion results in the generation of specific cell phenotypes. We have established stable HeLa cell lines that express siRNA-resistant forms of either GFP–CENP-A or CENP-A NH2-tail deletion mutant (GFP–ΔN–CENP-A) or GFP–H3–CENP-A (a swapped tail mutant with the tail of conventional H3 fused to the histone-fold domain of CENP-A). The expression of endogenous CENP-A was then suppressed by using siRNA treatment and the resulting cell phenotypes were observed and quantified by fluorescence microscopy. Importantly, all of the GFP–CENP-A fusions localize to the centromeres and the expression of any one of them in control cells, but not siRNA-treated cells, had no phenotypic effect, i.e., the cells behave identically to the naïve parental HeLa cells (Figs. 1B, 2A, and 3 and Fig. S1).
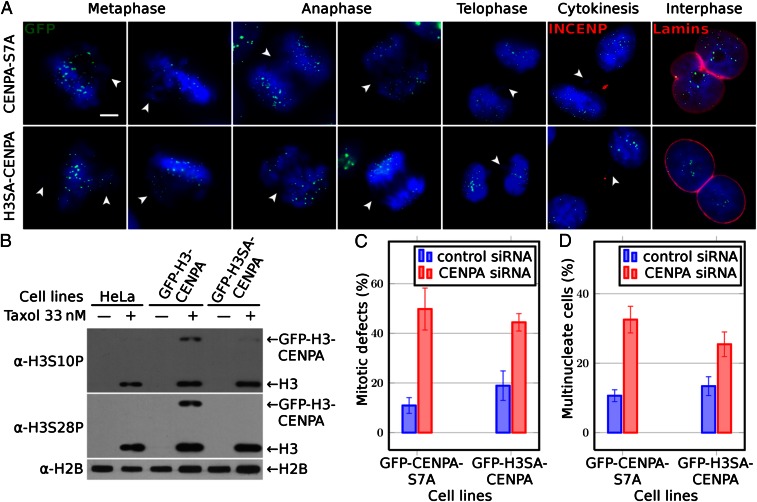
Fig. 2.
Both nonphosphorylatable NH2-tail mutants GFP–CENP-A–S7A and GFP–H3SA–CENP-A are unable to rescue the CENP-A knocked down cell phenotype. (A) Cell cycle microscopy visualization (after CENP-A suppression by siRNA treatment) of HeLa cells stably expressing siRNA-resistant either nonphosphorylatable NH2-tail GFP–CENP-A–S7A (first row) or GFP–H3SA–CENP-A (second row) mutants. GFP fluorescence was used for the visualization of CENP-A. An antibody against INCENP and antilamina antibody was used to detect the midbody during cytokinesis and the nuclear envelope in interphase cells, respectively. Blue, DNA; white arrowheads point to misaligned or lagging chromosomes and to chromatin bridges. (Scale bar, 5 µm.) (B) Phosphorylation status of both GFP–H3–CENP-A and GFP–H3SA–CENP-A fusions visualized by antibodies against histone H3 phosphorylated either at serine 10 or at serine 28; antihistone H2B antibody was used to show that equal amounts of proteins were loaded. Types of cells used are indicated (Upper). Arrows indicate the positions of the GFP fusions or histone H3. (C) Histograms showing the percentage of mitotic defects at 72 h posttransfection with siRNA against CENP-A in the indicated cell lines. (D) Same as C, but for the percentage of multinuclear cells. For each experiment at least 400 cells were counted. Data are means and SEM of five independent experiments.
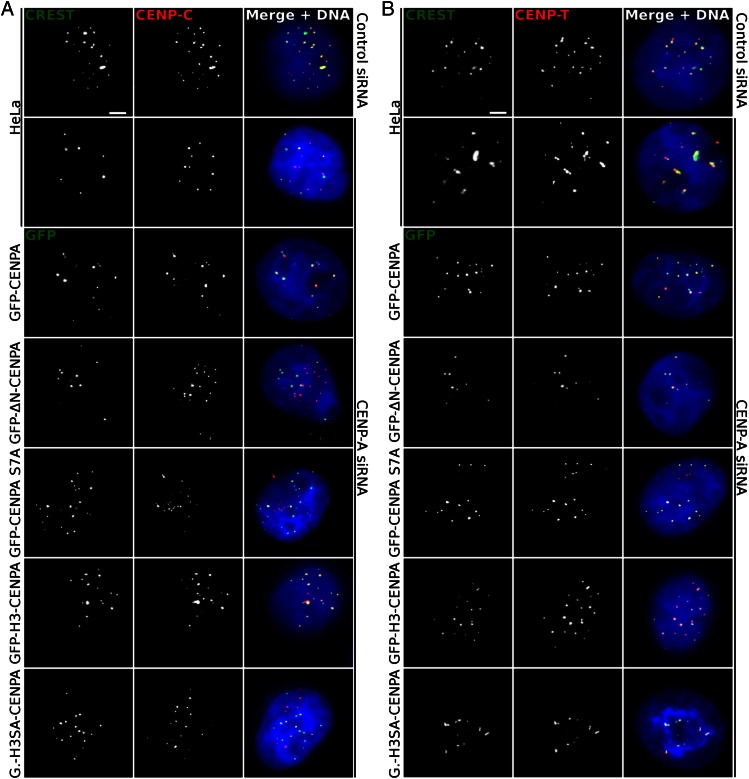
Fig. 3.
The absence of either the NH2 terminus of CENP-A or its phosphorylation affects the localization of CENP-C, but not that of CENP-T. (A) Localization of CENP-C in CENP-A–depleted cells stably expressing the indicated siRNA-resistant GFP–CENP-A fusions (rows 3–7). First two rows show the localization of CENP-C in naïve HeLa cells treated with control (first row) or CENP-A (second row) siRNA. Centromeres in naïve cells were stained with a CREST antibody. Blue, DNA. (Scale bar, 5 µm.) Note the complete delocalization of CENP-C from the centromeric chromatin. (B) Same as A, but for CENP-T.
SiRNA treatment led to very efficient suppression of CENP-A expression: more than 85–90% of the endogenous CENP-A was depleted in each of the cell lines used and, as expected, the expression of the siRNA-resistant GFP–CENP-A fusions was unaffected (Fig. 1C). In agreement with the reported data (5), the depletion of CENP-A in naïve HeLa cells has deleterious effects on mitosis and cytokinesis (Fig. 1B). Multiple mitotic and cytokinesis defects, such as chromosome misalignment, presence of lagging chromosomes, chromatin bridges, multiple nuclei in interphase, etc. were observed in cells depleted of CENP-A (Fig. 1B). The presence of stably expressed GFP–CENP-A in the siRNA treated HeLa cells was sufficient to completely rescue the severe mitotic (Fig. 1 B and D) and cytokinesis defects (Fig. 1 B and E). Importantly, no rescue was detected in CENP-A siRNA-treated HeLa cells, stably expressing the GFP–ΔN–CENP-A deletion mutant (Fig. 1 B, D, and E). These data illustrate the crucial role of the CENP-A NH2 tail in mitosis in human cells.
A complete rescue was found, however, in HeLa cells depleted of endogenous CENP-A, which stably express the swapped tail mutant GFP–H3–CENP-A (Fig. 1 B, D, and E). This result demonstrates that the NH2 tail of histone H3 can function identically to the NH2 tail of CENP-A in mitosis in human cells if it is targeted to the centromeric chromatin.
Phosphorylation of the NH2 Terminus of CENP-A Is Required for Its Mitotic and Cytokinetic Functions.
Taken together the data described above show that neither the amino acid sequence composition nor the length of the NH2 terminus of human CENP-A are essential for its mitotic and cytokinesis function. Examination of the alignments of the CENP-A NH2 termini from different species shows that they all contain potentially phosphorylatable serines (Fig. 1A). We hypothesized that serine phosphorylation of CENP-A is a general feature required for mitosis. To analyze this, we established HeLa cell lines stably expressing either a siRNA-resistant nonphosphorylatable GFP–CENP-A–S7A mutant (serine 7 was substituted with alanine) (10), or a siRNA-resistant nonphosphorylatable GFP–H3SA–CENP-A swapped-tail mutant (in which the two phosphorylatable serines in the H3 tail of the fusion mutant, S10 and S28, were substituted with alanines). These cell lines were treated with CENP-A siRNA to determine the capacity of the mutants to rescue the CENP-A depletion phenotypes (Fig. 2). When the stable cell lines were synchronized in mitosis with taxol, the GFP–H3–CENP-A fusion (which contains phosphorylatable serines in its NH2 terminus) was phosphorylated (Fig. 2B). In contrast, the GFP–H3SA–CENP-A mutant was, as expected, not phosphorylated (Fig. 2B). Importantly, neither GFP–CENP-A–S7A nor GFP–H3SA–CENP-A were able to rescue the severe cell phenotype due to the depletion of endogenous CENP-A (Fig. 2 A, C, and D). Thus, the absence of phosphorylation of the GFP fusions was responsible for their inability to fulfill their mitotic roles. We conclude that upon phosphorylation the tail of CENP-A acquires novel properties essential for mitotic progression.
One interesting property became apparent when one compares the phosphorylation levels of the GFP–H3–CENP-A chimera in relation to those of the endogenous H3 (Fig. 2B). The number of molecules of GFP–H3–CENP-A chimera in the transfected cells is extremely low compared with total wild-type H3 and yet the levels of phosphorylation at S10 and at S28 in the chimera appear to be comparable to those of H3. This suggests that the NH2 terminus of H3 is much more heavily phosphorylated when it is located in the centromeric chromatin as opposed to the bulk of the chromatin.
Phosphorylation of the CENP-A NH2 Terminus Determines the CENP-C Centromeric Localization.
CENP-C and CENP-T, two members of the Constitutive Centromere Associated Network (CCAN), form the chromatin-based platform required for vertebrate kinetochore assembly (4). In addition, depletion of CENP-A from DT40 chicken cells leads to mislocalization of inner kinetochore components including CENP-C (5).
Thus, based on the already published data we made the hypothesis that phosphorylation of the CENP-A NH2 tail might be required for CENP-C localization at centromeres. To test this, we first determined how expression of the GFP fusions affects the localization of CENP-C and CENP-T in our HeLa stable cell lines depleted of endogenous CENP-A. We found that depletion of CENP-A by siRNA resulted in a complete mislocalization of CENP-C in interphase cells; the CENP-C labeling did not colocalize with the calcinosis, Reynaud's syndrome, esophageal dismotility, sclerodactyly, telangiectasia (CREST) centromeric staining (Fig. 3A). Note that the delocalized noncentromeric CENP-C shows a punctuated pattern, a result in agreement with the reported data (5). Expression of the siRNA-resistant GFP–CENP-A rescued the colocalization of CENP-C with GFP–CENP-A (Fig. 3A). In contrast to the wild-type CENP-A, the expression of neither the NH2 tail CENP-A deleted mutant (GFP–ΔN–CENP-A) nor the other two nonphosphorylatable CENP-A mutants (GFP–CENP-A–S7A and GFP–H3SA–CENP-A) were able to rescue the mislocalization pattern of CENP-C. Indeed, in all three cases CENP-C does not colocalize with the centromeres (Fig. 3A). These data demonstrate that the CENP-A NH2 terminus and its phosphorylation are required for the centromere localization and function of CENP-C. The importance of the phosphorylation per se of the NH2 terminus was underlined even more by the observation that the phosphorylation of the H3 tail of the swapped tail mutant GFP–H3–CENP-A was necessary and sufficient for the rescue of the CENP-C localization and function.
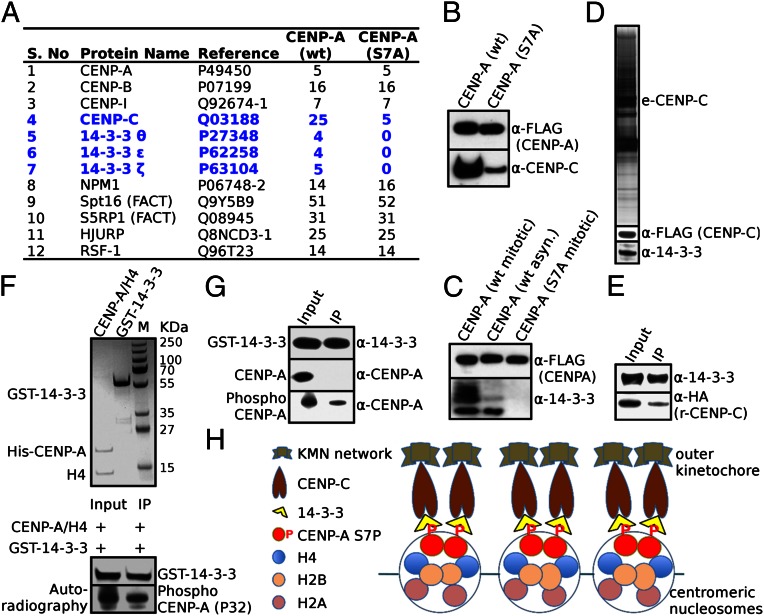
It should be noted that we have been able to study the localization of CENP-C in interphase cells only, because the antibody that was used to detect CENP-C failed to mark centromeres even in control untreated mitotic cells. However, CENP-A is phosphorylated in mitosis, and thus one should expect CENP-A phosphorylation to be essential for the CENP-C centromeric localization in mitosis. To test this, we have used biochemistry approaches and have quantitatively analyzed the composition of both CENP-A and the nonphosphorylatable CENP-A–S7A mitotic nucleosomal complexes. Briefly, we used mitotic stable HeLa cell lines expressing double HA-FLAG tagged CENP-A (e-CENP-A) or identically tagged CENP-A with serine 7 mutated to alanine (e-CENP-A–S7A) to isolate the respective CENP-A nucleosomal complexes (e-CENP-A.com). Mass spectrometry analysis showed, in agreement with the reported data (14), that the mitotic wild-type complex contained proteins from the constitutive centromere associated network (CENP-B, CENP-I, and CENP-C), the CENP-A histone chaperones Holliday Junction Recognition Protein (HJURP) and Nucleophosmin (NPM1), the chromatin remodeler Remodeling and Spacing Factor 1 (RSF-1), as well as several other proteins (Fig. 4A). Importantly, the number of CENP-C peptides identified by mass spectrometry decreased dramatically (from 25 to 5) in the mutated CENP-A–S7A mitotic nucleosomal complex compared with the wild-type one (Fig. 4A). In agreement with this, the Western blotting analysis revealed that the amount of CENP-C associated with the mutated CENP-A–S7A nucleosomal complex was very strongly decreased compared with wild-type CENP-A nucleosomal complex (Fig. 4B). We conclude that phosphorylation of CENP-A is also required for the centromeric localization of CENP-C in mitosis.
Fig. 4.
The 14-3-3 proteins act as bridges between phosphorylated CENP-A and CENP-C in mitosis. (A) The mitotic WT e-CENP-A nucleosomal complex, but not the mutated e-CENP-A–S7A nucleosomal complex, contains 14-3-3 proteins. Nucleosomal complexes containing HA-FLAG epitope-tagged wild-type CENP-A or CENP-A mutated at serine 7 were isolated by double immunoaffinity from either cycling or mitotic stable HeLa cells expressing either CENP-A or the CENP-A–S7A mutant. Proteins present in both the mitotic WT CENP-A and in the CENP-A–S7A mutant complexes together with the number of identified peptides are indicated. CENP-C and the 14-3-3 proteins are shown in blue. (B) Western blot analysis of CENP-C associated with CENP-A or CENP-A–S7A mitotic nucleosomal complexes. Identical amounts of the two complexes were analyzed by SDS/PAGE, blotted, and detected with anti–CENP-C antibody (Lower). The same blot was also probed with anti-FLAG antibody to detect CENP-A to show that equal amounts of complex had been loaded (Upper). (C) Western blot analysis of the 14-3-3 proteins associated with wild-type CENP-A nucleosomal complexes (isolated from either mitotic or nonsynchronized cells) or with mutated CENP-A–S7A nucleosomal complex (Lower). Pan-anti–14-3-3 antibodies were used. An anti-FLAG antibody was used to detect CENP-A to show that equal amounts of complex had been loaded (Upper). (D) The 14-3-3 proteins are members of the soluble CENP-C complex. HA-FLAG epitope-tagged CENP-C (e-CENP-C.com) soluble complex was isolated from a stable HeLa cell line expressing e-CENP-C. (Top) SDS/PAGE of the complex. (Middle) Western blot analysis of the complex using anti-FLAG antibody (for detection of e-CENP-C). (Bottom) Western blot analysis of the complex with pan-anti–14-3-3 proteins. (E) Pull-down experiments show that CENP-C binds to 14-3-3 proteins. Recombinant HA-tagged CENP-C isolated from insect cells was mixed with GST–14-3-3-ζ proteins. Glutathione agarose beads were used to pull down the GST–14-3-3-ζ proteins. CENP-C associated with the 14-3-3 proteins in the pull down was revealed by Western blotting using anti-HA antibody, whereas 14-3-3 were detected by pan-anti–14-3-3 antibodies. Input, 1/10 of the 14-3-3-ζ/HA-tagged CENP-C mixture used for the pull down. (F and G) The 14-3-3 proteins bind to phosphorylated, but not to unphosphorylated CENP-A. Purified to homogeneity (F, Upper) unphosphorylated or phosphorylated CENP-A–H4 tetramers were mixed with purified GST–14-3-3-ζ (F, Upper, and G) and incubated with glutathione agarose beads. The CENP-A attached to the beads (immunoprecipitated) was analyzed by autoradiography (F, Lower) or by Western blotting using anti–CENP-A antibodies (G). (H) Schematics describing how phosphorylated CENP-A and CENP-C act in concert with 14-3-3 proteins in the assembly and maintenance of active kinetochores. IP, immunoprecipitated.
As for CENP-T, the depletion of CENP-A does not affect its centromeric distribution pattern (Fig. 3B). This is in agreement with the reported data showing that CENP-T and CENP-C use distinct pathways for assisting the assembly of active kinetochores (15).
14-3-3 Proteins Act as Mitotic Bridges Required for the Assembly of the Phospho–CENP-A Nucleosome/CENP-C Complex.
How does the phosphorylated tail of CENP-A function in mitosis? One plausible hypothesis is that CENP-A phosphorylation may directly or indirectly, through some intermolecular bridges, stabilize the association of CENP-C with the centromere in mitosis. If such protein bridges exist, they should mediate a mitosis-specific interaction between CENP-A and CENP-C and thus would be associated with the CENP-A mitotic nucleosomal complex. And indeed, mass spectrometry analysis identified the phosphobinding proteins 14-3-3-θ, -ε, and -ζ associated with the e-CENP-A complex (Fig. 4A). Importantly, all three isotypes of 14-3-3 proteins were absent from the mutated mitotic e-CENP-A–S7A complex (Fig. 4A). These results were further confirmed by Western blot analysis (Fig. 4C). In addition, the Western blot analysis showed that the amount of 14-3-3 proteins in the complex isolated from asynchronous cells was much smaller than that present in the complex isolated from mitotic cells (Fig. 4C), further confirming the specific association of these proteins with mitotic centromeric CENP-A chromatin. Because 14-3-3 are phosphobinding proteins (16), this suggests that these proteins may act as intermolecular protein bridges that couple phosphorylated CENP-A with CENP-C in mitosis. If this was the case, CENP-C should also associate with 14-3-3 proteins in vivo. To address this, we have purified by immunochromatography the CENP-C complex (e-CENP-C) from HeLa cell lines stably expressing HA and FLAG double-tagged CENP-C (17) and, as expected, we found 14-3-3 proteins to be members of the e-CENP-C complex (Fig. 4D). This result is in agreement with a previous report showing that 14-3-3 proteins bind to the N-terminal part of Drosophila CENP-C in vivo (18).
The above described in vivo data for the bridging role of 14-3-3 proteins were further supported by a series of in vitro pull-down experiments using highly purified components (Fig. 4 E–G). The data reveal that recombinant purified from insect cells CENP-C binds directly to 14-3-3 proteins (Fig. 4E). A direct interaction was also observed between phosphorylated CENP-A and 14-3-3 proteins. The detected interaction was highly specific and phosphorylation dependent, because neither unphosphorylated CENP-A nor histone H4 were found to bind to 14-3-3 (Fig. 4 F and G). Taken as a whole, the data reveal that 14-3-3 proteins act as key mediator factors required for the assembly of the tripartite phospho–CENP-A/14-3-3/CENP-C complex, which is essential for mitotic progression.
Discussion
In this work we have studied the role of the CENP-A NH2 terminus and its phosphorylation in mitosis. We addressed this problem by using stable cell lines expressing siRNA-resistant either mutated CENP-A or CENP-A/H3 swapped tail chimeras. Importantly, the controlled expression of the different mutants has no effect on the stable HeLa cell lines, i.e., they behave identically to the parental naïve HeLa cells. This has allowed us to carry out the experiments in endogenous CENP-A depleted cells, a sine qua non condition for understanding the implication of the CENP-A NH2 terminus and its phosphorylation in mitosis.
We have first shown that the tail of CENP-A was required for mitosis in human cells, a result in agreement with the data in yeast (12). The same finding, i.e., the requirement of “some kind” of CENP-A NH2 terminus for proper mitosis proceeding, was also recently reported in plants (13). The data for both nonphosphorylatable GFP–CENP-A–S7A and GFP–H3SA–CENP-A fusions illustrate that the phosphorylation per se and not the amino acid composition and sequence of the CENP-A tail is required for mitosis. These results could explain why the NH2 termini of different species are completely divergent in length, amino acid composition, and sequence, but always contain serine residues (Fig. 1A). We speculate that during mitosis the NH2 tail of CENP-A is phosphorylated in the different species [probably at different site(s) along their tail] and that this phosphorylation is required for proper mitosis proceeding.
Earlier studies on the role of S7 phosphorylation of CENP-A by Aurora B resulted in contradictory results. In one study, the nonphosphorylatable mutant of CENP-A did not interfere with kinetochore formation, spindle assembly, or cell cycle progression, but a notable delay at the terminal stage of cytokinesis, probably at the abscission, was detected (10). In contrast, in a later study, expression of the same CENP-A–S7A mutant resulted in defective kinetochore function (11). However, an important caveat applying to both of these studies is the fact that the endogenous CENP-A was still present.
We further analyzed in detail the role of the CENP-A NH2 terminus phosphorylation in mitosis. Our in vivo data revealed that the centromeric localization of CENP-C was determined by the NH2 terminus phosphorylation of CENP-A. Immunoaffinity purification of the wild-type CENP-A and nonphosphorylatable CENP-A–S7A nucleosomal complexes together with a series of biochemistry experiments demonstrated the key role of the 14-3-3 proteins in maintaining the association of phosphorylated CENP-A nucleosomes with CENP-C and the assembly of a tripartite phosphorylated CENP-A nucleosomes/14-3-3 proteins/CENP-C complex in mitosis. The schematic scenario presented in Fig. 4H suggests a tentative mechanism for the assembly and function of this complex. The heavily phosphorylated population of CENP-A molecules acts as a sink to recruit 14-3-3 molecules to centromeres in the beginning of mitosis. Once recruited to centromeres, 14-3-3 proteins interact simultaneously with both CENP-C and the phosphorylated NH2 terminus of CENP-A and stabilize the already existing CENP-A nucleosome/CENP-C interaction (19). This allows CENP-C to be stably attached to the inner centromeres and to serve as a scaffold for the building of a functional kinetochore. This scenario implies a much higher level of CENP-A phosphorylation compared with that of histone H3 to recruit the 14-3-3 proteins specifically to the centromeres and not to bulk chromatin. This appears to be the case, because even though cells contain amounts of GFP–H3–CENP-A comparable to that of endogenous CENP-A (Fig. 1C), which is estimated to be several orders of magnitude smaller than the amount of conventional H3 (20), they nevertheless exhibited an NH2 terminus phosphorylation level comparable to that of endogenous H3 (Fig. 2B). Higher levels of phosphorylation may be achieved by the local activity of kinases, such as Aurora B, or by local inhibition of phosphatases (PP1s or PP2A), or both.
Materials and Methods
Plasmids.
The full-length human cDNA clones of CENP-A and CENP-C were purchased from Invitrogen. The complete coding sequence from each clone was subcloned into the XhoI–NotI sites of the pREV–HTF retroviral vector. CENP-A serine 7 mutant (S7A) was constructed by PCR-based mutagenesis and subcloned into pREV–HTF retroviral vector containing the Flag-HA. For protein expression in insect cells, the cDNA of CENP-C was cloned into pFastBac, which encodes HA tag at the N terminus. The human histones CENP-A/H4 were cloned in a homemade bicistronic pET28b vector as described previously (17). SiRNA-resistant GFP–CENP-A fusions were constructed from a CENP-A coding sequence from ref. 21 and cloned into a pBABE-puro vector (Addgene; 1764).
Cell Culture and Synchronization.
The cells were maintained in standard DMEM media containing 10% (vol/vol) FBS, 1% penicillin and streptomycin, and 1% glutamine at 37 °C in a 5% (vol/vol) CO2 atmosphere. For generation of stable HeLa cell lines expressing Flag-HA epitope-tagged CENP-A.wt, CENP-A-S7A, or CENP-C, the cells were transfected with calcium phosphate. The HeLa cell lines stably expressing e-CENP-A (WT and S7A) were synchronized in mitosis by treatment with thymidine–nocodazole. Cell lines stably expressing the various GFP–CENP-A fusions were established by retroviral infection with Moloney murine leukemia viruses (MMLVs) produced by amphotropic Phoenix packaging cells (22).
Tandem Affinity Purification.
The chromatin extracts were prepared from stable HeLa cell lines expressing either CENP-A, CENP-A–S7A, or CENP-C proteins fused to FLAG- and HA-epitope tags. The protein complexes were purified from asynchronous or mitotic cells by double immunoaffinity purification procedure with anti-Flag M2 antibody-conjugated agarose (Sigma), followed by anti-HA purification. Interacting partners of purified complexes (CENP-A.com, CENP-A–S7A.com, and CENP-C.com) were identified by an ion-trap mass spectrometer.
Recombinant Protein Purification.
Recombinant histones CENP-A/H4 were expressed in Escherichia coli strain BL21-CodonPlus-RIL (Stratagene) for 3 h at 25 °C in the presence of 0.5 mM isopropyl-d-thiogalactopyranoside (IPTG). Bacterial cells from 1-L cultures were then harvested and resuspended in 30 mL lysis buffer containing 1 M NaCl, 0.4 M ammonium acetate, 50 mM Tris⋅HCl pH 7.65, 2 mM DTT, 0.2 mM PMSF, 10% (wt/vol) glycerol, 0.01% Nonidet P-40, and 20 mM imidazole. The supernatants containing the soluble proteins were subjected to chromatography with Ni-NTA resin (0.5 mL; Qiagen) preequilibrated with lysis buffer; elution was performed with 150 mM imidazole.
Bacterially expressed GST-tagged 14-3-3-ζ protein was purified as described elsewhere (23). Recombinant baculoviruses encoding the full-length human HA-tagged–CENP-C were generated. The N-terminal HA fusion CENP-C was expressed in Sf9 insect cells for 48 h. The soluble proteins were purified on HA-agarose beads by standard procedure.
For in vitro interaction, the recombinant GST-tagged-14-3-3-ζ proteins were prebound to gluthatione sepharose 4B beads (Amersham) and then incubated with either HA–CENP-C, phosphorylated His–CENP-A, or nonphosphorylated His–CENP-A for 1 h at room temperature. After the beads were washed five times with washing buffer (20 mM Tris⋅HCl pH 7.65, 250 mM NaCl, 0.01% Triton X-100, and 5 mM EDTA) and once with low salt buffer (20 mM Tris⋅HCl pH 7.65, 150 mM NaCl, 0.01% Triton X-100). The eluted proteins were separated by 12% (wt/vol) SDS/PAGE and either visualized by Coomassie brilliant blue (CBB) staining or blotted and probed with appropriate antibodies.
RNA Interference.
Endogenous CENP-A expression was silenced by transient transfection with CENP-A siRNAs (Dharmacon). Transfections were carried out in six-well plates with 100 nM of siRNA mixed with 12 µL of HiPerfect (Qiagen), following the provider’s instructions. The following day, the transfection medium was replaced by fresh medium and cells were allowed to grow for 2 d before being fixed or collected.
Image Acquisition.
All microscopy was performed on fixed cells with a Zeiss Axio Imager Z1 microscope with a Plan-Apochromat 63× objective. GFP, cyanine-3, and Hoechst 33342 were used as fluorochromes. Images were acquired with a Zeiss Axiocam camera piloted with the Zeiss Axiovision 4.8.10 software. Acquired images were exported as grayscale 8-bit tagged image file format (TIFF) files, which were loaded into the GIMP 2.8.2 image manipulation software, where false colorization and merging of channels were performed with the “colorify” tool and the “lighten only” layer mode, respectively.
Supplementary Material
Acknowledgments
We thank R. Charton for participation in the initial experiments, Drs. W. Earnshaw and I. Cheeseman for the generous gift of an anti-CENP-C antibody, and Dr. C. Seiser for the generous gift of a bacterial strain expressing 14-3-3-ζ protein. This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Institut National du Cancer (INCa_4496 and INCa_4454), Agence Nationale de la Recherche (VariZome, Contract ANR-12-BSV8-0018-01; Nucleoplat, Contract NT09_476241; and CHROME, Project ANR-12-BSV5-0017-03 Blanc SVSE 5), and the Association pour la Recherche sur le Cancer (ARC SFI20101201822).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302955110/-/DCSupplemental.
References
- 1.Doyen C-M, et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25(18):4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci USA. 1991;88(9):3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henikoff S, Dalal Y. Centromeric chromatin: What makes it unique? Curr Opin Genet Dev. 2005;15(2):177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Buscaino A, Allshire R, Pidoux A. Building centromeres: Home sweet home or a nomadic existence? Curr Opin Genet Dev. 2010;20(2):118–126. doi: 10.1016/j.gde.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Régnier V, et al. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol. 2005;25(10):3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136(3):501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22(21):7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, et al. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20(18):7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravi M, et al. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 2011;7(6):e1002121. doi: 10.1371/journal.pgen.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155(7):1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunitoku N, et al. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell. 2003;5(6):853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- 12.Morey L, Barnes K, Chen Y, Fitzgerald-Hayes M, Baker RE. The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot Cell. 2004;3(6):1533–1543. doi: 10.1128/EC.3.6.1533-1543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravi M, et al. The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics. 2010;186(2):461–471. doi: 10.1534/genetics.110.120337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8(5):458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 15.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145(3):410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aitken A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin Cell Dev Biol. 2011;22(7):673–680. doi: 10.1016/j.semcdb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107(4):1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21(5):399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189(7):1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoda K, et al. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97(13):7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka Y, et al. Expression and purification of recombinant human histones. Methods. 2004;33(1):3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol. 2001;Chapter 10:10–17C. doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- 23.Winter S, et al. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27(1):88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.