Abstract
Myosin filaments of muscle are regulated either by phosphorylation of their regulatory light chains or Ca2+ binding to the essential light chains, contributing to on–off switching or modulation of contraction. Phosphorylation-regulated filaments in the relaxed state are characterized by an asymmetric interaction between the two myosin heads, inhibiting their actin binding or ATPase activity. Here, we have tested whether a similar interaction switches off activity in myosin filaments regulated by Ca2+ binding. Cryo-electron microscopy and single-particle image reconstruction of Ca2+-regulated (scallop) filaments reveals a helical array of myosin head-pair motifs above the filament surface. Docking of atomic models of scallop myosin head domains into the motifs reveals that the heads interact in a similar way to those in phosphorylation-regulated filaments. The results imply that the two major evolutionary branches of myosin regulation—involving phosphorylation or Ca2+ binding—share a common structural mechanism for switching off thick-filament activity in relaxed muscle. We suggest that the Ca2+-binding mechanism evolved from the more ancient phosphorylation-based system to enable rapid response of myosin-regulated muscles to activation. Although the motifs are similar in both systems, the scallop structure is more tilted and higher above the filament backbone, leading to different intermolecular interactions. The reconstruction reveals how the myosin tail emerges from the motif, connecting the heads to the filament backbone, and shows that the backbone is built from supramolecular assemblies of myosin tails. The reconstruction provides a native structural context for understanding past biochemical and biophysical studies of this model Ca2+-regulated myosin.
Keywords: scallop muscle, molluscan muscle, thick-filament structure, 3D reconstruction, muscle regulation
Contractile activity in muscle is switched on and off by protein subunits on the thick (myosin-containing) and thin (actin-containing) filaments (1). Regulation via myosin is based on either Ca2+-dependent phosphorylation of the myosin regulatory light chains (RLCs) (this mode of regulation occurs in vertebrate smooth muscle and some invertebrate striated muscles) or Ca2+ binding to the essential light chains (ELCs) (occurring in some invertebrate striated muscles) (2–6). In some muscles (vertebrate striated and some invertebrate striated), phosphorylation modulates activity but is not required for muscle activation (2, 7, 8). Electron-microscopic studies of vertebrate smooth muscle myosin molecules have revealed that, in the relaxed (dephosphorylated) state, the two myosin heads in a molecule interact with each other asymmetrically so that the actin-binding region of one (the “blocked” head) is blocked by interaction with the converter domain and ELC of the other (the “free” head). It is thought that this switches off actin-binding and ATPase activity of the blocked and free heads, respectively (9, 10), contributing to muscle relaxation. Phosphorylation of the RLC releases the two heads so that they can hydrolyze ATP and interact with actin.
In striated muscle, myosin molecules do not function as monomers, but are assembled into bipolar polymers (filaments), in which the tails form the filament backbone and the heads lie on the surface in helical arrays (11, 12). It was initially unknown whether the interacting-head conformation of isolated molecules also characterized native myosin filaments. This question was answered by cryo-electron microscopy (cryo-EM) and 3D reconstruction of filaments isolated from relaxed tarantula striated muscle, where a similar head arrangement was seen (13). This suggests that phosphorylation-regulated filaments in vivo are switched off by a similar mechanism to that proposed for isolated molecules. The filaments also showed interactions between the subfragment 2 (S2) portion of the myosin tail and the blocked head, and between heads at different axial levels of the filament (13).
The striking similarity between the head organization in isolated myosin molecules from vertebrate smooth muscle (9, 10) and in native myosin filaments from invertebrate striated muscle—two widely divergent systems—suggests that this organization is highly conserved in phosphorylation-regulated/modulated systems (13), and probably evolved before the divergence of vertebrates from invertebrates more than 600 million years ago (14). The observation of similar head–head interactions in other myosin filaments and molecules regulated or modulated by phosphorylation supports this view (15–17). Our goal here has been to answer the evolutionary question: does this interacting-head motif also characterize filaments of the other major branch of the myosin regulatory tree, in which Ca2+ binding to the myosin head switches on contraction (5, 6). Although support for this possibility has come from the observation of single myosin molecules (18), previous studies of filaments have suggested a very different structure, in which the heads are splayed apart from each other (19–21). Using cryo-EM and 3D reconstruction of scallop myosin filaments, we demonstrate that the interacting-head motif does indeed characterize relaxed Ca2+-regulated filaments, directly demonstrating the evolutionary importance of this structure. A pseudoatomic filament model produced by atomic fitting to the reconstruction provides a 3D structural context for understanding previous biochemical and biophysical studies of Ca2+-regulated myosin (reviewed in ref. 6).
Results
EM of Frozen-Hydrated Scallop Myosin Filaments.
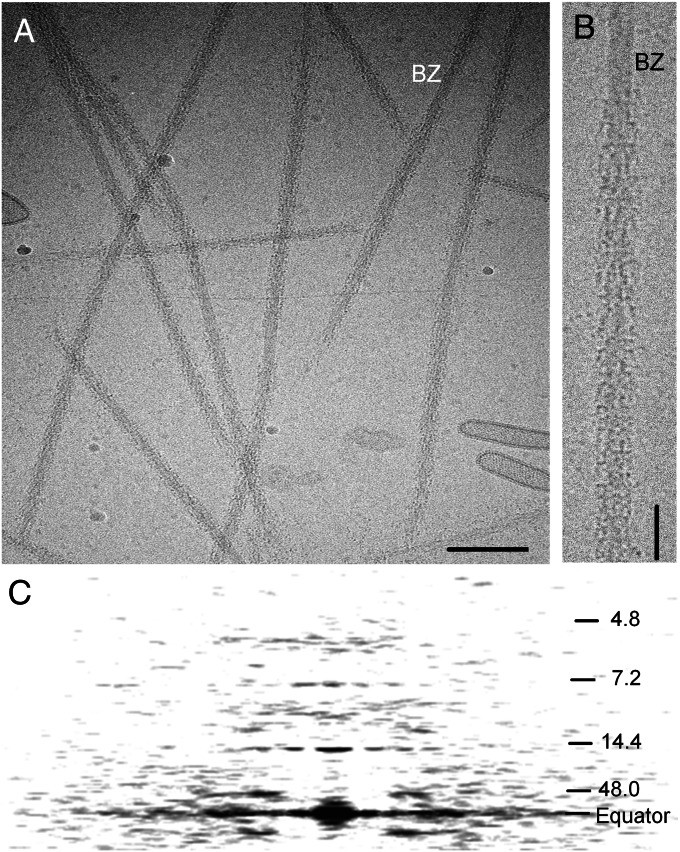
Thick filaments were purified under relaxing conditions from chemically skinned striated adductor muscle of the sea scallop Placopecten magellanicus (regulated by Ca2+ binding to the myosin heads). When observed by cryo-EM, most filaments showed relatively weak features, the most prominent being cross-striations at ∼14-nm intervals (Fig. 1 A and B, and Fig. S1 A and B). An averaged power spectrum of the 16 filament halves used in the reconstruction showed layer lines extending to the 30th order (4.8 nm) of a 144-nm filament repeat (19–21) (Fig. 1C and Fig. S2B). The reflections were similar in radial and axial position to those in X-ray patterns of living scallop muscle [Fig. S2 A and B, and Table S1 (19, 22)], suggesting that the native structure was well preserved in the cryo images. The strongest reflections were an off-meridional layer line at a spacing of 48 nm, corresponding to the axial distance between the main long-pitch myosin helices (20), and meridional reflections at 14.4 and 7.2 nm—the first and second orders of the 14.4-nm axial spacing of myosin heads (Fig. 1C and Fig. S2B).
Fig. 1.
Cryo-EM of scallop thick filaments. (A) Cryo-electron micrograph of purified filaments. BZ, bare zone. (Scale bar: 200 nm.) (B) Enlargement of part of one filament, bare zone at top. (Scale bar: 50 nm.) (C) Average power spectrum of the filaments used in the reconstruction, showing layer lines indexing on repeat of ∼144 nm (19, 20). The main helical tracks give rise to a layer line at the third order of 144 nm (∼48 nm) and to a meridional reflection at ∼14.4 nm (10th order), corresponding to the axial spacing between crowns of myosin heads. The weak 29-nm transverse periodicity reported in negatively stained filaments (20) was only occasionally apparent in our images, in agreement with previous cryo-EM of scallop filaments (21), and with the weakness of reflections at the third and fifth orders (∼5.8 and 9.6 nm) of this spacing in the power spectrum (cf. ref. 19).
Three-Dimensional Reconstruction of the Scallop Myosin Filament.
Three-dimensional reconstruction was carried out using a single-particle approach on the 16 best filament halves (from a total of 279). Selection was based on filament straightness, appearance after helical filtration, and individual reconstruction quality. Inclusion of more filaments (of lower quality) caused the reconstruction to deteriorate, presumably due to poorer head order in the additional filaments. The resolution of the reconstruction (based on a Fourier shell correlation of 0.5) was ∼5.0 nm.
The reconstruction revealed the organization of myosin heads into sevenfold symmetric “crowns” spaced 14.4 nm apart (20) (Fig. 2 and Movie S1). A clockwise 15.6° rotation between successive crowns, proceeding away from the viewer, generates seven steeply angled, coaxial, right-handed helices, creating a structure with an overall repeat of 144 nm (19–21). A density projection of the reconstruction showed features similar to those in the original micrographs (Fig. S1 B–D). The power spectrum of the reconstruction was also similar to that of the filaments (Fig. S2 B and C). The outside diameter of the reconstruction was ∼42 nm (21), similar to the diameter directly measured in cryo-EM images (23). All of these observations support the validity of the reconstruction.
Fig. 2.
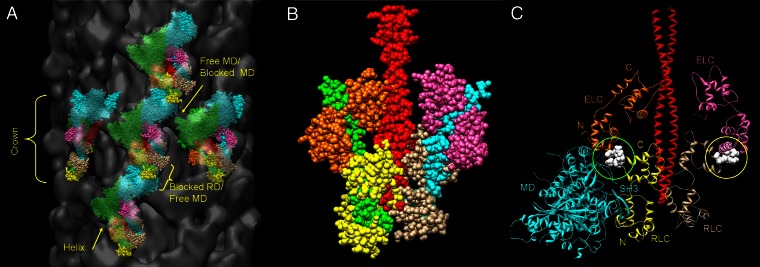
Three-dimensional reconstruction and atomic fitting of scallop filament. (A) Longitudinal view of filament segment containing five crowns, showing paired myosin head motifs (tilted “J”) in sevenfold symmetric crowns, 14.4 nm apart; three of the seven, steeply angled, right-handed helices are arrowed, and one is marked with a dashed line (Movie S1). Bare zone direction toward top; reconstruction low-pass filtered to 5-nm resolution (the calculated resolution of the reconstruction). (B) Fitting of space-fill atomic structures of scallop myosin head domains and S2 to one motif. The majority of the mass lies across the top of the “J” (the combined MDs) and is oriented almost along the right-handed helices (repeat ∼48 nm), consistent with the prominence of the 48-nm layer line in the filament power spectrum (Fig. 1C) and in X-ray patterns of scallop muscle (19). The stem and curl of the J represent the two light chain domains. Green, cyan: blocked and free MDs, respectively; orange, pink: blocked and free ELCs; yellow and beige: blocked and free RLCs; red, S2. See also Movies S2, S3, and S4.
Arrangement of Myosin Heads.
The most striking feature of the reconstruction was a repeating motif resembling the tilted “J” structure seen in phosphorylation-regulated filaments (Figs. 2 and 3A) (13, 16). In these filaments, the motif is created by the asymmetric, intramolecular interaction between the two heads of each myosin molecule. Our results suggest that a similar structure is present in Ca2+-regulated filaments. However, unlike the arthropod filaments, the heads form a shell of density ∼4 nm above the filament backbone, rather than closely contacting it [Fig. 4 A and B (21)], and the J-motifs are more steeply tilted and azimuthally closer together than in the arthropods (Figs. 2 and 3A).
Fig. 3.
Intermolecular and intramolecular contacts between myosin heads. (A) Fitting of scallop myosin head and S2 atomic structures to multiple motifs, showing apparent intermolecular contacts between blocked- and free-head MDs in the same crown, and between free-head MD in one crown and blocked-head RD in the next. Color scheme is as in Fig. 2. See also Movie S1. (B) Atomic fitting of the light chain domains suggests close proximity of the blocked- and free-head RLCs to each other (yellow, beige, respectively) at both their C and N lobes, and possible contact of the RLCs with S2 (red) in the interacting-head motif. (C) α-Carbon chain fit of free-head MD in one crown and RD of adjacent myosin in the same helix in next crown closer to the bare zone (HCs in regulatory domain omitted for clarity). The free-head MD in the lower crown comes close to the Ca2+-binding residues of the blocked-head ELC (green-circled white spheres) in the next. The Ca2+-binding site on the free head (yellow circle) is not involved in intermolecular interaction. C, N: C and N lobes of RLC and ELC.
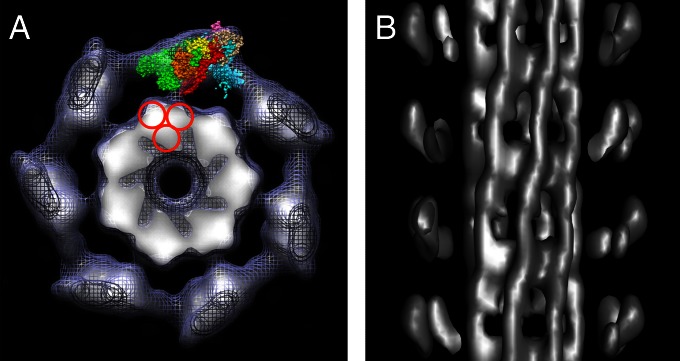
Fig. 4.
Backbone structure of scallop thick filament. (A) Transverse combined surface mesh and density projection of one myosin head crown, viewed toward bare zone. The filament backbone consists of seven high-density (white) features (subfilaments). Density at high radius is dominated by end-on views of blocked and free motor domains of each head pair; one motif has been fitted with head and S2 atomic structures as in Fig. 2. The red circles indicate possible arrangement of 4-nm–diameter subfilaments making up the larger subfilament. (B) Longitudinal surface view at high contour cutoff (from subset of filaments showing backbone features especially well) reveals subfilaments running with the same right-handed twist as the myosin head helices; heads at front of filament removed to expose subfilaments. Bare zone is toward top.
Atomic Fitting.
We gained insight into the molecular organization of the myosin heads by docking atomic structures of scallop myosin head and S2 tail domains into the reconstruction (Figs. 2 and 3, and Movies S1, S2, S3, and S4). The structures used were the motor domain (MD) of the prepower stroke state head [Protein Data Bank (PDB) ID code 1QVI (24)], the regulatory domain (RD) in the low-Ca2+ state [PDB ID code 3JTD (25)], and S2 [PDB ID code 3BAS (26)]. The fitting revealed multiple contacts between the myosin heads within a pair, between pairs within a crown, and between heads from different crowns. The head and S2 domains also came into close proximity. Although some of these contacts closely resembled those in smooth muscle, tarantula, and Limulus, others were different and may represent specializations of the Ca2+-regulatory mechanism.
Intramolecular contacts.
The regions of contact between the free and blocked scallop motor domains are essentially the same as those in the tarantula atomic model (13, 27). Thus, the actin-binding interface of the blocked head is close to the converter domain and ELC of the free head (9, 13, 27) (Fig. 2B). In addition, as in tarantula, the N-terminal segment of S2 is close to the actin-binding cleft of the blocked MD (Fig. S3 and Movie S3), and the RLC N-lobes are also close to each other (13, 27) (Figs. 2B and 3B). A possible difference from tarantula is the close proximity of the scallop RLC C-lobes to each other and their apparent contact with S2 (Fig. 3 B and C, and Movie S3), neither of which appears to occur in tarantula. These contacts suggest possible paths for intramolecular communication that are absent in phosphorylation-regulated structures. However, the RLC domains of the tarantula model (13) were based on a skeletal model (9), and it has since been reported that a scallop model produces a better connection to S2 in this region for smooth muscle (28). Therefore, it is possible that the use of a scallop model may also introduce changes in the tarantula fit.
Intermolecular contacts.
In addition to these intramolecular contacts, the head motifs also appear to make intermolecular contacts around the circumference of a crown as well as between crowns along the long-pitch helices (Fig. 3A). The proximity of head-pairs within a crown creates a continuous ring of contacts that appears to involve the SH3 domain of each blocked MD and the upper 50K region of the adjacent free MD. Such contacts do not occur in tarantula, where the motifs are widely separated within a crown. Along the long-pitch helices, contacts appear to occur between the actin-binding loop of the free MD and the blocked RD of the next head closer to the bare zone—specifically the Ca2+-binding site in the N-lobe of the ELC and the C-lobe of the RLC, near its junction with the ELC (Fig. 3C). The SH3 domain of the free MD is also close to the N-lobe of the blocked RLC (Fig. 3C). In the tarantula structure, close contact occurred between S2 from one motif and the converter and SH3 domains of the blocked head in the next motif closer to the bare zone (13). In the scallop, the heads are above the backbone surface (Fig. 4), and these intermolecular contacts do not occur.
Filament Backbone.
The reconstruction also reveals substructure in the filament backbone. In transverse view, the backbone has a polygonal appearance (Fig. 4A) created by an outer layer of seven repeating structures, in which the highest density (at the surface) tapers to a lower density toward the filament center. The central core of the filament shows relatively low protein density. In longitudinal view at high contour cutoff level, these structures appear to run as right-handed helices with the same pitch as the seven helices of myosin heads (Fig. 4B), thus pairing each with one strand of head motifs (21). When lower densities are included, the appearance becomes more complex (21) and is difficult to interpret. This appearance of “subfilaments” is consistent with previous observations (21, 29) and supports the general concept of the assembly of myosin molecules into higher order structures within thick-filament backbones. The subfilaments in the scallop are larger than the 4-nm–diameter subfilaments (containing three molecules in cross-section) seen in the arthropods (13, 30). It is possible that those in the scallop are built from such smaller (4-nm–diameter) subfilaments (Fig. 4B), but detailed interpretation remains speculative (29).
Discussion
Evolutionary Implications of the Reconstruction.
Our 3D reconstruction shows that, in native thick filaments of the scallop, a species regulated by direct binding of Ca2+ to the ELC on the myosin heads, the inactive (switched-off) state is characterized by intramolecular contact between the motor domains of the two heads. It seems likely that a similar structure is also present in other species (31) regulated by Ca2+ binding. The contact appears to involve a similar interface to that used in switching off phosphorylation-regulated myosin filaments (13, 16) and molecules (9, 10, 17). The key conclusion from these observations, therefore, is that both major branches of the myosin regulatory tree—one regulated by RLC phosphorylation and the other by Ca2+ binding to the ELC—appear to depend on a similar structural mechanism for switching off myosin activity, despite their different triggers. This is supported by in vitro observations of isolated scallop myosin molecules in the off state, which show a folded-back (32), interacting-head (18) structure similar to that seen in the reconstruction, and from mutational studies suggesting an asymmetric structure for scallop myosin (33). A similar structure may also occur in unregulated (vertebrate striated) myosin filaments (15). Our findings therefore provide further critical evidence that this off-state structure is highly conserved and most likely evolved more than 600 million years ago, before vertebrates and invertebrates diverged (13). This would suggest a common structural mechanism for switching off myosin II across most of the animal kingdom.
Relation to Previous Reconstructions.
There have been three previous 3D reconstructions of scallop thick filaments. In two, based on negative stain data (20, 34), the heads were close to the filament backbone, but not individually resolved. In a preliminary cryo-EM reconstruction (21), the heads were shown to be above the backbone (as found here), suggesting that the appearance in negative stain was an artifact. It was concluded that the heads had different conformations from each other and were splayed apart axially (21), although the reconstruction did not unambiguously define the two heads. Our reconstruction resolves the heads, and atomic fitting demonstrates that they lie close together, rather than splayed apart. Fig. S4 shows how the different reconstructions can be reconciled.
Myosin Head Interactions and the Mechanism of Regulation.
Because atomic structures of scallop S1 and S2 are available, we were able to build an atomic model of the scallop filament containing exclusively scallop conformations and sequences (Figs. 2B and 3). The hybridization and homology modeling that was required in atomic fitting of previous reconstructions was therefore avoided (13, 16, 27). In addition, the head domains were in the appropriate biochemical state [MD in ADP.Vi state (1QVI), RD in low-Ca2+ state (3JTD)] for the relaxed filament structure to which they were fitted. The resultant atomic model is supported by fluorescence resonance energy transfer measurements indicating that the average distance between residues 50 on the two RLCs of scallop myosin is at least 5.0 nm (35), consistent with our measured distance (also 5 nm). Cross-linking studies indicate that these RLC locations can sometimes approach as close as 0.2 nm (36), implying large excursions of the heads from their average position, consistent with the disorder seen in our cryo-images (21) and with the weakness of myosin reflections beyond 5-nm resolution in X-ray diffraction patterns of scallop muscle (19, 22).
Most of the intramolecular and intermolecular interactions involved in regulation are likely to be ionic, as regulation of scallop filaments is profoundly affected by ionic strength (37). The intramolecular interactions observed in both phosphorylation- and Ca2+-regulated filaments suggest a mechanism for switching off activity in which contact between the two heads and between the blocked head and S2 contribute to myosin inhibition by physically restricting myosin head mobility (9, 13). Formation of these interactions may depend on specialized features of these myosins. Crystallographic studies show that the region of S2 near its junction with the heads is unstable in regulated myosins, consistent with the suggestion that uncoiling of approximately two heptads may be required to enable the heads to adopt their asymmetric, interacting positions in the off state (26, 38–40). X-ray studies of the scallop RD suggest that formation of these interactions may also depend on increased flexibility of the regulatory domain heavy chain (HC) that results from reduction in the interaction of the ELC with the RLC upon loss of Ca2+ (25). Our fitting suggests close contact between the two RLCs and between the RLCs and S2 (Fig. 3C), which may stabilize these flexible regions of S2 and the RD in the relaxed filament (Fig. 3C), forming a possible off-state “regulatory complex.” Interaction of the N lobes of the two RLCs could modulate the mechanical characteristics of the head/rod junctional region and provide a structural pathway for communicating activation/relaxation signals between heads (6). Stiffening of the RD upon Ca2+ binding (25) may disrupt RLC–RLC and RLC–S2 interactions, in turn reducing the stabilization of the unwound N-terminal region of S2 (26) and leading to the increased freedom of movement of the heads that occurs when scallop myosin is activated (32). Thus, activation may result not from transmission of a directed conformational change along the myosin heads from the RLC region to the head–head interface, but from increased thermal motion of the heads made possible by an increase in flexibility of the head/rod junction.
Although the intramolecular interactions between heads appear similar in Ca2+- and phosphorylation-regulated filaments, intermolecular contacts, within crowns and along helical tracks (Fig. 3A), are different and may represent specializations of the Ca2+-regulatory system. These contacts—not present in single molecules (18)—may further shut down myosin activity and could contribute to the structural mechanism underlying the “super-relaxed” state thought to be present in scallop and other species, in which ATP turnover by myosin filaments is highly inhibited (41). Proximity of the free head MD in one crown to the ELC Ca2+-binding residues of the blocked head in the next crown (Fig. 3C) may impede Ca2+ binding to the blocked head. Upon activation, Ca2+ may bind initially to the free heads, where access is unimpaired. If this weakens their various interactions, these heads may become more mobile, allowing Ca2+ to bind more readily to the blocked heads, thus leading to cooperative activation of the filament as a whole (6, 42). A similar sequence of free and blocked head activation has been proposed for tarantula filaments based on the differential accessibility of myosin light chain kinase to phosphorylation sites on the free- and blocked-head RLCs (43).
The significant degree of head disorder seen in individual scallop filament images and implied by the low resolution of scallop muscle X-ray diffraction patterns suggests that the relaxed interactions between heads are relatively weak. The additive effect of multiple, weak interactions appears to keep the filament switched off (13), and may also be a requirement for the rapid activation that occurs when Ca2+ levels rise (44). When scallop filaments are activated by Ca2+, the myosin heads become disordered on the millisecond timescale (23), consistent with breaking of the head–head and other interactions, allowing free head movement and interaction with actin. The rapid reordering of heads that occurs upon relaxation (45) suggests that the structure and interactions that we have described are essential features of the relaxed state.
Relation to Vertebrate Smooth Muscle Thick Filaments.
The thick filaments of vertebrate smooth muscle have a nonhelical, “side-polar” structure in which heads on opposite sides have opposite polarity (46, 47). Although the heads in relaxed filaments presumably have similar intramolecular interactions to those in molecules (9), this has not yet been demonstrated, owing to the lability of these filaments, and potential intermolecular interactions remain unknown. The close relationship between vertebrate smooth and scallop striated muscle myosin (6, 18, 25, 28, 48), together with certain structural similarities between scallop and smooth muscle filaments, suggests that the scallop reconstruction may hold clues to molecular organization in the side-polar structure. The packing of myosin heads within a crown in the scallop is much tighter than in other striated muscle filaments (13, 15, 16), leading to azimuthal contacts between neighboring blocked and free motor domains that are absent from these other systems (Fig. 3A). Geometric considerations (47, 49, 50) suggest similarly tight packing in side-polar filaments, which might thus involve similar azimuthal interactions to those in scallop. Tomographic reconstruction of smooth muscle filaments will be required to test this prediction.
Conclusions
The results of this study fill a crucial gap in our understanding of the evolution of muscle regulation. Our reconstruction suggests that the shutting down of myosin filament activity that leads to relaxation is accomplished through a similar structural mechanism in both major branches of the myosin regulatory tree (phosphorylation and Ca2+ binding). Not only is the interacting head motif similar in both systems at the filament (13, 16) and single-molecule (18) level, but crystallographic studies of the RD suggest that the mechanism of activation may also be similar, although with different triggers (25). Recent genome-mining studies suggest that phosphorylation of the RLC by myosin light chain kinase is the most ancient mechanism for regulating actomyosin interaction (51). This mechanism must have sufficed for early, slowly contracting muscles, and persists today in nonmuscle, smooth muscle, and primitive striated muscle contractile systems (51). However, the slow speed of such an enzymatic reaction would not be adequate for striated muscles dependent on a rapid twitch response. It thus appears likely that the relaxed head–head interaction may have evolved in molluscs [and other species regulated by Ca2+ binding to myosin (31)] as a rapidly activatable adaptation of the early, phosphorylation-dependent structure. Other species have solved the rapid response problem by switching regulation to the thin filaments, where troponin rapidly binds and releases Ca2+ (1). In these systems, the relatively slow response brought about by enzymatically induced RLC phosphorylation has been retained but is used here to modulate contractility on a longer timescale (2).
Materials and Methods
EM.
Scallops (Placopecten magellanicus) were obtained from the Marine Biological Laboratory and stored in a marine aquarium at 12 °C. Myosin filaments were purified from the striated portion of the adductor muscle by detergent skinning of small fiber bundles in relaxing medium [100 mM NaCl, 3 mM MgCl2, 5 mM MgATP, 1 mM EGTA, 5 mM Pipes, 5 mM NaH2PO4, 1 mM NaN3 (pH 7.0)], followed by brief homogenization, centrifugation, and resuspension (23). Specimens were prepared for cryo-EM by applying 6 μL of filament suspension to a holey carbon grid, blotting to a thin film, and then plunging into liquid ethane (23). Grids were examined at 120 kV under low-dose conditions in a Philips CM120 cryo-electron microscope (FEI) using a Gatan 626 DH cryoholder at approximately −184 °C. Images of filaments suspended in vitreous ice over holes in the carbon film were recorded under low-dose conditions on Kodak S0163 film at a magnification of 35,000× and defocus of ∼1.5 µm. A second image, used to determine filament quality, was then obtained at 4.6-µm defocus.
Image Processing.
EM negatives were scanned on an Imacon FlexTight Precision scanner (Hasselblad) to give a pixel size of 0.51 nm in the original image. Each half-filament was oriented with the bare zone at the top to ensure correct relative polarity. Fast Fourier transforms and their averages were computed with ImageJ (W. S. Rasband, ImageJ, National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/; 1997–2011). Three-dimensional reconstruction was carried out on individual filament halves by single-particle methods using the SPIDER software package (52), assuming myosin head crowns with a rotational symmetry of 7, an axial rise of 14.4 nm between crowns, and a repeat of 48.0 nm (19–21) (SI Materials and Methods). Owing to possible perturbations in the myosin helix (20), only regions at least 150 nm from the bare zone and the filament tip were used. Individual reconstructions were aligned using University of California, San Francisco (UCSF) Chimera (53), and then averaged in SPIDER. The final average was based on images from 16 filament halves, selected from an original 279 on the basis of straightness, appearance after helical filtration (clear 14.4-nm myosin head cross-striation and 48-nm-spaced oblique lines, indicating helical preservation), and individual reconstruction quality. The reconstruction contained information from 402 filament segments, each 72.5 nm long, with a stagger of 14.4 nm between segments from the same filament. The total number of unique pairs of myosin heads that went into the reconstruction was 3,260. Reconstructions were rendered using Chimera (53).
Computational Fitting.
Docking of atomic models into the reconstruction was carried out manually within Chimera (53). The limited (∼5.0-nm) resolution of the reconstruction precluded unambiguous fitting of individual motor and regulatory domains in a single step [which had been possible when fitting the higher-resolution tarantula and Limulus reconstructions (13, 16)]. A two-step procedure was therefore used. Preliminary fitting was carried out using hybrid atomic models for the complete interacting-head motif derived from smooth muscle heavy meromyosin (PDB ID code 1I84) (9) or tarantula myosin filaments (PDB ID code 3DTP) (27). The best fit of the interacting motor domains (obtained with 3DTP) was then used as a template for fitting the MDs of the prepower stroke scallop head [1QVI (24)]. The Ca2+-free scallop RD [3JTD (25)] was then fitted into the reconstruction density of each head while ensuring that its N-terminal HC correctly matched up to the HC of each MD, and that its C-terminal HC correctly joined the α helices at the N terminal of the scallop S2 tail structure [3BAS (26)]. Minor adjustments were then made to optimize the fit, resulting in a final homogeneous atomic model containing only scallop structures (SI Materials and Methods).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants R01 AR034711 (to R.C.) and P01 HL059408 (to D. Warshaw). EM was carried out in the Core Electron Microscopy Facility of the University of Massachusetts Medical School. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at UCSF (supported by NIH Grant P41 RR-01081).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218462110/-/DCSupplemental.
References
- 1.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Sweeney HL, Bowman BF, Stull JT. Myosin light chain phosphorylation in vertebrate striated muscle: Regulation and function. Am J Physiol. 1993;264(5 Pt 1):C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 3.Trybus KM. Role of myosin light chains. J Muscle Res Cell Motil. 1994;15(6):587–594. doi: 10.1007/BF00121066. [DOI] [PubMed] [Google Scholar]
- 4.Sellers JR. Phosphorylation-dependent regulation of Limulus myosin. J Biol Chem. 1981;256(17):9274–9278. [PubMed] [Google Scholar]
- 5.Szent-Györgyi AG. Calcium regulation of muscle contraction. Biophys J. 1975;15(7):707–723. doi: 10.1016/S0006-3495(75)85849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chantler PD. In: Scallops: Biology, Ecology and Aquaculture. Shumway SE, Parsons GJ, editors. Amsterdam: Elsevier; 2006. pp. 231–320. [Google Scholar]
- 7.Craig R, Padrón R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105(3):1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stull JT, Kamm KE, Vandenboom R. Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch Biochem Biophys. 2011;510(2):120–128. doi: 10.1016/j.abb.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci USA. 2001;98(8):4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess SA, et al. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372(5):1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7(3):281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 12.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16(2):204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Woodhead JL, et al. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436(7054):1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 14.Peterson KJ, et al. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci USA. 2004;101(17):6536–6541. doi: 10.1073/pnas.0401670101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci USA. 2008;105(7):2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao FQ, Craig R, Woodhead JL. Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J Mol Biol. 2009;385(2):423–431. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19(8):3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung HS, et al. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci USA. 2008;105(16):6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wray JS, Vibert PJ, Cohen C. Diversity of cross-bridge configurations in invertebrate muscles. Nature. 1975;257(5527):561–564. doi: 10.1038/257561a0. [DOI] [PubMed] [Google Scholar]
- 20.Vibert P, Craig R. Electron microscopy and image analysis of myosin filaments from scallop striated muscle. J Mol Biol. 1983;165(2):303–320. doi: 10.1016/s0022-2836(83)80259-9. [DOI] [PubMed] [Google Scholar]
- 21.Vibert P. Helical reconstruction of frozen-hydrated scallop myosin filaments. J Mol Biol. 1992;223(3):661–671. doi: 10.1016/0022-2836(92)90982-p. [DOI] [PubMed] [Google Scholar]
- 22.Millman BM, Bennett PM. Structure of the cross-striated adductor muscle of the scallop. J Mol Biol. 1976;103(3):439–467. doi: 10.1016/0022-2836(76)90212-6. [DOI] [PubMed] [Google Scholar]
- 23.Zhao FQ, Craig R. Ca2+ causes release of myosin heads from the thick filament surface on the milliseconds time scale. J Mol Biol. 2003;327(1):145–158. doi: 10.1016/s0022-2836(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 24.Gourinath S, et al. Crystal structure of scallop myosin S1 in the pre-power stroke state to 2.6 Å resolution: Flexibility and function in the head. Structure. 2003;11(12):1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Himmel DM, Mui S, O’Neall-Hennessey E, Szent-Györgyi AG, Cohen C. The on-off switch in regulated myosins: Different triggers but related mechanisms. J Mol Biol. 2009;394(3):496–505. doi: 10.1016/j.jmb.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JH, et al. An unstable head-rod junction may promote folding into the compact off-state conformation of regulated myosins. J Mol Biol. 2008;375(5):1434–1443. doi: 10.1016/j.jmb.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alamo L, et al. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384(4):780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann BA, et al. Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation. J Mol Biol. 2012;415(2):274–287. doi: 10.1016/j.jmb.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellani L, Vibert P. Location of paramyosin in relation to the subfilaments within the thick filaments of scallop striated muscle. J Muscle Res Cell Motil. 1992;13(2):174–182. doi: 10.1007/BF01874154. [DOI] [PubMed] [Google Scholar]
- 30.Wray JS. Structure of the backbone in myosin filaments of muscle. Nature. 1979;277(5691):37–40. doi: 10.1038/277037a0. [DOI] [PubMed] [Google Scholar]
- 31.Lehman W, Szent-Györgyi AG. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J Gen Physiol. 1975;66(1):1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stafford WF, et al. Calcium-dependent structural changes in scallop heavy meromyosin. J Mol Biol. 2001;307(1):137–147. doi: 10.1006/jmbi.2000.4490. [DOI] [PubMed] [Google Scholar]
- 33.Colegrave M, Patel H, Offer G, Chantler PD. Evaluation of the symmetric model for myosin-linked regulation: Effect of site-directed mutations in the regulatory light chain on scallop myosin. Biochem J. 2003;374(Pt 1):89–96. doi: 10.1042/BJ20030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.AL-Khayat HA, Morris EP, Squire JM. The 7-stranded structure of relaxed scallop muscle myosin filaments: Support for a common head configuration in myosin-regulated muscles. J Struct Biol. 2009;166(2):183–194. doi: 10.1016/j.jsb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Chantler PD, Tao T, Stafford WF., 3rd On the relationship between distance information derived from cross-linking and from resonance energy transfer, with specific reference to sites located on myosin heads. Biophys J. 1991;59(6):1242–1250. doi: 10.1016/S0006-3495(91)82339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bower SM, Wang Y, Chantler PD. Regulatory light-chain Cys-55 sites on the two heads of myosin can come within 2Å of each other. FEBS Lett. 1992;310(2):132–134. doi: 10.1016/0014-5793(92)81313-b. [DOI] [PubMed] [Google Scholar]
- 37.Nyitrai M, Stafford WF, Szent-Györgyi AG, Geeves MA. Ionic interactions play a role in the regulatory mechanism of scallop heavy meromyosin. Biophys J. 2003;85(2):1053–1062. doi: 10.1016/S0006-3495(03)74544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, et al. Visualization of an unstable coiled coil from the scallop myosin rod. Nature. 2003;424(6946):341–345. doi: 10.1038/nature01801. [DOI] [PubMed] [Google Scholar]
- 39.Tama F, Feig M, Liu J, Brooks CL, 3rd, Taylor KA. The requirement for mechanical coupling between head and S2 domains in smooth muscle myosin ATPase regulation and its implications for dimeric motor function. J Mol Biol. 2005;345(4):837–854. doi: 10.1016/j.jmb.2004.10.084. [DOI] [PubMed] [Google Scholar]
- 40.Blankenfeldt W, Thomä NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci USA. 2006;103(47):17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci USA. 2010;107(1):430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chantler PD, Sellers JR, Szent-Györgyi AG. Cooperativity in scallop myosin. Biochemistry. 1981;20(1):210–216. doi: 10.1021/bi00504a035. [DOI] [PubMed] [Google Scholar]
- 43.Brito R, et al. A molecular model of phosphorylation-based activation and potentiation of tarantula muscle thick filaments. J Mol Biol. 2011;414(1):44–61. doi: 10.1016/j.jmb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rall JA. Mechanics and energetics of contraction in striated muscle of the sea scallop, Placopecten magellanicus. J Physiol. 1981;321:287–295. doi: 10.1113/jphysiol.1981.sp013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao FQ, Craig R. Millisecond time-resolved changes occurring in Ca2+-regulated myosin filaments upon relaxation. J Mol Biol. 2008;381(2):256–260. doi: 10.1016/j.jmb.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craig R, Megerman J. Assembly of smooth muscle myosin into side-polar filaments. J Cell Biol. 1977;75(3):990–996. doi: 10.1083/jcb.75.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu JQ, Harder BA, Uman P, Craig R. Myosin filament structure in vertebrate smooth muscle. J Cell Biol. 1996;134(1):53–66. doi: 10.1083/jcb.134.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellers JR, Chantler PD, Szent-Györgyi AG. Hybrid formation between scallop myofibrils and foreign regulatory light-chains. J Mol Biol. 1980;144(3):223–245. doi: 10.1016/0022-2836(80)90088-1. [DOI] [PubMed] [Google Scholar]
- 49.Squire JM. General model of myosin filament structure. 3. Molecular packing arrangements in myosin filaments. J Mol Biol. 1973;77(2):291–323. doi: 10.1016/0022-2836(73)90337-9. [DOI] [PubMed] [Google Scholar]
- 50.Tonino P, Simon M, Craig R. Mass determination of native smooth muscle myosin filaments by scanning transmission electron microscopy. J Mol Biol. 2002;318(4):999–1007. doi: 10.1016/S0022-2836(02)00191-2. [DOI] [PubMed] [Google Scholar]
- 51.Steinmetz PR, et al. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487(7406):231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.