Significance
Human congenital diaphragmatic defects are almost as common as cystic fibrosis, cause neonatal mortality, and often result in multisystem morbidity throughout childhood. Research investigating mechanisms of development is needed to understand the pathogenesis of disease. In this report, we identified a model caused by a mutation in kinesin motor family gene kinesin family member 7. We determined that this gene is required to regulate cell proliferation, patterning, and differentiation in the embryonic diaphragm. We ascertained several functions of retinoid signaling in diaphragm development and gained insight into how perturbations in this signaling network promote the pathogenesis of congenital diaphragmatic hernia.
Keywords: Gli transcription factors, myotendonous junction, tendon differentiation
Abstract
Congenital diaphragmatic hernia (CDH) is a common birth defect that results in a high degree of neonatal morbidity and mortality, but its pathological mechanisms are largely unknown. Therefore, we performed a forward genetic screen in mice to identify unique genes, models, and mechanisms of abnormal diaphragm development. We identified a mutant allele of kinesin family member 7 (Kif7), the disorganized diaphragm (dd). Embryos homozygous for the dd allele possess communicating diaphragmatic hernias, central tendon patterning defects, and increased cell proliferation with diaphragmatic tissue hyperplasia. Because the patterning of the central tendon is undescribed, we analyzed the expression of genes regulating tendonogenesis in dd/dd mutant embryos, and we determined that retinoic acid (RA) signaling was misregulautted. To further investigate the role of Kif7 and RA signaling in the development of the embryonic diaphragm, we established primary mesenchymal cultures of WT embryonic day 13.5 diaphragmatic cells. We determined that RA signaling is necessary for the expression of tendon markers as well as the expression of other CDH-associated genes. Knockdown of Kif7, and retinoic acid receptors alpha (Rara), beta (Rarb), and gamma (Rarg) indicated that RA signaling is dependent on these genes to promote tendonogenesis within the embryonic diaphragm. Taken together, our results provide evidence for a model in which inhibition of RA receptor signaling promotes CDH pathogenesis through a complex gene network.
The diaphragm is an essential organ in mammals that is required for respiratory and nonrespiratory functions. In humans, congenital diaphragmatic hernia (CDH) is a life-threatening birth defect that occurs at a frequency of 1:2,000–1:3,000 live births (1) and results in a high mortality rate (2). CDH may occur as an isolated diaphragmatic defect or part of a larger syndrome with other congenital defects in multiple systems, including musculoskeletal, cardiac, and CNS (3, 4). In the mouse, the diaphragm is believed to develop from transient mesenchymal tissue often referred to as the pleuroperitoneal folds or posthepatic mesenchymal plate on embryonic days (E) 11.5 and E12.5 (5–7). This tissue is thought to fuse ventrally with a mesenchymal structure separating the thorax and abdomen called the septum transversum (8), producing a complete diaphragm. The mature fetal diaphragm is made up of muscular regions composed of hypaxial-derived skeletal muscle, an amuscular region called the central tendon, and the crural diaphragm (7, 9). Because diaphragmatic hernias are believed to occur through a mechanism that affects nonmuscle mesenchymal cells (10), it is necessary to investigate the molecular basis controlling diaphragmatic mesenchymal cell proliferation, patterning, and differentiation.
The Hedgehog (Hh) and retinoic acid (RA) signaling pathways are critical regulators of organogenesis and mesenchymal cell fate determination in mammals (11, 12), and both have been implicated in CDH pathogenesis. In humans, mutations in the Hh transporter Dispatched homolog 1 (13), the Sonic hedgehog (SHH) coreceptor low density lipoprotein-related protein 2 (LRP2) (14), and stimulated by retinoic acid gene 6 (STRA6) (15) are associated with syndromic CDH. In mice, diaphragmatic defects are associated with mutations in the Hh-associated zinc finger transcription factors GLI-Kruppel family member GLI2 (Gli2) and GLI-Kruppel family member GLI3 (Gli3) (16) and the RA nuclear receptors alpha and beta (Rara, Rarb) (17). Administration of the herbicide nitrofen to pregnant rodents is thought to cause diaphragmatic hernias, in part through down-regulation of RA receptor signaling (18). How these genes regulate normal diaphragm development in mice and humans is unknown.
Kinesin family member 7 (Kif7) was recently identified as an essential component of the Hh signaling pathway. Kif7 encodes a motor protein that functions downstream of the G protein-coupled transmembrane receptor, Smoothened (Smo), and interacts with zinc finger transcription factors Gli2 and Gli3. These transcription factors are the primary effectors of the Hh signaling pathway (11). Kif7 is a negative regulator of Hh signaling in early embryonic development, and loss of Kif7 promotes ligand-independent activation of the Gli target genes (19, 20).
In this study, we investigated the role of Kif7 and RA signaling in the development and cellular differentiation of the embryonic diaphragm. We found that Kif7 is required for central tendon development and necessary for RA-mediated induction of tendon markers. We also show that RA signaling promotes the development of the embryonic diaphragm through regulation of Lrp2, nuclear receptor subfamily2, group F, member 2 (Nr2f2), platelet derived growth factor receptor, alpha polypeptide (Pdgfra), Wilms tumor-1 (Wt1), and zinc finger protein, multitype 2 (Zfpm2) gene networks.
Results
Loss of Kif7 Results in Diaphragmatic Hernia, a Central Tendon Patterning Defect, and Increased Accumulation of Wt1-Expressing Cells in the Primordial Diaphragm.
To identify unique models and mechanisms of abnormal diaphragm development, we performed a forward N-ethyl-N-nitrosourea (ENU) mutagenesis screen designed to recover mouse autosomal recessive mutations affecting the development and patterning of the embryonic diaphragm. Because both the central tendon and the muscular diaphragm can be disrupted in human disease (21), we performed our screening to detect abnormalities in both areas. Our screen used a three-generation strategy, in which our mutagenized strain (A/J) was outcrossed to FVB/NJ (22). We screened ∼2,500 E16.5 embryos from 32 independent families and identified a mouse line with both left-sided posterior diaphragmatic hernia and central tendon patterning defects characterized by muscle expansion into the central tendon domain (Fig. 1A; a detailed description of diaphragmatic anatomy is shown in Fig. S1A). The diaphragm defects were present in 125/125 mutant embryos examined. These embryos also possessed other phenotypes associated with syndromic congenital diaphragmatic hernia, such as neural tube defects, abnormal cardiac position, ventricular septal defects, skeletal defects, and pulmonary hypoplasia (Fig. S1 C–J) (23).
Fig. 1.
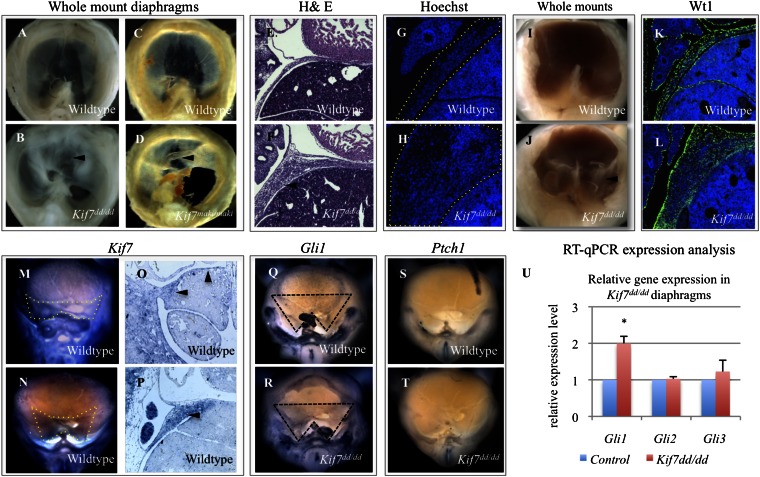
Loss of Kif7 results in diaphragmatic hernia, a central tendon patterning defect, and increased accumulation of Wt1-expressing cells in the primordial diaphragm. (A and B) Whole-mount images of E17.5 control and Kif7dd/dd diaphragms. (C and D) Whole-mount images of E17.5 control and Kif7maki/maki diaphragms. The defect in central tendon patterning is marked with arrowhead. (E and F) H&E-stained sagittal sections of E14.5 control and Kif7dd/dd diaphragms show thickened diaphragmatic tissue in Kif7dd/dd embryos (arrowhead). (G and H) Hoechst nuclear stain of E13.5 embryos shows that thickened diaphragmatic tissue is because of increased cell number in Kif7dd/dd embryos (diaphragms outlined with dotted lines). (I and J) Immature diaphragms from E13.0 WT and Kif7dd/dd embryos show an early bilateral accumulation of extra diaphragmatic tissue in mutant embryos (arrowhead). (K and L) Wt1 (green) and Hoechst (blue) immunofluorescent staining of E13.5 control and Kif7dd/dd diaphragms show an accumulation of Wt1+ cells in the mutant diaphragm. (M and N) Whole-mount in situ hybridization shows Kif7 expression in the posterior immature diaphragm (inside dotted lines) at (M) E11.5 and (N) E12.5. (O and P) Section in situ hybridization of Kif7 in (O) E11.5 and (P) E12.5 immature diaphragms (arrowheads point to diaphragm expression). (Q–T) Whole-mount in situ hybridization in E12.5 control and Kif7dd/dd diaphragms. Note that there is increased Gli1 expression in the posterior Kif7dd/dd diaphragm (region delineated by dotted line), whereas there is no Ptch1 expression in either the WT or mutant. (U) Expression analysis for Gli1, Gli2, and Gli3 in E13.5 diaphragms of control and Kif7dd/dd diaphragms. Data are means, with error bars representing SE; n = 3 independent experiments, each with three replicates (*P < 0.05, t test).
We mapped the mutation using a whole-genome SNP array to identify regions of mutagenized strain homozygosity retained in mutant embryos. Candidate gene sequencing in the mapped interval revealed that the disorganized diaphragm allele (dd) was a unique mutation in Kif7. The mutation is an A-T transversion at position 2002 in the Kif7 cDNA (NM_010626) (Fig. S2A), and it is predicted to generate a nonsense mutation producing a 100 kDa protein instead of the predicted 150 kDa WT protein. To confirm that the dd mutation generates a truncated protein, an N-terminal GFP Kif7 fusion construct was generated for the WT Kif7 and the Kif7dd A-T mutation. Western blot analysis of transfected WT GFP:Kif7 and GFP:Kif7dd showed that the A-T mutation observed in Kif7dd/dd embryos generates a truncated protein of predicted size (Fig. S2B). Genotyping was established using a restriction fragment-length polymorphism 210 bases from the mutation, and the line was backcrossed to FVB/NJ mice for 11 generations with continued retention of the phenotype. This backcross assured us that our phenotype was associated with the Kif7 point mutation and not other parts of the originally mutagenized genome. We then obtained embryos homozygous for another independent mutant allele of Kif7 (matariki or maki), an ENU-generated mutation in the Kif7 motor domain (L130P) (19). Kif7maki/maki mutant embryos also possessed diaphragmatic hernias and central tendon patterning defects. Whole diaphragms showing identical phenotypes in the Kif7dd/dd and Kif7maki/maki embryos are shown in Fig. 1 B and D. Both have left-sided communicating diaphragmatic hernias (communication between the abdominal and thoracic cavities) and tendon patterning defects, indicating that Kif7 is required for normal diaphragm development.
Examination of histology in WT and Kif7dd/dd E14.5 embryos showed diffuse defects in morphogenesis (Fig. 1 E and F) with extremely thickened tissue. A count of cell number after nuclear fluorescent staining (Hoechst) determined that the abnormal accumulation of diaphragmatic tissue in Kif7dd/dd embryos was associated with an increase in cell number rather than cell size (Figs. 1 G and H and 2). To identify the severity of earlier tissue patterning defects in Kif7dd/dd mutants, we examined whole-mount primordial diaphragms at E13.0. In WT embryos, the immature diaphragm appears as a thin sheet extending from the posterior body wall to the sternum. However, in Kif7dd/dd embryos, we observed abnormal accumulation of tissue in the posterior diaphragm (Fig. 1 I and J). To determine the cell type that contributes to the diaphragmatic tissue overgrowth, we examined the expression of myosin heavy chain (MyHC) and Wilms tumor-1 (Wt1) in sections of Kif7dd/dd embryos; MyHC labels diaphragmatic skeletal muscle, and Wt1 is expressed in the early mesothelium and throughout the nonmuscle mesenchymal cells of the diaphragm (24, 25). We determined that the skeletal muscle fiber distribution and orientation were normal at E13.5, whereas the nonmuscular Wt1+ cells accumulated throughout the mutant diaphragms (Fig. 1 K and L). Wt1 is known to be required for normal diaphragm development (26), but the function of this gene in this organ is unknown. Taken together, these findings suggest that Kif7 regulates the patterning and/or proliferation of Wt1+ cells in the developing diaphragm.
Fig. 2.
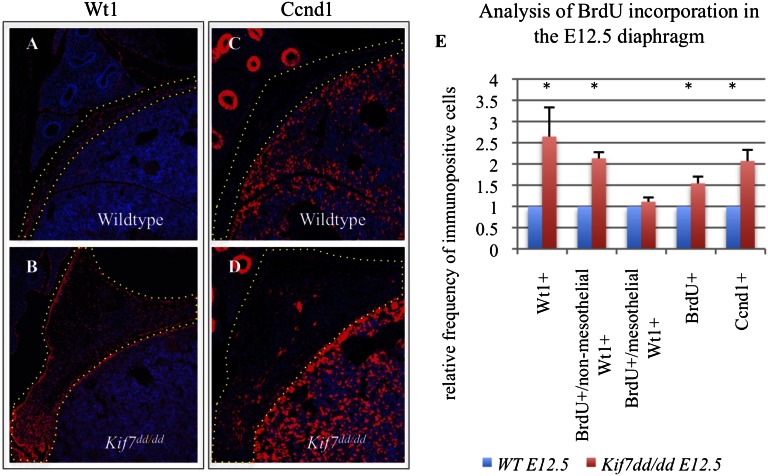
Kif7 is a negative regulator of cell proliferation in the primordial diaphragm. (A and B) Wt1 and (C and D) Ccnd1 immunofluorescent staining (red) in sagittal sections of E12.5 control and Kif7dd/dd embryos. (E) Comparison of relative frequency of numbers of Wt1+ cells, Wt1+/BrdU+ nonmesothelial mesenchymal cells, Wt1+/BrdU+ mesothelial cells, BrdU+ cells, and Ccnd1+ within the domains marked by the dotted line in A–D in E12.5 control and Kif7dd/dd diaphragms. Data represent mean relative frequency of cell number, and error bars represent SE; n = 4 independent experiments (*P < 0.05, t test).
Kif7 plays a role in the Shh signaling pathway, and although diaphragm defects have been reported in the Hh pathway effectors Gli2 and Gli3 mutant embryos (13, 16), neither expression of Hh ligands nor receptors has been identified during the initial stages of diaphragm development (13). To further investigate the potential role of Hh signaling, we examined the expression of Kif7, Gli2, Gli3, and the Hh target genes Gli1 and Ptch1 in the developing diaphragm. At E11.5, Kif7 is expressed in both the anterior and posterior diaphragm (Fig. 1 M and O), and by E12.5, Kif7 expression becomes very prominent in the posterior diaphragm (Fig. 1 N and P). This finding is interesting, because the posterior diaphragm is the site at which we observed both diaphragmatic hernias and accumulations of Wt1+ cells. We evaluated Gli expression in both WT and mutant diaphragms and found that Gli1 transcripts, but not Gli2 or Gli3, were elevated in the E12.5 diaphragms of mutant embryos (Fig. 1 Q, R, and U). Consistent with previous reports, the expression of Ptch1, the SHH receptor, was not detected.
Kif7 Is a Negative Regulator of Cell Proliferation in the Primordial Diaphragm.
We reasoned that the bilateral accumulation of Wt1+ cells could arise from either changes in cell proliferation or a patterning defect resulting in isolated accumulations of cells. To distinguish between the two possibilities, we counted the number of Wt1+ cells and measured BrdU incorporation in Wt1+ cells of the E12.5 diaphragms. We determined that Kif7dd/dd mutants exhibited a twofold increase in the number of Wt1+ cells, an increase in total cell proliferation, and an increase in the number of proliferating Wt1+ nonmesothelial mesenchymal cells (Fig. 2 A and B). Cell proliferation was unchanged in Wt1+ diaphragmatic mesothelial cells. These results illustrate that Kif7 negatively regulates the proliferation of Wt1+ nonmesothelial mesenchymal cells of the immature diaphragm, and the loss of Kif7 results in diaphragmatic tissue hyperplasia.
Kif7 is a key regulator of Hh signaling in neural tube (19), limb (20), bone (27), and skin development (28), and Hh signaling promotes cell proliferation in several systems by regulating the expression of the cell cycle regulating gene, Cyclin D1 (Ccnd1) (11). Hence, we examined the expression and counted the number of Ccnd1+ cells in the E12.5 diaphragms of Kif7dd/dd mutant embryos, and we determined that the number of Ccnd1+ cells was increased approximately twofold compared with cells in littermate controls (Fig. 2 C–E). Taken together, these findings suggest that Kif7 regulates cell proliferation in the developing diaphragm, in part through negative regulation of Ccnd1.
Kif7 Is Necessary for the Patterning, Differentiation, and Induction of Scleraxis-Positive Tendon Progenitors in the Central Tendon.
The primary function of tendons within the musculoskeletal system is to connect muscle to bone. However, in the diaphragm, the central tendon is responsible for joining together anterior, lateral, and posterior sections of diaphragmatic skeletal muscle. The myotendonous junction (MTJ) delineates the muscular and central tendon domains and is the site responsible for anchoring the muscle fibers of the diaphragm with the central tendon. Tendon development is believed to begin when SRY Box 9 (Sox9) -positive mesenchymal cells become specified to become tendon progenitor cells by induction of Scleraxis (Scx). The tendon progenitor cells then assemble and differentiate into tenocytes, and eventually, they connect muscle to bone (29). The processes controlling the induction and specification of diaphragmatic central tendon tenocytes have not been established. To identify the origin of the central tendon patterning phenotype observed in Kif7dd/dd mutants, we examined the tendon cell lineage within the developing diaphragm. Tenascin C (Tnc), a well-described marker of early tenocytes (29, 30), is expressed strongly at the MTJ of WT E17.5 diaphragms, whereas skeletal muscle MyHC labels muscle fibers (Fig. 3 A and C). Expression of Tnc was decreased in the diaphragm of Kif7dd/dd embryos. In these mutant embryos, the muscle fibers bypass the MTJ, grow into the central tendon domain, and make ectopic connections with the cural diaphragm (Fig. 3 B and D).
Fig. 3.
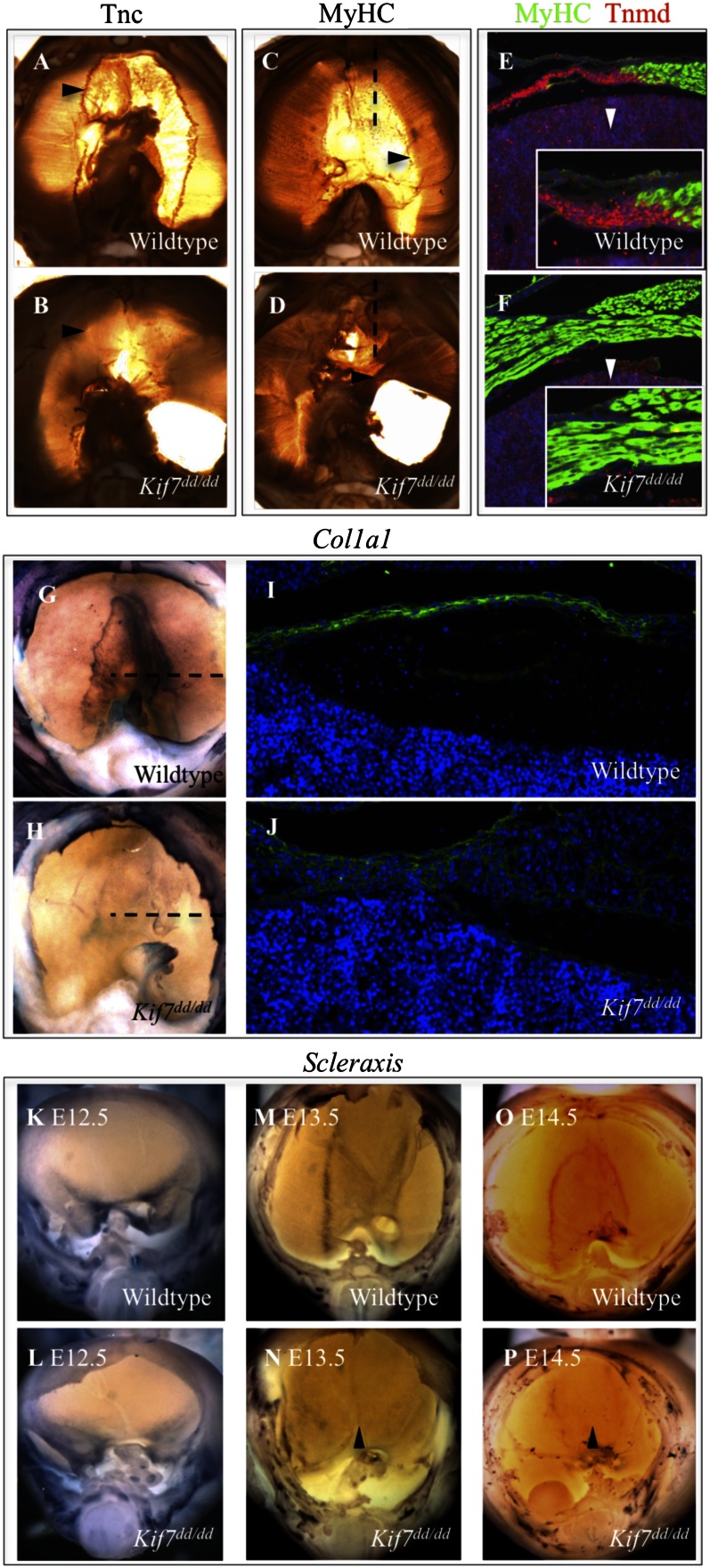
Kif7 is necessary for the patterning, differentiation, and induction of Scx-positive tendon progenitors in the central tendon. (A–D) Whole-mount immunohistochemical-stained E17.5 control and Kif7dd/dd diaphragms. Mutants show loss of Tnc expression at the MTJ (arrowheads in A and B) and an overgrowth of muscle fibers (arrowheads in C and D). (E and F) MyHC (green) and Tnmd (red) coimmunofluorescent staining in sagittal sections of the anterior diaphragm at the plane marked by dashed line in C and D in E17.5 control and Kif7dd/dd embryos. Insets show a zoomed-in view of the MTJ in control and mutant diaphragms (arrowheads). (G and H) Col1a1 whole-mount in situ hybridization at E15.5. (I and J) Col1a1 immunofluorescent staining in frontal sections of the diaphragm at the plane marked by the dashed line in G and H in E15.5 control and Kif7dd/dd embryos. (K–P) Scx whole-mount in situ hybridization at E12.5, E13.5, and E14.5 in control and Kif7dd/dd diaphragms. Arrowheads show reduced induction of Scx in the central tendon domain of the diaphragm in Kif7dd/dd embryos relative to littermate controls.
Because differentiating tendons may play an important role in arresting muscle fiber migration and elongation at the MTJ (29), we examined the expression of the transmembrane protein, Tenomodulin (Tnmd), a differentiation marker of tenocytes (31). Tnmd and MyHC coimmunofluorescent staining clearly identified the MTJ at E17.5 in WT embryos (Fig. 3E). In Kif7 mutants, the Tnmd-expressing domain was drastically reduced, and expression at the expected MTJ was undetectable (Fig. 3F). Because collagen is a major component of connective tissue such as tendons, we examined the expression of Collagen type 1, α-1 (Col1a1), the gene encoding the major component of type 1 collagen. Col1a1 is expressed throughout the developing central tendon of control embryos at E15.5; however, its expression was significantly decreased in Kif7dd/dd diaphragms (Fig. 3 G–J). Together, these findings show that the central tendon exhibits reduced differentiation in Kif7dd/dd embryos.
Next, we examined the expression of Scx throughout diaphragm development. Scx is a basic helix-loop-helix transcription factor that is expressed in tendon progenitor cells and required for tenocyte maturation (31, 32). Scx was expressed in the posterior diaphragm at E12.5; however, the pattern of expression was slightly abnormal in the mutant (Fig. 3 K and L). By E13.5, normal diaphragms expressed Scx throughout the central tendon, with the strongest expression defining the MTJs (Fig. 3 M and O). In mutants, Scx expression was dramatically decreased and occurred outside of the domain of the central tendon, suggesting compromise of both patterning and induction of Scx in Kif7dd/dd diaphragms (Fig. 3 M and N). The specification of the central tendon domain is complete by E14.5 (Fig. 3O), and mutants show almost no discernable central tendon, MTJs, or Scx expression (Fig. 3P, arrowhead marks the central tendon midline). These findings show that Kif7 is required for the patterning and induction of Scx-positive tendon progenitor cells and that, in the absence of Kif7, tendon progenitor cells have impaired differentiation into Tnc- and Tnmd-positive central tendon tenocytes. Collectively, these defects prevent the arrest of muscle fiber elongation at the MTJ and allow the diaphragm’s central tendon to become overmuscularized.
Rara, Rarb, and Rarg Regulate the Expression of Scx and Tnc in Diaphragmatic Cells.
Recent studies performed in primary cells derived from mouse limb bud mesenchyme have shown that pharmacological inhibition of the RA-metabolizing enzyme, Cytochrome P450 26B1 (Cyp26b1), results in elevated RA signaling and increased expression of Scx and Tnc. Additionally, Cyp26b1 null mouse embryos possess Scx patterning defects in their limb buds (33). To determine whether RA signaling was perturbed in diaphragms of Kif7dd/dd embryos, we quantified transcripts of Cyp26b1, Cytochrome P450 26A1 (Cyp26a1), Cellular retinoic acid binding protein 2 (Crabp2), retinaldehyde dehydrogenase (Aldh1a2), and the retinoic acid nuclear receptors Rara, Rarb, and Rarg in E13.5 diaphragms of Kif7dd/dd embryos and littermate controls. Mutant diaphragms had increased expression of the RA-synthesizing enzyme Aldh1a2 and RA-signaling molecules Crabp2, Cyp26a1, and Cyp26b1 compared with controls, whereas expression of RA nuclear receptors Rara, Rarb, and Rarg was normal (Fig. S3A). The genes Aldh1a2, Crabp2, Cyp26a1, Cyp26b1, Rara, Rarb, and Rarg all contain RA response elements and are components and transcriptional targets of the RA-signaling pathway (12). Because a complex of Meis homeobox 2 and Pre B-cell leukemia homeobox 1,2 (Pbx1,2) regulates the expression of Aldh1a2 and may be important for diaphragm development (12, 34), we assayed the expression of these genes to determine if they were also up-regulated in mutant diaphragms. Meis homeobox 2 and Pbx1,2 were expressed normally in mutant diaphragms (Fig. S3A). We then wanted to determine if Aldh1a2 was expressed in the central tendon domain of the diaphragm and if its expression was altered in Kif7dd/dd embryos. We determined that Aldh1a2 was expressed at this site in the E13.5 differentiating diaphragm, and its expression was elevated in Kif7dd/dd embryos (Fig. S3 D and E). These findings show that RA is synthesized in the central tendon domain of the differentiating E13.5 diaphragm and that RA signaling is perturbed in the developing diaphragms of Kif7dd/dd embryos.
RA signaling regulates cell fate determination and differentiation through transcriptional control of large gene networks, and perturbations in RA signaling are associated with diaphragm defects in mice and humans (12, 15, 17). The expressions of the RA nuclear receptors and several components of the RA pathway have been examined in the developing diaphragm; however, the function of these genes during diaphragm development is unknown (35). To assess whether RA signaling is necessary for the induction of Scx and Tnc in the diaphragm and gain insight into how Kif7 and other genes known to be required for normal diaphragm development interact with the RA-signaling pathway, we investigated the function of RA signaling in confluent cultures of E13.0 diaphragmatic cells. The composition of disassociated diaphragmatic cultures was examined, and cultures contained Scx- and Tnc-positive tenocytes, Zfpm2- and Wt1-expressing nonmuscle mesenchymal cells, transcription factor 4 (Tcf4)-positive muscle connective tissue fibroblasts, and uroplakin 3B (Upk3b)-positive mesothelial cells, an expression profile similar to what has been shown in the developing diaphragm (24, 25, 36–38). In this system, we could modulate RA nuclear receptor-dependent signaling (RA-RAR) by addition of either all trans-RA (ATRA) to activate the RA nuclear receptors Rara, Rarb, and Rarg or a pan-RAR antagonist (BMS493) to inhibit basal signaling (reviewed in refs. 12 and 39).
Treatment of WT cells with ATRA led to elevated expression of the tendon markers Scx and Tnc, the RA-signaling molecules Crabp2 and Cyp26b1, and the CDH-associated genes Pdgfra, Wt1, and Zfpm2 (Table 1). RA treatment also promoted decreased expression of Sox9, the RA-synthesizing molecule Aldh1a2, and the Hh/Gli target gene Gli1. Inhibition of the RA nuclear receptors with BMS493 led to reduced expression of Tnc, the RA-signaling molecules Cyp26b1 and Rarb, and the CDH-associated genes Nr2f2, Pdgfra, Wt1, Zfpm2, and Lrp2 (Table 1).
Table 1.
Rara, Rarb, and Rarg regulate Scx and Tnc expression in diaphragmatic cells
| Marker type and gene | Treatment |
|||||
| ATRA | BMS493 | Rara KD | Rarb KD | Rarg KD | Rara Rarb KD | |
| RA-signaling molecules | ||||||
| Rara | NSC | NSC | 0.33 | NSC | NSC | 0.40 |
| Rarb | 8.56 | 0.09 | 1.75 | 0.34 | 0.52 | 0.43 |
| Rarg | 1.85 | NSC | NSC | NSC | 0.11 | NSC |
| Aldh1a2 | 0.80 | 1.34 | NSC | NSC | NSC | NSC |
| Crabp2 | 5.25 | NSC | NSC | NSC | 0.15 | 0.63 |
| Cyp26b1 | 104.9 | 0.18 | NSC | NSC | NSC | NSC |
| Tendon markers | ||||||
| Sox9 | 0.33 | 3.17 | NSC | NSC | NSC | NSC |
| Scx | 1.62 | NSC | NSC | NSC | 0.43 | NSC |
| Tnc | 1.89 | 0.49 | 0.62 | NSC | NSC | 0.40 |
| Genes required for diaphragm development | ||||||
| Nr2f2 | NSC | 0.76 | NSC | NSC | NSC | 0.74 |
| Pdgfra | 1.61 | 0.43 | NSC | NSC | 0.52 | NSC |
| Wt1 | 1.83 | 0.49 | NSC | NSC | 0.52 | 0.68 |
| Zfpm2 | 1.26 | 0.59 | NSC | NSC | NSC | 0.76 |
| Gli1 | 0.37 | 1.79 | NSC | NSC | 0.47 | 2.75 |
| Gli2 | NSC | NSC | NT | NT | NT | NT |
| Gli3 | NSC | NSC | NT | NT | NT | NT |
| Lrp2 | NSC | 0.40 | NSC | NSC | NSC | NSC |
| Kif7 | NSC | NSC | NT | NT | NT | NT |
Relative fold change in gene expression in independently treated cultures of primary E13 diaphragmatic cells. RT-qPCR analysis of RA-signaling molecules, tendon markers, and genes associated with abnormal diaphragm development in cultures of primary diaphragmatic cells isolated from WT embryos. Expression changes that were deemed significant (P < 0.05, t test) are plotted as mean relative fold change over control (bold). Expression changes that were deemed not significant (P > 0.05, t test) are plotted as no significant change (NSC). Genes not tested are represented as NT. KD, knockdown.
Inhibition of RA signaling in WT diaphragmatic cells was associated with increased expression of the RA-synthesizing enzyme Aldh1a2 and decreased expression of the RA-metabolizing enzyme Cyp26b1. Kif7 mutant diaphragms have elevated expression of both of these genes, suggesting that the RA pathway is compensating for a defect in RA-RAR signaling. We found that inhibition of the RA nuclear receptors caused increased expression of Gli1, a Gli2 and Gli3 target gene, whereas activation inhibited Gli1 expression. These results suggest that RA-RAR signaling may negatively regulate Gli2 and Gli3 activity in the developing diaphragm.
We next wanted to determine which RA nuclear receptors specifically mediated the induction of Scx, Tnc, Nr2f2, Pdgfra, Wt1, Zfpm2, and Lrp2 in diaphragmatic cells. Hence, we knocked down the expression of Rara, Rarb, Rarg, and Rara and Rarb together under basal conditions using gene-specific siRNAs. We determined that Rara is necessary for Tnc expression and that reduced expression of Rara resulted in increased expression of Rarb. Depletion of Rarg transcripts revealed that Rarg is necessary for the expression of Scx, Pdgfra, Wt1, Rarb, Crabp2, and Gli1. Knockdown of both Rara and Rarb together resulted in decreased expression of Tnc, Nr2f2, Wt1, and Zfpm2 and resulted in a strong up-regulation of Gli1 (Table 1). These data support a model in which RA signaling promotes the differentiation of the central tendon through cooperation of Rara, Rarb, and Rarg.
Kif7 Is Necessary for the RA-Mediated Induction of Scx and Tnc.
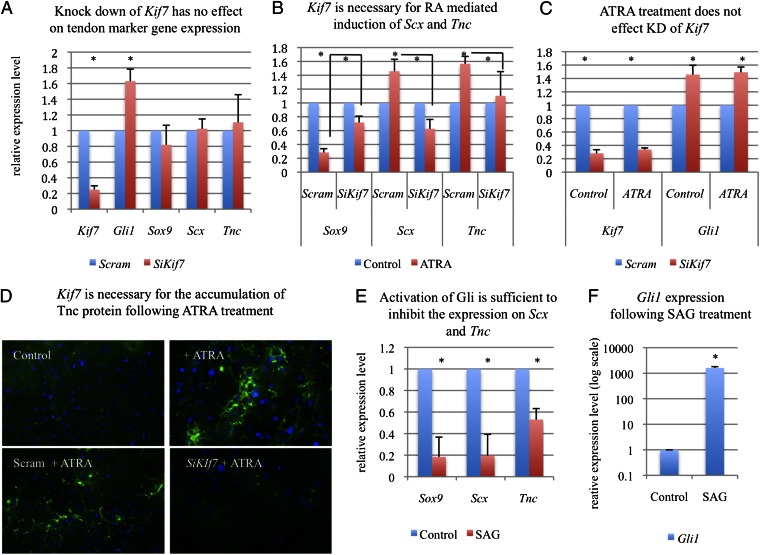
To determine if Kif7 regulates diaphragmatic tendon cell differentiation, we transfected siRNAs against Kif7 into E13 diaphragmatic cells and then assayed the expression of Sox9, Scx, and Tnc. Knockdown of Kif7 had no effect on the expression of these genes; however, it did promote an increase in Gli1 expression (Fig. 4A). Because we observed that RA signaling was perturbed in the developing diaphragm of Kif7dd/dd embryos and that RA-RAR signaling was necessary for Scx expression, we hypothesized that Kif7 was required for RA-mediated induction of Scx and Tnc in diaphragmatic cells. To test this hypothesis, we knocked down Kif7 and then treated diaphragmatic cells with RA for 48 h. RA treatment of control cells (transfected with scrambled siRNA) resulted in an up-regulation of Scx and Tnc and a down-regulation of Sox9 (Fig. 4B). When this experiment was repeated using cells transfected with Kif7-specific siRNA, RA treatment did not change transcript expression levels of Scx, Tnc, or Sox9 (Fig. 4B). Because these experiments evaluated total transcript levels in a mixed cell population, we wanted to determine whether RA signaling promoted the accumulation of Tnc protein within individual tenocytes and whether Kif7 was required for this event. We treated nontransfected, scrambled siRNA-transfected, or Kif7 siRNA-transfected diaphragmatic cell cultures with RA for 72 h and examined the expression of Tnc. Indeed, RA treatment promoted increased accumulation of Tnc protein in WT and scrambled control cells but not Kif7-depleted cells (Fig. 4D). Because altered knockdown efficiency between treatment groups could confound results, Kif7 transcript levels were ascertained in both control and RA-treated groups. Transcript expression was the same between groups, and both groups showed the equal induction of Gli1 expression (Fig. 4C).
Fig. 4.
Kif7 is necessary for RA-mediated induction of Scx and Tnc in primary diaphragmatic cells. (A) RT-qPCR analysis of tendon markers Sox9, Scx, and Tnc in primary E13 diaphragmatic cells transfected with either a scrambled control siRNA or Kif7-specific siRNAs. (B) RT-qPCR analysis of tendon markers in diaphragmatic cells after transfection with either a scrambled control siRNA or Kif7-specific siRNAs and then treatment with 500 nM ATRA for 48 h. (C) RT-qPCR analysis of Kif7 and Gli1 expression in diaphragmatic cells after siRNA transfection with either the scrambled control siRNA or Kif7-specific siRNAs and then treatment with ATRA or vehicle control. Note that there is equal knockdown of Kif7 under both ATRA-treated and control conditions. (D) Tnc immunostaining (green) in diaphragmatic cells after treatment with ATRA, transfection with either a scrambled control siRNA or Kif7-specific siRNAs, and then treatment with 500 nM ATRA for 72 h. (E and F) RT-qPCR analysis of Sox9, Scx, Tnc, and Gli1 expression in diaphragmatic cells after treatment with 150 nM Hh/Smo agonist SAG or vehicle control. All data are means, with error bars representing SE; n = 3–4 independent experiments performed in triplicate (*P < 0.05, t test).
Activation of Gli Is Sufficient to Inhibit the Expression of Scx and Tnc in Diaphragmatic Cells.
In the chick, overexpression of Shh inhibits Scx induction in the developing sclerotome (32). Because Kif7dd/dd mutant diaphragms have both elevated Gli1 expression and decreased Scx induction, we wanted to determine whether misactivation of Gli would alter the expression of Scx and Tnc in diaphragmatic cells. Hence, we treated diaphragmatic cells with the Smo agonist (SAG) for 48 h and examined the expression of Gli1, Sox9, Scx, and Tnc transcripts by quantitative RT-PCR (RT-qPCR) (Fig. 4 E and F). Treatment with SAG led to both a robust induction of Gli1 and a down-regulation of Sox9, Scx, and Tnc expression. These findings show that ligand-independent activation of Gli is sufficient to inhibit the expression of Scx and Tnc in diaphragmatic cells.
Discussion
In this study, we identified a mouse model of syndromic congenital diaphragmatic hernia with a mutation in a gene encoding a cilia-associated kinesin motor protein, Kif7. This discovery allowed us to assign several functions to this important gene and investigate mechanisms of diaphragm development. Kinesin family member genes have been associated with a wide range of diseases (40); however, they have not been recognized in defects of the diaphragm. In our study, we used two independent mutant Kif7 mouse lines to identify diaphragmatic phenotypes that are highly similar to phenotypes seen in some human patients. Our findings both clarify and bring to the forefront a topic of confusion in diaphragm development research (that is, the contribution of potential Hh signaling in diaphragm development). Although a subset of compound Gli mutant mice has diaphragmatic defects (16), neither phenotypic details nor mechanisms has been described. At the same time, evidence for the expression of Hh pathway ligands or receptors in the developing diaphragm has been negative or incomplete (13). In this study, we found that the loss of Kif7 leads to the inappropriate activation of Gli signaling in the developing diaphragm, which was seen by elevated expression of the Gli target gene, Gli1. Previously, Kif7 was shown to negatively regulate the activity of Gli2 and Gli3 transcription factors, and in the absence of Smo (and thus, Hh signaling), the loss of Kif7 can lead to ligand-independent activation of the Hh pathway downstream of Smo (19). Together, these data may suggest that Kif7/Gli cooperate to promote diaphragm development in the absence of the Hh/Ptch signal, possibly through Gli repressor, and that the diaphragm defects present in these mutants are caused by impaired differentiation of the mesenchymal cells within either the primordial diaphragm or the parental cells that give rise to this structure. Future investigations using the Kif7 dd mouse line will allow us to determine the precise role of a Hh pathway in diaphragm development.
We have provided a description of the patterning and organization of the diaphragmatic central tendon, and we determined that Kif7 is required for the differentiation of this structure. To verify this role and investigate the complex requirement of multiple genes, we developed methodology for dissection and culture of primordial diaphragm cells. We determined that RA is a necessary and sufficient inducer of tendon marker gene expression and that Kif7, Rara, Rarb, and Rarg are required for this process. This tendon structure is often compromised in human congenital diaphragmatic hernia (21); however, the causative mechanisms are unclear. It is interesting that the Kif7 mutant diaphragm has a hernia, tissue hyperplasia, and decreased mesenchymal cell differentiation. Previously, diaphragmatic hernias have been associated with a decrease in the mass of the primordial diaphragm (5, 10); however, it is possible that these defects may also arise from increased proliferation with decreased differentiation, because both were observed in our model and both may occur in defects of organogenesis associated with mutations in cilia-related genes (41).
In humans, KIF7 is located 2 Mb from a common 15q26.1–26.2 cytogenetic hotspot for syndromic congenital diaphragmatic hernia. Neither evaluation of candidate gene expression in the primordial diaphragmatic tissue (25) nor resequencing of hotspot candidate genes has identified a probable mechanism or human gene–disease association (42). KIF7 should be considered as a human CDH candidate gene, because it is in close proximity to this region, and transcriptional regulation of genes may occur through long-distance enhancers (43). In other patient cohorts, KIF7 mutations have been associated with developmental defects of the CNS, craniofacial structures, and limb abnormalities (44, 45). Because KIF7 is a large gene with multiple functions, there is potential to associate many different genetic aberrations to a variety of human phenotypes. An association of syndromic CDH with KIF7 mutations would provide evidence that a subset of syndromic diaphragm defects may, indeed, be cilia-related, perhaps justifying investigation as to whether some comorbidities in these patients could be related to respiratory or primary ciliary dysfunction.
Materials and Methods
Mouse Strains.
A/J (JAX 000646) and FVB/NJ (JAX 001800) mice were purchased from Jackson Laboratory. Kif7Maki/Maki (MGI: 4355980) mutant embryos were provided by Kathryn V. Anderson (Developmental Biology Program, Sloan Kettering Institute, New York, NY). All animal studies were performed in accordance with the guidelines set by the University Committee on Animal Resources at the University of Rochester Medical Center.
Identification of the dd Mutation.
ENU mutagenesis injections, breeding, and screening were performed using techniques developed to identify autosomal recessive mutations that cause human birth defect phenotypes (22, 36). Screening was conducted on third-generation embryos from 32 G1 families derived from 12 injected G0 A/J mice. Genomic DNA from six phenotypically mutant embryos was used for mapping with the Illumina mouse medium density SNP chip. The dd mutation mapped to chromosome 7 between SNPs rs3719311 (35.0 Mb) and rs3673653 (114.2 Mb). Sequencing of candidate gene exons and flanking genome in the region revealed one homozygous mutation in the Kif7 gene. The dd mutation is an A-to-T nonsense mutation that created a premature stop in exon 8. To confirm that the dd mutation generates a truncated protein, an N-terminal GFP-tagged fusion construct was generated by cloning both the WT Kif7 and Kif7dd/dd cDNAs into the pEGFP-C3 vector (Clontech). Primer sequences used were as follows: forward-CGGGTACCGATGGGGCTGGAGGCCC and reverse-GCGGGGCGTCTAGAGTACAAGGGGT. Either pEGFP-Kif7 or pEGFP-Kif7dd/dd was transfected into HEK293T cells using X-tremeGENE HP (Roche). Cell extracts were analyzed by Western blots using standard methods.
Establishment of Primary Mesenchymal Diaphragmatic Cultures.
Whole diaphragms were microdissected from E13.5 FVB/NJ embryos. Tissue was resuspended in 1× trypsin EDTA (Gibco), minced into 1 mM pieces, and proteolytically digested for 30 min at 37 °C to generate single-cell suspensions. Cells were seeded at 5.0 × 104 cells/mL in 12-well dishes and cultured in DMEM (Gibco) with 10% (vol/vol) FBS (HyClone) and 1× antibiotic-antimycotic (Gibco) until confluent. Cultures were then treated independently with vehicle control, 150 nM SAG (Santa Cruz), 500 nM ATRA (Sigma), 1 μM BMS493 (Sigma), 100 nM Rara siRNA, 100 nM Rarb siRNA, 100 nM Rarg siRNA, or 75 nM Rara and 75 nM Rarb siRNAs (Dharmacon) together. RT-qPCR and siRNA transfections were performed using standard techniques. Details are provided in SI Materials and Methods.
Skeletal Preparations, Histology, in Situ Hybridization, and Immunostaining.
E18.5 mouse skeletal preparations were performed using standard techniques. For histology, embryos were fixed in 4% (vol/vol) paraformaldehyde, embedded in paraffin wax, and sectioned at 7 μM. Sections were stained with H&E or processed for in situ hybridization using standard methods. Whole-mount in situ hybridization and immunostaining were performed on fixed whole diaphragms using the methods previously described in refs. 41 and 46. A description of how the Kif7 in situ plasmid was created and a list of antibodies and dilutions used in this report can be found in SI Materials and Methods.
Detection of Cell Proliferation.
To detect cell proliferation within the primordial diaphragm, pregnant females were injected i.p. on E12.5 with 50 μg/g body weight BrdU (Sigma); 1 h later, dams were killed, and embryos were harvested and processed for coimmunofluorescent staining for Wt1 and BrdU. In control and mutant embryos, comparable sections were selected between the right posthepatic mesenchymal plate and the medial diaphragm at the esophageal mesentery. Mesothelial Wt1+ and nonmesothelial Wt1+ mesenchymal cell counts were recorded separately in at least six consecutive sections from each embryo. Immunopositive cells were counted in at least three embryos using ImageJ software. Cell counts were normalized to control counts to provide a relative rate of cell proliferation, and statistical significance was determined as described below.
Statistical Analysis.
All experiments were performed in at least three biological replicates, and statistical analysis was performed by the two-tail t test. Data are plotted as mean ± SE and were considered statistically significant at levels of P < 0.05.
Supplementary Material
Acknowledgments
We thank Nian Zhang and Laurel Baglia for technical advice and comments regarding this manuscript. We also thank Kathryn V. Anderson and Mu He for providing Kif7Maki/Maki mice and giving advice regarding our work. G.L.C. is supported as a doctoral student by National Institutes of Health T32 Training Grant HL066988. This work was supported by a Buswell Fellowship Award at The University of Rochester School of Medicine and Dentistry and National Institutes of Health Grant R01 HL085459 (to K.G.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222797110/-/DCSupplemental.
References
- 1.Beurskens N, Klaassens M, Rottier R, de Klein A, Tibboel D. Linking animal models to human congenital diaphragmatic hernia. Birth Defects Res A Clin Mol Teratol. 2007;79(8):565–572. doi: 10.1002/bdra.20370. [DOI] [PubMed] [Google Scholar]
- 2.Menon SC, et al. Clinical characteristics and outcomes of patients with cardiac defects and congenital diaphragmatic hernia. J Pediatr. 2013;162(1):114–119. doi: 10.1016/j.jpeds.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 3.Ackerman KG, Pober BR. Congenital diaphragmatic hernia and pulmonary hypoplasia: New insights from developmental biology and genetics. Am J Med Genet C Semin Med Genet. 2007;145C(2):105–108. doi: 10.1002/ajmg.c.30133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady PD, et al. Recent developments in the genetic factors underlying congenital diaphragmatic hernia. Fetal Diagn Ther. 2011;29(1):25–39. doi: 10.1159/000322422. [DOI] [PubMed] [Google Scholar]
- 5.Iritani I. Experimental study on embryogenesis of congenital diaphragmatic hernia. Anat Embryol (Berl) 1984;169(2):133–139. doi: 10.1007/BF00303142. [DOI] [PubMed] [Google Scholar]
- 6.Mayer S, Metzger R, Kluth D. The embryology of the diaphragm. Semin Pediatr Surg. 2011;20(3):161–169. doi: 10.1053/j.sempedsurg.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am J Med Genet C Semin Med Genet. 2007;145C(2):109–116. doi: 10.1002/ajmg.c.30128. [DOI] [PubMed] [Google Scholar]
- 8.Yuan W, et al. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc Natl Acad Sci USA. 2003;100(9):5217–5222. doi: 10.1073/pnas.0730709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allan DW, Greer JJ. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J Comp Neurol. 1997;382(4):459–468. doi: 10.1002/(sici)1096-9861(19970616)382:4<459::aid-cne3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Babiuk RP, Greer JJ. Diaphragm defects occur in a CDH hernia model independently of myogenesis and lung formation. Am J Physiol Lung Cell Mol Physiol. 2002;283(6):L1310–L1314. doi: 10.1152/ajplung.00257.2002. [DOI] [PubMed] [Google Scholar]
- 11.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 12.Rhinn M, Dollé P. Retinoic acid signalling during development. Development. 2012;139(5):843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 13.Kantarci S, et al. Characterization of the chromosome 1q41q42.12 region, and the candidate gene DISP1, in patients with CDH. Am J Med Genet A. 2010;152A(10):2493–2504. doi: 10.1002/ajmg.a.33618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantarci S, et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat Genet. 2007;39(8):957–959. doi: 10.1038/ng2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasutto F, et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80(3):550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim PC, Mo R, Hui Cc C. Murine models of VACTERL syndrome: Role of sonic hedgehog signaling pathway. J Pediatr Surg. 2001;36(2):381–384. doi: 10.1053/jpsu.2001.20722. [DOI] [PubMed] [Google Scholar]
- 17.Lohnes D, et al. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120(10):2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- 18.Chen MH, MacGowan A, Ward S, Bavik C, Greer JJ. The activation of the retinoic acid response element is inhibited in an animal model of congenital diaphragmatic hernia. Biol Neonate. 2003;83(3):157–161. doi: 10.1159/000068932. [DOI] [PubMed] [Google Scholar]
- 19.Liem KF, Jr, He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA. 2009;106(32):13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung HO, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2(76):ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 21.Ackerman KG, et al. Congenital diaphragmatic defects: Proposal for a new classification based on observations in 234 patients. Pediatr Dev Pathol. 2012;15(4):265–274. doi: 10.2350/11-05-1041-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stottmann RW, Beier DR. Using ENU mutagenesis for phenotype-driven analysis of the mouse. Methods Enzymol. 2010;477:329–348. doi: 10.1016/S0076-6879(10)77017-8. [DOI] [PubMed] [Google Scholar]
- 23.Holder AM, et al. Genetic factors in congenital diaphragmatic hernia. Am J Hum Genet. 2007;80(5):825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clugston RD, et al. Teratogen-induced, dietary and genetic models of congenital diaphragmatic hernia share a common mechanism of pathogenesis. Am J Pathol. 2006;169(5):1541–1549. doi: 10.2353/ajpath.2006.060445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clugston RD, Zhang W, Greer JJ. Gene expression in the developing diaphragm: Significance for congenital diaphragmatic hernia. Am J Physiol Lung Cell Mol Physiol. 2008;294(4):L665–L675. doi: 10.1152/ajplung.00027.2008. [DOI] [PubMed] [Google Scholar]
- 26.Kreidberg JA, et al. WT-1 is required for early kidney development. Cell. 1993;74(4):679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 27.Hsu SH, et al. Kif7 promotes hedgehog signaling in growth plate chondrocytes by restricting the inhibitory function of Sufu. Development. 2011;138(17):3791–3801. doi: 10.1242/dev.069492. [DOI] [PubMed] [Google Scholar]
- 28.Li ZJ, et al. Kif7 regulates Gli2 through Sufu-dependent and -independent functions during skin development and tumorigenesis. Development. 2012;139(22):4152–4161. doi: 10.1242/dev.081190. [DOI] [PubMed] [Google Scholar]
- 29.Schweitzer R, Zelzer E, Volk T. Connecting muscles to tendons: Tendons and musculoskeletal development in flies and vertebrates. Development. 2010;137(17):2807–2817. doi: 10.1242/dev.047498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pryce BA, et al. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development. 2009;136(8):1351–1361. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol. 2006;298(1):234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113(2):235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 33.Dranse HJ, Sampaio AV, Petkovich M, Underhill TM. Genetic deletion of Cyp26b1 negatively impacts limb skeletogenesis by inhibiting chondrogenesis. J Cell Sci. 2011;124(Pt 16):2723–2734. doi: 10.1242/jcs.084699. [DOI] [PubMed] [Google Scholar]
- 34.Russell MK, et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci USA. 2012;109(8):2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clugston RD, Zhang W, Alvarez S, de Lera AR, Greer JJ. Understanding abnormal retinoid signaling as a causative mechanism in congenital diaphragmatic hernia. Am J Respir Cell Mol Biol. 2010;42(3):276–285. doi: 10.1165/rcmb.2009-0076OC. [DOI] [PubMed] [Google Scholar]
- 36.Ackerman KG, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1(1):58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew SJ, et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138(2):371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanamori-Katayama M, et al. LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS One. 2011;6(10):e25391. doi: 10.1371/journal.pone.0025391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Germain P, et al. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006;58(4):712–725. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- 40.Mandelkow E, Mandelkow EM. Kinesin motors and disease. Trends Cell Biol. 2002;12(12):585–591. doi: 10.1016/s0962-8924(02)02400-5. [DOI] [PubMed] [Google Scholar]
- 41.Gerbe F, et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192(5):767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klaassens M, et al. Congenital diaphragmatic hernia and chromosome 15q26: Determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet. 2005;76(5):877–882. doi: 10.1086/429842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lettice LA, et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 44.Putoux A, et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet. 2011;43(6):601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dafinger C, et al. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121(7):2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.