Abstract
Germ-line mutations in PALB2 lead to a familial predisposition to breast and pancreatic cancer or to Fanconi Anemia subtype N. PALB2 performs its tumor suppressor role, at least in part, by supporting homologous recombination-type double strand break repair (HR-DSBR) through physical interactions with BRCA1, BRCA2, and RAD51. To further understand the mechanisms underlying PALB2-mediated DNA repair and tumor suppression functions, we targeted Palb2 in the mouse. Palb2-deficient murine ES cells recapitulated DNA damage defects caused by PALB2 depletion in human cells, and germ-line deletion of Palb2 led to early embryonic lethality. Somatic deletion of Palb2 driven by K14-Cre led to mammary tumor formation with long latency. Codeletion of both Palb2 and Tumor protein 53 (Trp53) accelerated mammary tumor formation. Like BRCA1 and BRCA2 mutant breast cancers, these tumors were defective in RAD51 focus formation, reflecting a defect in Palb2 HR-DSBR function, a strongly suspected contributor to Brca1, Brca2, and Palb2 mammary tumor development. However, unlike the case of Brca1-mutant cells, Trp53bp1 deletion failed to rescue the genomic instability of Palb2- or Brca2-mutant primary lymphocytes. Therefore, Palb2-driven DNA damage control is, in part, distinct from that executed by Brca1 and more similar to that of Brca2. The mechanisms underlying Palb2 mammary tumor suppression functions can now be explored genetically in vivo.
Keywords: mouse model, familial breast cancer
Partner and Localizer of BRCA2 (PALB2) is a breast cancer susceptibility gene. Its product was identified as a major interacting protein of the BReast CAncer susceptibility gene product 2, BRCA2 (1). This interaction is required for the repair of DNA double strand breaks (DSBs) by homologous recombination (HR) because PALB2 is necessary for the chromatin association of BRCA2 and its partner, RAD51 (1). RAD51 is the central recombinase in HR, and it participates in D-loop formation and strand displacement (2). PALB2 also plays a BRCA2-independent role in the HR process by enhancing RAD51 function (3, 4).
PALB2 interacts with both BRCA1 and BRCA2 and mediates the long-known interaction between these proteins (5, 6). Loss of PALB2 does not affect BRCA1 recruitment to irradiation-induced foci (IRIF) but abrogates colocalization of BRCA2 and RAD51 at these structures (1, 5). Genetic analyses have shown that, like BRCA2, a member of Fanconi anemia complementation group D1, PALB2 is also the Fanconi anemia complementation group N protein (FANCN) (7, 8). PALB2 is also a breast cancer suppressor protein in its own right (9–12). Unlike BRCA1 and BRCA2 mutant tumors, only some PALB2-associated breast cancers have undergone loss of PALB2 heterozygosity (LOH) (9, 10). This finding implies that a reduction of PALB2 gene copy number might be sufficient to allow breast cancer development in some, but not all, settings. Why this difference exists is an open question.
Breast cancer in PALB2-mutated families is of intermediate penetrance, unlike that in BRCA1/2 families (10, 12). Although PALB2 mutations are rarer than BRCA1/2 mutations, available clinical data suggest that heterozygous, germ-line PALB2 mutations do not precisely phenocopy either BRCA1 or BRCA2 cancer predisposition syndromes (9, 10). This finding is consistent with the notion that PALB2 biological functions extend beyond simply enabling BRCA1–BRCA2 complex formation. PALB2 also interacts with MRG15 (also known as MORF4L1) (13), a subunit of histone acetyl transferase/deacetylase complexes, and with KEAP1, a major regulator of the antioxidant transcription factor NRF2 (also known as NFE2L2) (14). In addition, PALB2 contains a highly conserved, chromatin-associated domain (ChAM) for which no binding partners are known (15). The contribution of these PALB2 binding partners and of the ChAM domain to the BRCA1-PALB2-BRCA2 HR machinery and/or to PALB2’s cancer suppression function is unclear. Thus, it is conceivable that PALB2 exerts multiple functions that extend beyond its known role in HR-mediated double strand break repair.
To date, it has been difficult to study the molecular pathogenesis of PALB2 breast cancer in detail because of the lack of a genetically engineered mouse model that recapitulates the human disease. Thus, we have generated a model of Palb2 breast cancer in the mouse and have documented its most salient properties. An analogous model was recently generated by others (16).
Results and Discussion
Targeting the Mouse Palb2 Gene and Generation of Palb2-Deficient ES Cells.
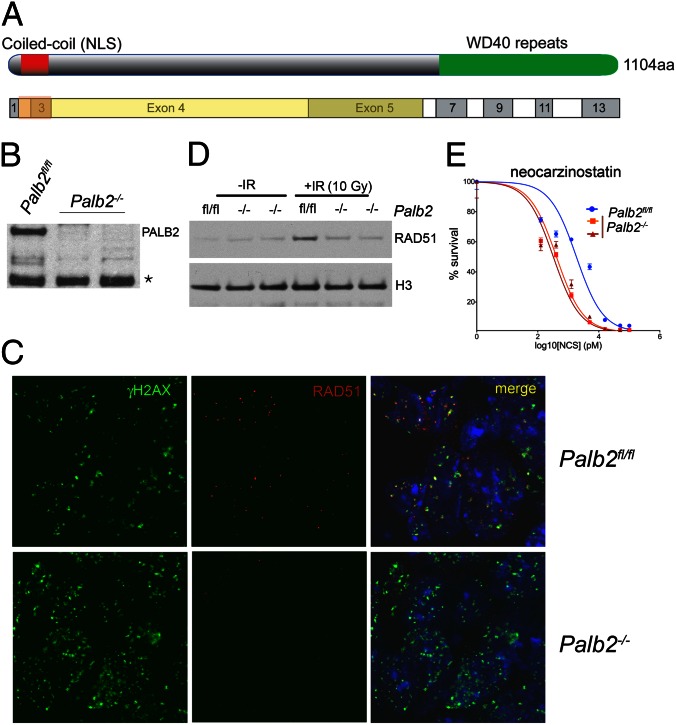
To generate a Palb2 allele that could be conditionally inactivated upon Cre recombinase expression, we inserted loxP sites flanking exons 2 and 3 of the Palb2 gene (Fig. S1 A and B). These exons encode a putative nuclear localization signal sequence and the PALB2 coiled-coil domain (Fig. 1A, Fig. S1A). The latter mediates the PALB2 interaction with BRCA1 (5, 6). Deletion of these exons would result in out-of-frame reading of exon 4 and premature termination of the PALB2 translation before the BRCA2-interacting, seven-bladed WD40-type β-propeller domain (Fig. 1A, Fig. S1A). Due to premature truncation of the Palb2 ORF, the resulting transcript is also a candidate for degradation via nonsense-mediated decay.
Fig. 1.
Conditional gene targeting of mouse Palb2. (A) Schematic representation of Palb2 domains and the exons from which they are encoded. The yellow area corresponds to the frameshifted ORF that results from recombination of the inserted loxP recombination sites. (B) Western blot analysis for PALB2 isolated in chromatin-enriched extracts (S420) of three independent ES cell lines. The full-length mouse PALB2 protein is ∼120 kDa. A nonspecific background band is indicated by an asterisk and can be used as an internal loading control. (C) Recruitment of RAD51 to DSBs marked by γH2AX IRIF 2 h after exposure of Palb2fl/fl and Palb2−/− ES cells to to 5 Gy of ionizing radiation (IR). (D) Western blot analysis of chromatin-bound (S420) RAD51 in Palb2fl/fl and Palb2−/− ES cells that received 10 Gy of IR and the respective unirradiated control cells. Histone H3 was used as a loading control. (E) Dose–response curves of Palb2fl/fl and Palb2−/− ES cells after exposure to increasing concentrations of neocarzinostatin.
Targeting of the Palb2 locus and integration of both loxP recombination sites was confirmed by Southern blot analysis (Fig. S1C). Heterozygous ES cells (Palb2neo/+) were injected into blastocysts, and the resulting chimeras from two individual clones were bred to either Flp-deleter mice (to eliminate the Frt-flanked neomycin resistance cassette and generate a conditional allele) or Cre-deleter mice (to generate a conventional Palb2 KO allele). Germ-line transmission of the Palb2neo allele occurred from nearly all chimeras, and mice were successfully genotyped for the Flp- and the Cre-recombined alleles (Palb2fl or Palb2−, respectively).
We attempted to derive ES cells from the Palb2− allele. Three, independent Palb2 ES cell lines were derived from a single, heterozygous Palb2fl/− cross. Expression analysis of Palb2 mRNA by quantitative real-time RT-PCR (qRT-PCR) confirmed that one of these ES lines was Palb2fl/fl and the other two were Palb2−/− (Fig. S1D). The loss of full-length Palb2 expression from these lines was further confirmed by Western blotting, using a polyclonal anti-mouse PALB2 antibody raised against the N-terminal 200 residues of the mouse PALB2 protein (Fig. 1B). Therefore, Palb2 loss did not prevent ES cell derivation and subsequent survival. The three ES cell lines we derived were morphologically comparable, proliferated at the same rate as wild-type (WT) ES cells and were capable of differentiation into embryoid bodies.
ES cells that are deficient for Brca1 or Brca2 have been notoriously difficult to isolate and are severely compromised in their proliferation (17, 18). In keeping with these findings, Palb2−/− ES cells could not be derived from embryos carrying a conventional PALB2 gene-trap allele (19). Because PALB2 operates immediately upstream of BRCA2 and is required for BRCA2 localization at DNA double strand breaks, it is possible that the viability and robustness of our Palb2−/− ES cells were due to residual expression of a truncated PALB2 species. Such a polypeptide could, in theory, result from translation initiation downstream of the engineered Palb2 genomic deletion. No such truncated protein was detected with our polyclonal antibody (Fig. 1B).
To test whether the conditional gene targeting approach that was used had generated an allele that would be rendered null after Cre action, the response of Palb2fl/fl and Palb2−/− ES cells to DNA damaging agents that cause double strand breaks was analyzed. Normally, exposure of PALB2-proficient cells to ionizing radiation (IR) leads to the formation of phosphorylated histone H2A.X (γH2AX) nuclear foci and subsequent recruitment of BRCA1, BRCA2, and RAD51 to these structures. As expected, after exposure to IR, γH2AX and BRCA1 IRIF formation was unaffected in Palb2−/− cells (Fig. 1C, Fig. S2A). However, the recruitment of RAD51 was severely compromised (Fig. 1C). This defect was also evident at the biochemical level because no increase in chromatin loading of RAD51 after IR could be detected in nuclear extracts of IR-treated Palb2−/− cells (Fig. 1D).
Because biallelic PALB2 mutations in humans cause Fanconi anemia, a hallmark of which is increased sensitivity to DNA cross-linking agents such as mitomycin C (MMC), the sensitivity of Palb2−/− ES cells to MMC as well other DNA damaging agents was assayed. Both Palb2−/− ES lines displayed increased sensitivity to MMC, IR, and the radiomimetic drug, neocarzinostatin (Fig. 1E, Fig. S2 B and C). These findings further imply that these Palb2−/− cells are functional KOs for Palb2 because they are compromised in multiple, known Palb2-associated functions. Thus, upon Cre-mediated recombination in vivo, the aforementioned conditional Palb2 allele appears to be converted to a Palb2-null allele.
Loss of Palb2 in the Germ Line Results in Early Embryonic Lethality.
Germ-line deletion of Brca1 or Brca2 results in early embryonic lethality (17, 20, 21). Although Palb2−/− ES cells displayed no apparent growth defects compared with Palb2fl/fl controls, Palb2 loss could still be deleterious in differentiated progeny cells, and thereby negatively affect mouse development. Indeed, we were unable to obtain Palb2−/− mice from heterozygous crosses (Fig. S3A), consistent with previous reports (19, 22). Dissection of embryos from timed pregnancies revealed that Palb2-null embryos could be recovered only up to E12.5, but even then at sub-Mendelian ratios. These embryos repeatedly exhibited severe malformations. At earlier time points, morphological aberrations of Palb2−/− embryos were less obvious. However, these mutant embryos were clearly smaller than WT or heterozygous littermate embryos (Fig. S3B), and some displayed exencephaly as well as malformations of the placental labyrinth and yolk sac-associated blood islets (Fig. S3 C–F). The fact that Palb2 nullizygosity resulted in embryonic lethality detectable at E8.5–E10.5 is consistent with earlier reports showing that homozygous Palb2-deficient mice also die during embryogenesis at ∼E8.5 (19, 22).
Embryonic lethality due to loss of Brca1 or Brca2 can be delayed by concomitant loss of P53 (encoded by Trp53) or the CDK inhibitor p21 (encoded by the Cdkn1a gene) (23), (24). Trp53 loss also delayed the lethality of Palb2 KO embryos, which otherwise exhibited increased p21 abundance (22). We therefore tested whether loss of p21 expression affects Palb2−/− embryonic lethality by generating Palb2; Cdkn1a−/− embryos. As expected, loss of p21 expression did delay embryonic lethality of Palb2 KO embryos by 2–3 d (Fig. S3A). However, all Palb2/Cdkn1a double KO embryos still displayed multiple malformations and impaired growth compared with Palb2 heterozygous or WT littermates and were eventually resorbed. Therefore, these Palb2−/− embryonic rescue effects were incomplete and failed to suppress embryonic lethality.
Because the establishment of the placenta and onset of embryonic hematopoiesis are critical steps in development that take place around the time of lethality of Palb2 embryos, we asked whether the lethality of Palb2 KO embryos could be bypassed by a WT placenta. To this end, we used the Palb2fl/fl and Palb2−/− ES cells we had generated to perform tetraploid complementation assays. In this assay, diploid KO ES cells are aggregated to tetraploid WT blastocysts to generate KO embryos that are supported by a WT placenta because the tetraploid WT blastomeres are still capable of forming a placenta but cannot contribute to the embryo.
We found that embryos derived from the Palb2−/− ES cells were already underdeveloped and malformed at E9.5 compared with their Palb2fl/fl counterparts (Fig. S3 I and J). At E12.5, embryos derived from Palb2fl/fl ES cells appeared normal whereas embryos from Palb2−/− ES cells had been resorbed (Fig. S3 K and L). Likewise, breeding of the Palb2fl/fl conditional allele to Meox2-Cre knock-in (KI) mice (in which Cre is expressed from the endogenous Meox2 locus only in the embryo proper and not in the placenta or extraembryonic tissues) (25) only yielded viable mice in which the Palb2 deletion was incomplete. Collectively, these findings indicate that Palb2 is an essential gene during development, and its deficiency in the embryo proper is incompatible with life. These findings are analogous to previous results showing that the lethality of Brca1−/− embryos could not be rescued by tetraploid complementation assay (20).
Palb2 Is a Breast Tumor Suppressor in Mice.
To assess the effect of Palb2 loss-of-function on mammary tumorigenesis, we crossed Palb2fl/fl mice with keratin 14 promoter-driven Cre (K14-Cre) transgenic mice (26). K14-Cre transgenic animals preferentially express Cre recombinase in the basal epithelium of the mammary ducts, as well as in skin and oral mucosa. K14-Cre has previously been used to model murine Brca1 and Brca2 mammary tumorigenesis (27).
PALB2, like BRCA1 and -2, appears to be a breast cancer suppressor in humans (9, 10, 28). Therefore, in an effort to develop a tractable system for studying how Palb2 operates in this regard, we set out to develop a Palb2 mouse breast cancer model. Mammary tumor formation initiated by BRCA1 or BRCA2 loss requires concomitant loss of functional p53 (encoded by Trp53 in mice) (27, 29). This observation was considered in efforts to establish a Palb2 model.
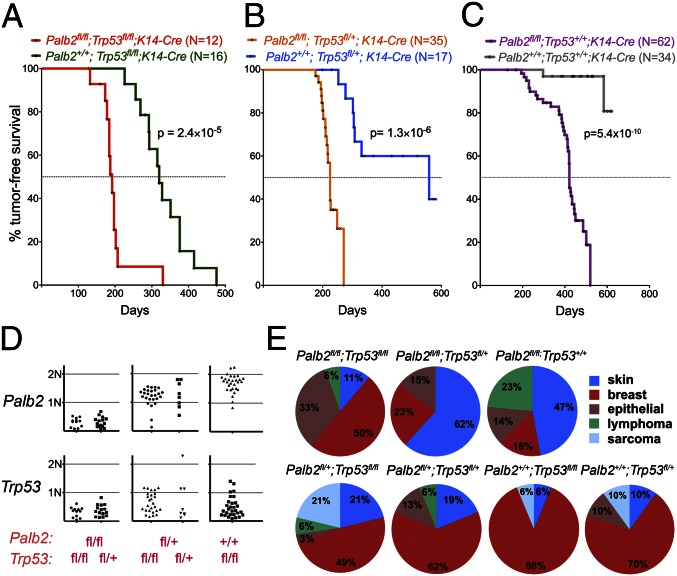
We first generated Palb2/Trp53 double conditional mice by crossing Palb2fl/fl; K14-Cre transgenic mice with Trp53 conditional mice. All mice that harbored the conditional alleles for Palb2 and/or Trp53 in the absence of K14-Cre were phenotypically normal, fertile, and capable of nursing their litters. During the period of tumor monitoring (up to 600 d after birth), Trp53fl/fl; K14-Cre female mice developed spontaneous mammary tumors with a frequency of ∼80% and a mean tumor-free interval (T1/2) of 320 d. By contrast, Palb2fl/fl; Trp53fl/fl; K14-Cre double conditional mice developed tumors much faster (T1/2 = 192 d, P = 2.4 × 10−5), indicating that Palb2 loss accelerates tumor formation on a Trp53-null background (Fig. 2A). These latencies are comparable with Brca2fl/f; Trp53fl/fl; K14-Cre and Brca1fl/fl; Trp53fl/fl; K14-Cre mice (T1/2 = 181 and 213 d, respectively) (27, 30).
Fig. 2.
Tumor formation in Palb2fl/fl conditional mice. (A–C) Kaplan–Meier curves display that Palb2 loss accelerates tumor formation both on a Trp53-conditional null background (A), on a Trp53-conditional heterozygous background (B), and on a Trp53 WT background (C). (D) Gene dosages of Palb2 (Upper) and Trp53 (Lower) in mammary tumors derived from Palb2/Trp53 double conditional mouse cohorts. The germ-line Palb2 and Trp53 genotypes of the mice are indicated in red below the graphs. (E) Spectrum of tumors arising in mouse cohorts with different combinations of Palb2 and Trp53 alleles. The genotypes of the mice are shown above the graphs.
Somatic loss of one Trp53 allele displayed, as expected, a haploinsufficient tumor suppressor phenotype (27, 30), given that Palb2fl/fl; Trp53fl/+; K14-Cre mice developed tumors significantly faster than Palb2fl/fl; Trp53+/+; K14-Cre mice (T1/2 = 225 d vs. 420 d, respectively, P = 2.5 × 10−12, Fig. S4C). Palb2 loss of function also accelerated tumor formation on a Trp53 heterozygous (fl/+) background, again reflecting the genetic interaction of these two genes (Fig. 2B).
K14-Cre-mediated loss of Palb2 and Trp53 led, predominantly, to tumor formation in breast, skin, and oral mucosa (Fig. 2E), as previously reported for K14-Cre-driven Brca1 and Brca2 cancers (27, 30–32). All of the mammary cancers were estrogen and progesterone receptor (ER/PR)-negative, basal-like (Fig. S4 D–F), much like the BRCA1 and -2 tumors generated by K14-Cre (30, 32). Although most of the tumors found in Palb2+/+;Trp53fl/fl;K14-Cre mice were breast carcinomas, Palb2/Trp53 compound KO mice displayed an expanded spectrum of tissues affected by tumors (Fig. 2E), suggesting that combined loss of PALB2 and P53, possibly due to some expression of Cre in other tissues, also results in tumor formation that is not restricted to the mammary gland.
Mice harboring conditional alleles for Palb2 and Trp53, but no K14-Cre transgene, and Palb2+/+; Trp53+/+; K14-Cre mice did not display overt tumor formation during the observation period (Fig. 2C), despite the intrinsic mutagenic activity of Cre in mammalian cells (31, 33). Therefore, tumor formation in the above-noted experiments is a product of targeted gene deletion.
All tumors from Palb2fl/fl; Trp53fl/fl; K14-Cre mice (n = 12) had lost both copies of Palb2 and Trp53 (Fig. 2D). Similarly, in all tumors from Palb2fl/fl; Trp53fl/+; K14-Cre mice (n = 15), the conditional Palb2 and Trp53 alleles were recombined. The WT copy of Trp53 was also lost in most tumors, probably through LOH.
Early reports describing a lack of PALB2 LOH in clinical tumor samples from heterozygous patients suggested that PALB2 could be a haploinsufficient tumor suppressor in humans whereas other reports showed that multiple PALB2 tumors revealed PALB2 LOH, implying that the PALB2 tumor formation process is not uniform (9, 34, 35). In our experimental setting, no haploinsufficiency for tumor suppression was observed for Palb2, as indicated by the comparable latency in Palb2fl/+; Trp53fl/fl; K14-Cre and Palb2+/+; Trp53fl/fl; K14-Cre tumor development (P = 0.46, Fig. S4A). Similarly, when compared on a Trp53fl/+; K14-Cre background, cohorts of Palb2+/+ and Palb2fl/+ mice developed tumors with similar latency and frequency (P = 0.96, Fig. S4B), implying that heterozygous Palb2 loss of function did not contribute to tumor formation.
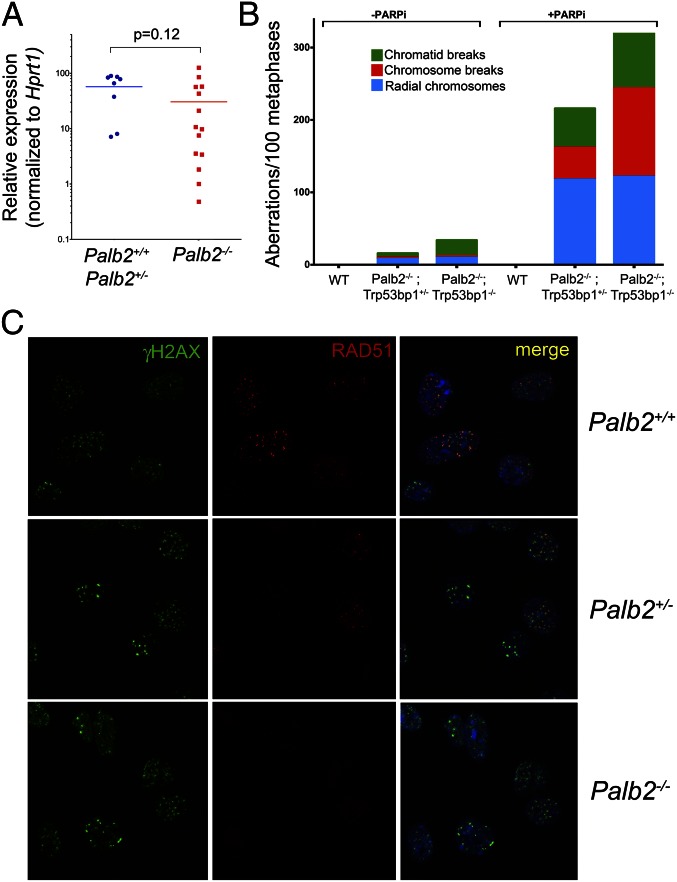
Moreover, Palb2 heterozygous mouse breast tumor lines displayed proper RAD51 localization at IRIF, consistent with preserved HR function (see Fig. 4C). By contrast, Palb2−/− breast tumor cell lines displayed the same defect in RAD51 accumulation observed in Palb2-null primary cells (see Fig. 4C). These findings suggest a role for HR deficiency in the genesis of Palb2 tumors, a state that is not compatible with retention of a functional copy of the gene. Moreover, an analysis of tumors that arose in Palb2fl/+; Trp53fl/fl; K14-Cre mice implies that a significant fraction of these tumors retained at least one copy of PALB2, suggesting that loss of one copy of the gene did not contribute to tumor formation in this model (Fig. 2D).
Fig. 4.
The HR defect in Palb2-deficient cells and tumors. (A) qRT-PCR for Trp53bp1 mRNA in freshly isolated tumor samples that are either Palb2-proficient (+/+ and +/−, n = 8) or Palb2-deficient (−/−, n = 14). Horizontal lines represent the average relative expression value and the P value associated to this comparison (Mann–Whitney U test) is indicated. (B) Acute chromosomal damage and genome instability observed in chromosome spreads following PARPi treatment that are not rescued by Trp53bp1 deletion in Palb2fl/fl;CD19-Cre B lymphocytes. (C) Established Palb2/Trp53-deficient breast tumor cell lines (example shown in the Bottom panels) reveal a defect in the recruitment of RAD51 to IRIF whereas Palb2 heterozygosity does not impair the proper IRIF localization of RAD51 in breast tumor lines (Middle panels). Both should be compared with a Palb2 WT control breast tumor line (Top panels).
Although we observed long latency tumors only in Palb2fl/fl; K14-Cre mice on a WT Trp53 background (T1/2 = 420 d), tumor formation was nonetheless highly significant compared with Palb2+/+; Trp53+/+; K14-Cre controls (P = 5.4 × 10−10, Fig. 2C). The majority of these tumors were small lesions in the head and neck, and a few were mammary tumors. All of these mammary tumors were Palb2−/−, and all displayed either mutations in or loss of Trp53 by LOH.
The finding that loss of Palb2 alone is sufficient to induce long latency tumor formation contrasts with most Brca1 and Brca2 mouse models in which significant numbers of these tumors could not be detected, unless Trp53 was codeleted (27, 30, 36, 37). One explanation for this finding is that somatic Palb2 loss might be better tolerated than somatic Brca1/2 loss on a Trp53 WT background. This hypothesis fits with the finding that Palb2 nullizygosity gives rise to a less severe phenotype in ES cells than biallelic Brca1 or Brca2 loss (17, 18, 27, 38).
Genomic Features of Palb2/Trp53-Deficient Mammary Tumors.
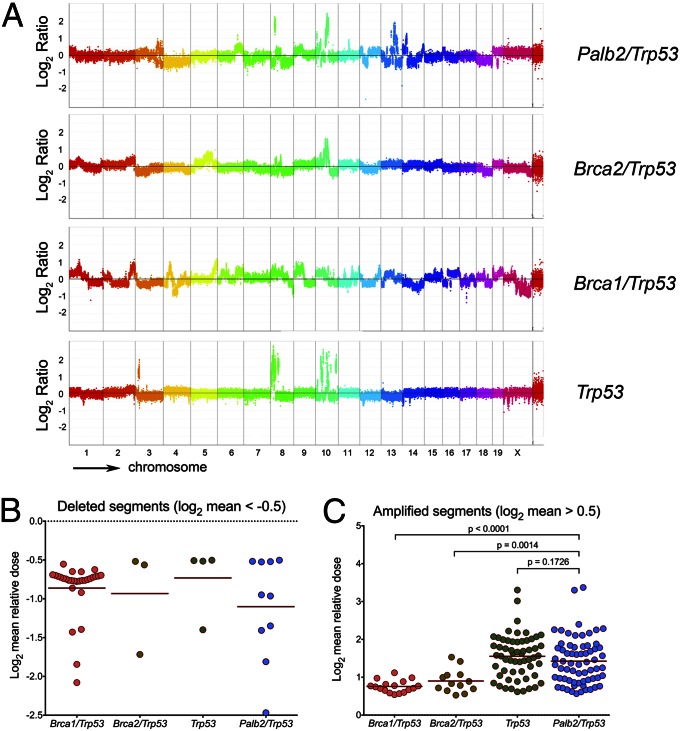
Genomic instability is a hallmark of human cancer, and it promotes tumor initiation and progression. Experimental mouse tumor models have recapitulated this aspect of human tumorigenesis (39). To gain insight into the genomic structures of the tumors that arose due to the loss of Palb2, we performed high-resolution comparative genomic hybridization (CGH) (40) analysis of Palb2/Trp53, Brca1/Trp53, Brca2/Trp53, and Trp53 only-deficient mammary tumors that arose in K14-Cre mice (Fig. 3 A–C).
Fig. 3.
CGH analysis of Palb2 tumors. (A) Control (spleen) and tumor DNAs were hybridized to whole genome arrays to determine regions of loss or gains in mouse breast tumor samples. Representative rainbow graphs for each tumor genotype showing log2 mean DNA relative dose ratio (tumor/spleen) across the entire genome are presented. Each dot represents the average signal from 10 or more consecutive probes (or a segment of ∼40 kb of genomic DNA). The amplitude of the data points above or below the midline indicates the extent of loss/gain in each segment, respectively. (B and C) Scatter plot graphs indicating the relative dose of the deleted segments (log2 < −0.5, B) and amplified segments (log2 > 0.5, C), each dot representing one segment. The number of dots represents the number of segments for all tumors of the relevant genotype, and the number of tumors analyzed (n) for Brca1/Trp53, Brca2/Trp53, Trp53-only, and Palb2/Trp53 tumors were 4, 5, 7, and 8, respectively. The horizontal lines are the average segment dose per genotype, and the P values displayed correspond to the result of the nonparametric Mann–Whitney–Wilcoxon signed-rank test, which followed the Kruskal–Wallis one-way analysis of variance (P < 0.0001).
Segmentation analysis of the CGH data was performed for each tumor to assess the number of genomic segments with deviating copy number changes (aka genomic segmentation), as a readout of genomic instability (41). Palb2/Trp53 and Brca1/Trp53 tumors displayed apparently greater genomic instability (genomic segmentation) compared with Brca2/Trp53 and Trp53-only tumors, but the difference was not statistically significant. However, when the relative dose of amplified segments (log2 dose ≥ 0.5) was analyzed, Palb2/Trp53 mammary tumors (n = 8) displayed a significantly higher average dose of amplified segments than either Brca1/Trp53 (n = 4; P < 0.0001, Fig. 3C) or Brca2/Trp53 tumors (n = 5; P = 0.0014, Fig. 3C). The low numbers of deletions (log2 dose ≤ -0.5) detected in Brca2/Trp53 and Trp53-only tumors precluded further analysis of this aspect of genomic instability (Fig. 3B).
No significant difference in focal genome amplification appeared when Palb2/Trp53 tumors were compared with Trp53-only tumors, indicating that Palb2/Trp53 tumors share a similar amplification-prone genomic profile with Trp53-only tumors, despite their marked difference in tumor formation kinetics. The inability of Palb2 loss to suppress focal genomic amplifications (unlike what was observed in Brca1/Trp53 and Brca2/Trp53 tumors) could be accounted for by three, alternative explanations.
First, these differences could be due to residual activity of the conditional Palb2 allele we generated, and other Palb2 loss of function mutations might trigger the formation of true Brca2 tumor phenocopies. Second, our allele is a functional null, as our studies suggest, but complete loss of Palb2 is similar to a BRCA2 hypomorphic phenotype rather than a complete loss of BRCA2 function. Alternatively, there are PALB2 functions that are, at least in part, nonoverlapping with the tumor suppressing functions of its BRCA2 partner protein. New experiments with additional Palb2 and Brca2 mutant mouse strains would be required to distinguish between these possibilities.
Finally, tumor heterogeneity likely affected the CGH profiles (Fig. S5) in ways that make it difficult to identify regions of chromosomal imbalances that were unique to Palb2/Trp53 tumors. Conceivably, a more comprehensive analysis with a much larger collection of tumor samples would reveal such regions.
Loss of 53BP1 Fails to Rescue the HR Defect Caused by PALB2 Deficiency.
Loss of 53BP1 can rescue the HR defect and lethality observed in either Brca1∆11/∆11 or Brca1-null cells and mice (42–45). Similarly, decreased expression of the P53 binding protein 1 (53BP1) was detected in triple negative breast cancers as well as human BRCA1 tumors (45). Therefore, we asked whether Trp53bp1 (which encodes mouse 53BP1) expression is reduced in Palb2/Trp53 KO tumors, and whether its absence rescues the HR defect associated with Palb2 loss. Quantitative RT-PCR analysis of Trp53bp1 mRNA in freshly isolated Palb2 breast tumor samples showed that levels of Trp53bp1 messenger varied considerably among the tumors that were analyzed. However, overall Trp53bp1 mRNA levels were not significantly different in Palb2-deficient and Palb2-proficient tumors (Fig. 4A).
To determine whether HR deficiency due to Palb2 loss is complemented by Trp53bp1 loss in primary cells (cultured primary splenic B cells), we generated Palb2fl/fl; CD19-Cre mice that were or were not deficient in Trp53bp1. Cultured primary splenocytes from these mice were then assayed for HR competence upon treatment with PARP inhibitors (PARPi), which selectively induces DNA damage and chromosomal aberrations in HR-deficient cells (46). Treatment with KU0058948 (PARPi) led to an accumulation of chromosomal and chromatid breaks, and radial structures were evident in chromosomal spreads from cultured Palb2fl/fl; Trp53bp1+/−; CD19-Cre primary splenocytes (Fig. 4B). The number of chromosomal aberrations observed was not reduced in Palb2fl/fl; Trp53bp1−/−; CD19-Cre splenocytes, implying that Trp53bp1 deletion did not complement the HR defect caused by Palb2 deficiency (Fig. 4B). Trp53bp1 deletion also failed to rescue the chromosomal aberrations found in spreads from PARPi-treated Brca2fl/fl; Trp53bp1−/−; CD19-Cre splenocytes (Fig. S6A), which appeared to be even more extensive than those observed in PARPi-treated Palb2fl/fl; Trp53bp1−/−; CD19-Cre cells (Fig. 4B). As has been previously described (43, 45), complete rescue of the DNA repair deficiency in Brca1fl/fl; Trp53bp1−/−; CD19-Cre splenocytes was observed (Fig. S6B).
Of note, both Palb2/Trp53bp1 and Brca2/Trp53bp1 compound KO cells displayed more chromosomal aberrations after PARPi exposure than Palb2 or Brca2 single mutants (Fig. 4B, Fig. S6A). Thus, whereas Brca1, Palb2, and Brca2 manifest closely related, even overlapping functions, loss of Palb2 or Brca2 also resulted in a different DNA damage response after Trp53bp1 elimination from that manifested by Brca1 KO cells, in which Trp53bp1 codeletion rescued the genomic instability observed after PARP inhibition.
These observations suggest that the contributions of PALB2 and BRCA2 to HR-based DSB repair are distinct from those of BRCA1 and cannot be complemented by 53BP1 loss. In keeping with existing evidence, PALB2 and BRCA2 may be de facto HR effectors that cannot be replaced or bypassed, except by artificially forcing the loading of RAD51 onto chromatin at/near DSB, which 53BP1 loss has not yet been shown to promote (45, 47–49). These observations, along with earlier results (4), also suggest that PARP inhibition might be a potential therapeutic regimen in PALB2-deficient tumors, as it is in BRCA1- and BRCA2-associated tumors (46).
Conclusions
In summary, we have shown that Palb2 is a breast tumor suppressor gene in mice as it is in humans and that it synergizes with Trp53 to suppress tumor formation. The outcome of dual Palb2/Trp53 nullizygosity in the mouse mammary gland is highly penetrant breast cancer. In keeping with the fact that PALB2 is also a breast tumor suppressor in humans, PALB2 might be viewed as a BRCA3-like allele. Moreover, tumorigenesis driven by Palb2 loss in the mouse is not entirely suppressed on a Trp53 WT germ-line background, unlike most Brca1 and Brca2 mouse models of breast tumorigenesis (50).
Despite many similarities to Brca2/Trp53 and Brca1/Trp53 breast tumors, Palb2 tumors displayed certain divergent genomic features that might be viewed as separating them from BRCA1 and -2 cancers. Specifically, we observed patterns of genomic aberrations that were different in Palb2/Trp53-derived tumors from those detected in Brca1/Trp53- or Brca2/Trp53-derived tumors. These data are consistent with the hypothesis that PALB2 possesses biological functions that extend beyond those of its major interactors, BRCA1 and BRCA2. Alternatively, the effect of Palb2 deletion may mimic a phenotype akin to partial loss of BRCA2, resulting in a less dramatic genomic instability profile in the relevant tumor cells.
Although the genomic instability of Brca1-deficient cells can be rescued by loss of Trp53bp1, deletion of the latter had, if anything, an adverse effect in Palb2 KO cells. In that context, Palb2 is more similar to Brca2, the absence of which leads to an HR defect that also cannot be rescued by Trp53bp1 deletion.
Haploinsufficiency for Palb2 tumor suppression was not detected in this model although one cannot rule out the possibility that it would be manifest in a different model system and/or with enlarged cohorts of experimental mice. For example, the tumors in this mouse model driven by K14-Cre were uniformly of the triple negative phenotype. This characteristic might well contribute to the absence of haploinsufficiency in our system, in the same way that BRCA1 mammary tumors derived from distinct cell populations display preferential patterns of consecutive LOH events along the tumorigenesis pathway (32, 51).
We believe that this mouse model will be useful in understanding how Palb2 serves its breast cancer suppression function.
Materials and Methods
ES Cell Derivation, Embryo Harvesting, and Tetraploid Complementation Assay.
Generation of the conditional allele for Palb2 and additional experimental details are described in SI Material and Methods. Oligo sequences used are described in Table S1. ES cell derivation was performed according to standard protocols (52). Pregnant mice from timed matings were killed at indicated time points by CO2 asphyxiation following institutional guidelines. Uterine horns and embryos were dissected under the microscope, and isolated embryos were directly used for digestion, DNA extraction, and genotyping. Negative selection of Palb2 KO embryos was analyzed according to the Hardy–Weinberg equilibrium model, using an online tool (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). Tetraploid complementation assays were performed as described (53). All experimental procedures involving mouse work were approved by the Dana-Farber Cancer Institute Institutional Care and Use Committee under Animal Protocol 07–011.
Tumorigenesis Studies.
Mouse cohorts were monitored for tumor formation biweekly. Mammary tumor formation was scored when a palpable tumor of 1.0 cm in its greatest diameter could be detected, as previously described (27, 30). Mice harboring tumors were humanely killed when the tumor diameter reached 2.0 cm in its greatest dimension. Mice that were otherwise severely diseased/distressed were also killed according to institutional guidelines. Mantel–Cox logrank test was applied for comparison of tumor-free survival of mouse cohorts.
Supplementary Material
Acknowledgments
We thank Drs. Ron DePinho and William Kaelin for Trp53fl/fl mice and Dr. Bing Xia for openly exchanging information on PALB2 KO mice with our laboratory. We also thank Drs. Kristine McKinney, Nana Naetar-Kerenyi, Patricia Dahia, and Stefan Muljo for critical reading of this manuscript; Dvora Ghitza and Dr. Klaus Rajewsky for help with the tetraploid complementation assays; Dr. Rene Maehr for V6.5 ES cells; Dr. Ronny Drapkin for the antibody developed against mouse BRCA1; and James Horner for ES cell microinjections. We also thank all members of the D.M.L. laboratory for cooperation, expertise, and reagent sharing, as well as fruitful discussions. Finally, we thank the staff from the Dana-Farber Cancer Institute Animal Resources Facility for excellent technical support and Anuradha Kohli and Nancy Gerard for outstanding administrative support. This work was supported by National Cancer Institute Grant P01CA80111, by a Specialized Program of Research Excellence grant in breast cancer research (2P50CA089393 to the Dana-Farber/Harvard Cancer Center), by the Susan G. Komen Foundation for the Cure (SAC110022), and grants from the Breast Cancer Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305362110/-/DCSupplemental.
References
- 1.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 2.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 3.Dray E, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17(10):1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buisson R, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17(10):1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106(17):7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 8.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 9.Tischkowitz M, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA. 2007;104(16):6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446(7133):316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 11.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324(5924):217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahman N, et al. Breast Cancer Susceptibility Collaboration (UK) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284(32):21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 15.Bleuyard JY, Buisson R, Masson JY, Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13(2):135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huo Y, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discovery. 2013 doi: 10.1158/2159-8290.CD-13-0011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10(14):1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 18.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386(6627):804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 19.Rantakari P, et al. Inactivation of Palb2 gene leads to mesoderm differentiation defect and early embryonic lethality in mice. Hum Mol Genet. 2010;19(15):3021–3029. doi: 10.1093/hmg/ddq207. [DOI] [PubMed] [Google Scholar]
- 20.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85(7):1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki A, et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11(10):1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 22.Bouwman P, et al. Loss of p53 partially rescues embryonic development of Palb2 knockout mice but does not foster haploinsufficiency of Palb2 in tumour suppression. J Pathol. 2011;224(1):10–21. doi: 10.1002/path.2861. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: Lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11(10):1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 24.Hakem R, de la Pompa JL, Elia A, Potter J, Mak TW. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16(3):298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 25.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26(2):113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127(22):4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 27.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 28.Erkko H, et al. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clin Cancer Res. 2008;14(14):4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- 29.Holstege H, et al. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res. 2009;69(8):3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loonstra A, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98(16):9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8(1):233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 34.García MJ, et al. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative Spanish breast cancer families. Breast Cancer Res Treat. 2009;113(3):545–551. doi: 10.1007/s10549-008-9945-0. [DOI] [PubMed] [Google Scholar]
- 35.Casadei S, et al. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71(6):2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22(1):37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig T, Fisher P, Ganesan S, Efstratiadis A. Tumorigenesis in mice carrying a truncating Brca1 mutation. Genes Dev. 2001;15(10):1188–1193. doi: 10.1101/gad.879201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12(2):191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 39.Cheon DJ, Orsulic S. Mouse models of cancer. Annu Rev Pathol. 2011;6:95–119. doi: 10.1146/annurev.pathol.3.121806.154244. [DOI] [PubMed] [Google Scholar]
- 40.Kallioniemi A, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 41.de Ronde JJ, et al. KC-SMARTR: An R package for detection of statistically significant aberrations in multi-experiment aCGH data. BMC Res Notes. 2010;3:298. doi: 10.1186/1756-0500-3-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35(4):534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bunting SF, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46(2):125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 47.Brown ET, Holt JT. Rad51 overexpression rescues radiation resistance in BRCA2-defective cancer cells. Mol Carcinog. 2009;48(2):105–109. doi: 10.1002/mc.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin RW, et al. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67(20):9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 49.Schild D, Wiese C. Overexpression of RAD51 suppresses recombination defects: A possible mechanism to reverse genomic instability. Nucleic Acids Res. 2010;38(4):1061–1070. doi: 10.1093/nar/gkp1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene. 2006;25(43):5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 51.Martins FC, et al. Evolutionary pathways in BRCA1-associated breast tumors. Cancer Discov. 2012;2(6):503–511. doi: 10.1158/2159-8290.CD-11-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanellopoulou C, et al. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci USA. 2009;106(4):1122–1127. doi: 10.1073/pnas.0812210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy A, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110(3):815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.