Abstract
The following three diagnostic algorithms were evaluated in comparison with the Illumigene assay as a stand-alone test for Clostridium difficile detection: glutamate dehydrogenase antigen screen (GDH) followed by toxin A/B antigen testing (Tox A/B) with the cell cytotoxicity assay for discordant specimens (algorithm 1), GDH followed by the Illumigene (algorithm 2), and GDH followed by Tox A/B with the Illumigene for discordant specimens (algorithm 3). A total of 428 stool specimens submitted to three clinical microbiology laboratories in Manitoba, Canada, for C. difficile detection between June 2011 and April 2012 were included in the study. The prevalence of C. difficile in the stool specimens was 14.7% (63/428) based on toxigenic culture (microbiologic reference standard). The sensitivity and specificity of the Illumigene for C. difficile detection were 73.0% and 99.7%, respectively. The corresponding sensitivities and specificities were 65.1% and 100.0% for algorithm 1, 68.3% and 100.0% for algorithm 2, and 69.8% and 100.0% for algorithm 3. Using algorithm 1, a cell cytotoxicity assay was required for toxin detection in 37% of positive tests, prolonging turnaround time. However, the predictive value of a positive test based on a clinical reference standard (all tests positive or cytotoxigenic culture positive and clinical disease on chart review) was slightly higher with algorithm 1 than with the Illumigene assay as a stand-alone test or as part of an algorithm (algorithms 2 and 3). Based on a reduction in turnaround time, simplicity, and acceptable sensitivity and specificity, we recommend algorithm 2 (screening with the GDH antigen test and confirmatory testing with the Illumigene).
INTRODUCTION
Clostridium difficile is an anaerobic, spore-forming Gram-positive bacillus that is capable of causing diarrhea mediated by the production of C. difficile toxins A and B (1–3). It is estimated that C. difficile infection (CDI) is responsible for 15 to 25% of cases of antibiotic-associated diarrhea (1). Severe CDI may result in the need for intensive care unit admission or even colectomy (4). Further, CDI is associated with an attributable mortality rate of approximately 5.7% (4). Related to both the frequency and severity of disease caused by C. difficile, it is imperative that diagnostic testing be sensitive, specific, and rapid, such that appropriate therapy may be instituted in a timely fashion (5).
For diagnosis of CDI, many clinical microbiology laboratories currently use enzyme immunoassays (EIAs) that detect C. difficile toxins (1). These assays have the advantage of a rapid turnaround time. However, they demonstrate suboptimal sensitivity (5, 6). Algorithms relying on detection of the C. difficile glutamate dehydrogenase antigen (GDH) with follow-up toxin testing for positive specimens with a cell cytotoxicity assay demonstrate improved sensitivity (5, 7). However, the turnaround time may be increased by up to 48 h with this approach (5, 7). Additionally, recent data suggest that the cell cytotoxicity assay for toxin detection also lacks sensitivity (7). C. difficile culture with subsequent toxin detection (toxigenic culture) is highly sensitive, but this method is time-consuming and very labor-intensive (5). More recently, a number of molecular assays have been developed for C. difficile detection (8–11). These assays offer improved sensitivity for C. difficile diagnosis and rapid turnaround for results (8–11).
In view of the large number of diagnostic tests available to clinical microbiology laboratories for the detection of C. difficile, uncertainty exists as to which test or series of tests should be employed. The purpose of this study was to compare an algorithmic approach to C. difficile diagnosis (GDH test, followed by confirmatory or toxin testing) with direct testing of stool specimens by a molecular platform (the Illumigene C. difficile assay; Meridian Bioscience, Cincinnati, OH). Three diagnostic algorithms were considered: GDH antigen screen followed by toxin A/B antigen testing, with the cell cytotoxicity assay for discordant specimens (algorithm 1); GDH antigen screen followed by the Illumigene (algorithm 2); and GDH antigen screen followed by toxin A/B antigen testing and the Illumigene for discordant specimens (algorithm 3).
MATERIALS AND METHODS
Study sites.
This study was performed at the Health Sciences Centre (Winnipeg, Manitoba), St. Boniface Hospital (Winnipeg, Manitoba), and Westman (Brandon, Manitoba) clinical microbiology laboratories. The Health Sciences Centre and St. Boniface Hospital microbiology laboratories provide general bacteriology services, including C. difficile diagnostic testing, for all seven major hospitals in Winnipeg. Westman Laboratory provides microbiology services to the city of Brandon and surrounding communities. These laboratories are all part of Diagnostic Services of Manitoba (DSM), and currently they all utilize the same protocol for C. difficile detection from patient clinical specimens. The study was carried out sequentially, first at Westman Laboratory, then at St. Boniface Hospital, and, finally, at the Health Sciences Centre, related to availability of the Illumigene instrument.
Specimens.
All diarrheal stools containing a minimum specimen volume of 5 ml received by the participating microbiology laboratories for C. difficile testing were eligible for inclusion. Exclusion criteria included formed stools, stool submitted for a patient with a positive C. difficile test result in the preceding 7 days, stool samples which spent >72 h in transit, and samples from patients less than 1 year of age. These criteria resulted in specimens being rejected by the microbiology laboratory, as per routine protocol. The planned sample size was 500 specimens (200 at St. Boniface Hospital, 200 at the Health Sciences Centre, and 100 at Westman Laboratory). Duplicate specimens were subsequently excluded from data analysis.
Diagnostic testing.
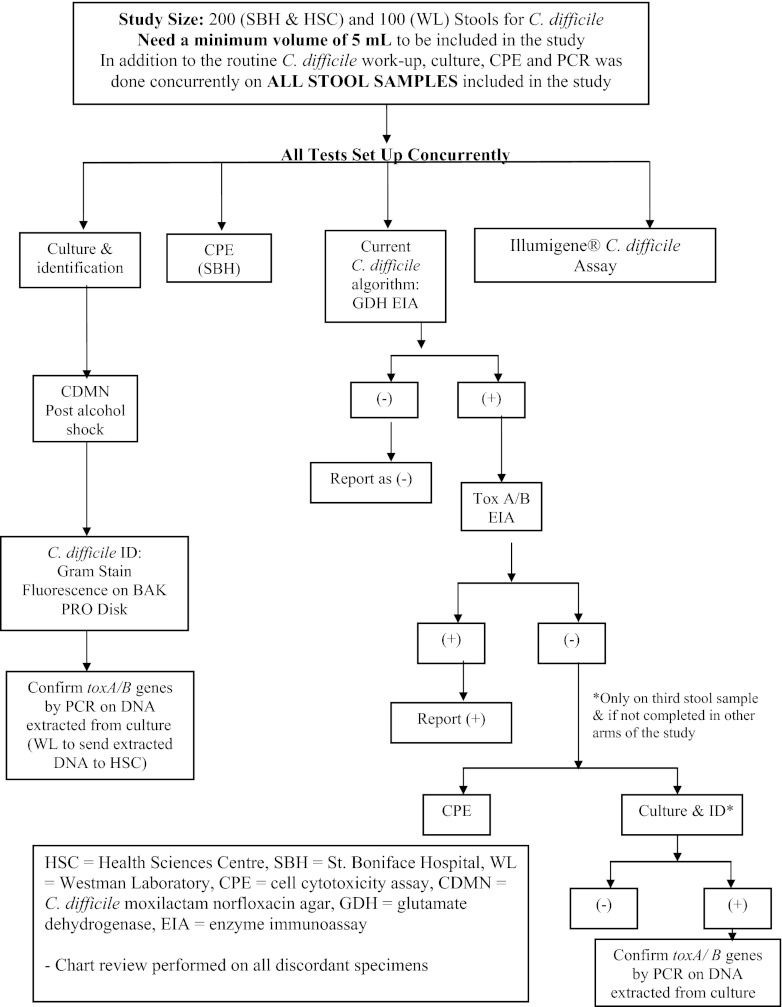
The stool testing protocol is summarized in Fig. 1. All stools were tested by the algorithm currently in place by DSM for C. difficile detection (algorithm 1). Stools are first screened with a GDH antigen test. Stools negative by the GDH test are reported as negative. Stools positive by the GDH test go on to have testing for C. difficile toxins A and B, first by an EIA and, as necessary, by the cell cytotoxicity assay. Patient results were reported based on testing from this protocol. An algorithmic approach involving screening of stool specimens using the GDH antigen test, followed by testing of positive specimens with the Illumigene or a toxin enzyme immunoassay followed by the Illumigene for discordant samples, was also evaluated (algorithms 2 and 3). All stools were additionally directly tested by the cell cytotoxicity assay, toxigenic culture, and the Illumigene assay.
Fig 1.
C. difficile study overview.
(i) Glutamate dehydrogenase antigen test.
The C. Diff Quik Chek test (Techlab Inc., Blacksburg, VA) was used to test for the presence or absence of C. difficile GDH antigen. This test is a rapid membrane EIA. Testing was performed as directed by the product monograph (12).
(ii) C. difficile toxin A and B testing.
The Tox A/B Quik Chek test (Techlab Inc.) is an EIA used for detection of C. difficile toxins A and B. This test was performed as directed by the product monograph on stools that tested positive with the C. Diff Quik Chek test (13).
(iii) Cell cytotoxicity assay.
Bartels cytotoxicity assay for Clostridium difficile toxin (Trinity Biotech Company, Carlsbad, CA) microtiter plates with viable human fibroblasts was used as the cell cytotoxicity assay (14). Briefly, stool samples are diluted 1:5 with sterile PBS buffer. The samples are then centrifuged and the supernatant is passed through a 0.45-μm filter. Forty microliters of the filtered sample is then added to two wells of the microtiter plate, with 40 μl of diluent added to one well and 40 μl of antitoxin added to the second. The microtiter plate is incubated at 34 to 37°C for up to 48 h. The plate is read at 24 and 48 h for the development of typical cytopathic effect. A positive result is indicated by the presence of typical cytopathic effect in the well without antitoxin and the absence of this effect in the well with antitoxin.
(iv) Illumigene assay.
The Illumigene assay detects a conserved 204-bp sequence in the 5′ portion of the toxin A gene using loop-mediated isothermal amplification technology. This assay was performed on all stool specimens as described in the product monograph (15). The Illumigene instrument was kindly provided on a trial basis by Meridian Bioscience (Cincinnati, OH) for the duration of the study.
(v) Toxigenic culture.
Culture for C. difficile was performed for all specimens using selective media. Briefly, approximately 1 ml of stool is mixed with an equal volume of 95% ethyl alcohol and allowed to stand at room temperature for 1 h. The specimen is centrifuged and the supernatant is removed. The sediment is then planted to C. difficile moxalactam-norfloxacin (CDMN) agar, prepared in-house using C. difficile agar base (Oxoid, Nepean, Ontario, Canada) and C. difficile selective supplement (Oxoid), and incubated anaerobically in an anaerobic jar at 35°C for up to 5 days. Isolated colonies suspected of being C. difficile are subcultured onto BAK medium (brucella blood agar with vitamin K and hemin), which is incubated anaerobically. C. difficile isolates are subsequently identified based on Gram stain appearance, fluorescence on BAK medium, and presence of the enzyme l-proline aminopeptidase. Isolates confirmed to be C. difficile are evaluated for the presence of C. difficile toxin A and/or B genes by PCR, using an in-house assay as previously described by Alfa et al. (16). Results of testing by toxigenic culture served as the microbiologic reference standard for C. difficile infection.
Chart review.
A limited retrospective chart review was performed for patients for whom the results of the different C. difficile assays were discordant (i.e., positive by toxigenic culture but negative by at least one other assay). The chart review was predominantly carried out by one of the authors (A.W.), who works as an infectious disease physician. This author was not blinded to the laboratory test results at the time of the chart review. The purpose of the chart review was to determine, based on the patient's clinical presentation, whether the patient truly had CDI or asymptomatic colonization with C. difficile. Data obtained on chart review included patient demographics (age and sex), risk factors for CDI (receipt of antibiotics in the preceding 8 weeks, receipt of cancer chemotherapy in the preceding 30 days, or current use of a proton pump inhibitor), clinical symptoms (presence of ≥3 unformed bowel movements a day or presence of clinically significant diarrhea, fever, elevated white cell count, elevated creatinine, and findings at colonoscopy when performed), recent use of laxatives, and response to treatment when provided. The determination of whether a patient had clinically significant CDI was made based on an assessment of risk factors, clinical presentation, and response to treatment when provided (or absence of adverse consequences when treatment was not provided). Ethics approval for the chart review was obtained from the University of Manitoba Bannatyne Campus Research Ethics Board.
Interpretation of results.
The microbiologic reference standard used in this study was the result obtained by toxigenic culture. With the toxigenic culture result (positive or negative) taken as the true result, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for the current algorithm in place at DSM laboratories and new algorithms using GDH testing followed by testing with the Illumigene assay (with or without the toxin A/B antigen test), the cell cytotoxicity assay, and the Illumigene assay. Comparison with the microbiologic reference standard represented the primary study outcome. As a secondary outcome, all assays and diagnostic algorithms were also compared against a clinical reference standard. The clinical reference standard was all tests positive for C. difficile or toxigenic culture positive with clinically significant disease demonstrated on chart review. The clinical reference standard was similarly used to determine the sensitivity, specificity, PPV, and NPV of the C. difficile assays and algorithms evaluated in this study. Confidence intervals were determined using the Wilson procedure with a correction for continuity. This analysis was performed using VassarStats, available at http://vassarstats.net/.
Cost analysis.
A brief cost analysis was performed for 4 testing models: Illumigene as a stand-alone test, algorithm 1, algorithm 2, and algorithm 3. The costs per test used in this analysis were as follows: Illumigene, $13.00 to $15.00 per test; GDH, $7.00 to $11.00 per test; toxin EIA, $7.00 to 11.00 per test; and cell cytotoxicity assay, $7.00 to $10.00 per test. Labor costs were not included in the analysis.
RESULTS
In total, 502 stool specimens submitted for C. difficile detection between June 2011 and April 2012 were evaluated. After the exclusion of duplicate specimens, 428 stool specimens remained for inclusion in the study. Using the results of toxigenic culture as the microbiologic reference standard for diagnosis of CDI, the overall positivity rate was 14.7% (63/428). The sensitivity, specificity, PPV, and NPV of the various diagnostic assays and algorithms evaluated are presented in Table 1. The sensitivity and specificity of the current DSM algorithm (algorithm 1) were 65.1% and 100.0%, respectively. Similar results were obtained for algorithms 2 and 3 (Table 1). Results obtained with the Illumigene assay as a stand-alone test compared favorably with all algorithms, with a sensitivity of 73.0% and a specificity of 99.7%. All diagnostic assays assessed demonstrated suboptimal sensitivity in comparison with toxigenic culture, although the predictive value of a positive test was greater than 95% in every case except with the stand-alone GDH antigen test. The poor positive predictive value of the GDH antigen test is not surprising, as this assay does not provide any information as to the presence of C. difficile toxins.
Table 1.
Summary of C. difficile diagnostic test performance results using toxigenic culture as the reference standard
| Test | % Sensitivity (no. positive/no. tested) (CIb) | % Specificity (no. negative/no. tested) (CI) | Positive predictive value (%) (CI) | Negative predictive value (%) (CI) |
|---|---|---|---|---|
| Illumigene C. difficile assaya | 73.0 (46/63) (60.1–83.1) | 99.7 (363/364) (98.2–100) | 97.9 (87.3–99.9) | 95.5 (92.8–97.3) |
| Cell cytotoxicity assay | 66.7 (42/63) (53.6–77.7) | 99.5 (363/365) (97.4–100) | 95.5 (83.3–99.2) | 94.5 (91.6–96.5) |
| GDH antigen test | 82.5 (52/63) (70.5–90.6) | 97.0 (354/365) (94.5–98.4) | 82.5 (70.5–90.6) | 97.0 (94.5–98.4) |
| Toxin A/B antigen test | 41.3 (26/63) (29.2–54.4) | No data | No data | No data |
| GDH antigen test followed by toxin A/B antigen test | 41.3 (26/63) (29.2–54.4) | 100.0 (365/365) (98.7–100) | 100.0 (84.0–100) | 90.8 (87.4–93.4) |
| GDH antigen test followed by cell cytotoxicity assay | 63.5 (40/63) (50.4–75.0) | 100.0 (365/365) (98.7–100) | 100.0 (89.1–100) | 94.1 (91.1–96.1) |
| GDH antigen test followed by toxin A/B antigen test (and cell cytotoxicity assay for discordant results) (algorithm 1) | 65.1 (41/63) (52.0–76.4) | 100.0 (365/365) (98.7–100) | 100.0 (89.3–100) | 94.3 (91.4–96.3) |
| GDH antigen test followed by Illumigene C. difficile assay (algorithm 2) | 68.3 (43/63) (55.2–79.1) | 100.0 (365/365) (98.7–100) | 100.0 (89.8–100) | 94.8 (92.0–96.7) |
| GDH antigen test followed by toxin A/B antigen test (and Illumigene C. difficile assay for discordant results) (algorithm 3) | 69.8 (44/63) (56.8–80.4) | 100.0 (365/365) (98.7–100) | 100.0 (90.0–100) | 95.1 (92.2–96.9) |
| Toxigenic culture | 100.0 (63/63) (92.8–100) | 100.0 (365/365) (98.7–100) | 100.0 (92.8–100) | 100.0 (98.7–100) |
For the Illumigene C. difficile assay, for invalid specimen results the result obtained on repeat testing was used for the calculations. If retesting was not performed, the specimen was excluded from the analysis (n = 1).
CI, 95% confidence interval.
Thirty-nine specimens yielded discordant results for at least one of the C. difficile assays under evaluation. A chart review was undertaken for the discordant samples to determine the clinical significance of the test results. It should be noted that in many cases, clinical symptoms and response to treatment, where provided, were not well documented in the patient chart. Based on the clinical presentation of the patient, the decision to treat (or not) by the attending physician, and the response to treatment where available, 29 of the discordant specimens were considered to represent clinically significant CDI. In two cases in which the hospital chart was not available, the patient was considered to have clinically significant infection based on the toxigenic culture result. The remaining 10 discordant specimens were not considered to represent a clinically important test result. Reanalyzing the data with the reference standard of all tests positive for C. difficile or toxigenic culture positive with clinically significant disease demonstrated on chart review (the clinical reference standard), the positivity rate was 12.4% (53/428). The sensitivity, specificity, PPV, and NPV of all assays and algorithms in comparison with the clinical reference standard are presented in Table 2. The Illumigene assay either as a stand-alone test or as part of an algorithm (algorithms 2 and 3) had a slightly lower positive predictive value than the algorithm currently in place at DSM microbiology laboratories (algorithm 1), related to the specificity of this test. While toxigenic culture remained the most sensitive test evaluated, the specificity of this assay was suboptimal, resulting in a positive predictive value of only 84.1%.
Table 2.
Summary of C. difficile diagnostic test performance results using presence of all tests positive or toxigenic culture positive with clinically significant disease by chart review as the reference standard
| Test | % Sensitivity (no. positive/no. tested) (CIb) | % Specificity (no. negative/no. tested) (CI) | Positive predictive value (%) (CI) | Negative predictive value (%) (CI) |
|---|---|---|---|---|
| Illumigene C. difficile assaya | 77.4 (41/53) (63.4–87.3) | 98.4 (368/374) (96.4–99.4) | 87.2 (73.6–94.7) | 96.8 (94.4–98.3) |
| Cell cytotoxicity assay | 75.5 (40/53) (61.4–85.8) | 98.9 (371/375) (97.1–99.7) | 90.9 (77.4–97.0) | 96.6 (94.1–98.1) |
| GDH antigen test | 84.9 (45/53) (71.9–92.8) | 95.2 (357/375) (92.4–97.0) | 71.4 (58.5–81.8) | 97.8 (95.6–99.0) |
| Toxin A/B antigen test | 49.1 (26/53) (35.2–63.0) | No data | No data | No data |
| GDH antigen test followed by toxin A/B antigen test | 49.1 (26/53) (35.2–63.0) | 100.0 (375/375) (98.7–100) | 100.0 (84.0–100) | 93.3 (90.3–95.4) |
| GDH antigen test followed by cell cytotoxicity assay | 71.7 (38/53) (57.4–82.8) | 99.5 (373/375) (97.9–99.9) | 95.0 (81.8–99.1) | 96.1 (93.6–97.7) |
| GDH antigen test followed by toxin A/B antigen test (and cell cytotoxicity assay for discordant results) (algorithm 1) | 73.6 (39/53) (59.4–84.3) | 99.5 (373/375) (97.9–99.9) | 95.1 (82.2–99.2) | 96.4 (93.9–97.9) |
| GDH antigen test followed by Illumigene C. difficile assay (algorithm 2) | 71.7 (38/53) (57.4–82.8) | 98.7 (370/375) (96.7–99.5) | 88.4 (74.1–95.6) | 96.1 (93.5–97.7) |
| GDH antigen test followed by toxin A/B antigen test (and Illumigene C. difficile assay for discordant results) (algorithm 3) | 73.6 (39/53) (59.4–84.3) | 98.7 (370/375) (96.7–99.5) | 88.6 (74.6–95.7) | 96.4 (93.8–97.9) |
| Toxigenic culture | 100.0 (53/53) (91.6–100) | 97.3 (365/375) (95.0–98.6) | 84.1 (72.3–91.7) | 100.0 (98.7–100) |
For the Illumigene C. difficile assay, for invalid specimen results the result obtained on repeat testing was used for the calculations. If retesting was not performed, the specimen was excluded from the analysis (n = 1).
CI, 95% confidence interval.
For the cost analysis, Illumigene as a stand-alone test was slightly more expensive than an algorithmic approach involving screening with the GDH antigen test, followed by confirmatory or toxin testing. All three algorithms evaluated were comparable in terms of overall cost in this study (data not shown).
DISCUSSION
In this study, the Illumigene assay was compared with several algorithms for C. difficile detection, using toxigenic culture as the microbiologic reference standard. The sensitivity of the Illumigene assay was slightly higher than the sensitivity of all three algorithms evaluated. Given the prevalence of C. difficile infection in the current study (14.7%), it is doubtful that the improved sensitivity would be of clinical significance. In 37% of cases in which C. difficile was detected by the current DSM algorithmic approach (algorithm 1), a cell cytotoxicity test was required to demonstrate the presence of toxin. This would be expected to delay reporting of results by at least 24 h. Hence, use of the Illumigene assay as a stand-alone test or in an algorithm with a GDH antigen screening test (algorithms 2 and 3) would be expected to positively impact turnaround time in result reporting. The trade-off is that specificity for clinical disease appears to be slightly lower with the Illumigene assay (again, as a stand-alone test or as part of an algorithm), resulting in a lower predictive value for a positive test in comparison with an algorithm that relies on the cell cytotoxicity assay for discordant specimens. Toxigenic culture remained the most sensitive assay for detection of toxigenic C. difficile. The main limitations to this method are the prolonged turnaround time, the labor involved, and the suboptimal specificity for detection of clinically significant disease. In fact, when analyzing the data in comparison with the clinical reference standard, the positive predictive value of toxigenic culture was found to be only 84.1%. Similar to other publications, the data presented here argue against use of a toxin EIA (toxin A/B antigen test) as the sole test for C. difficile detection, related to suboptimal sensitivity.
Somewhat surprisingly, the Illumigene assay was found to have a relatively low sensitivity, 73.0%, in comparison with toxigenic culture. Other studies in the literature have similarly evaluated the sensitivity of the Illumigene assay for toxigenic C. difficile detection. In contrast with the current study, the previously reported sensitivity has varied between 81.4% and 98% in comparison with either toxigenic culture or a composite of toxigenic culture and molecular testing (10, 11, 17, 18). The reason for this discrepancy is not entirely clear. It may in part be related to the choice of microbiologic reference standard utilized in this study (C. difficile culture for all specimens with molecular detection of the toxin A and/or B gene). The culture method utilized in this study involved alcohol shock of stools, followed by culture on CDMN agar. CDMN agar has been demonstrated to yield improved recovery of C. difficile from stool specimens in comparison with cycloserine-cefoxitin-fructose agar (CCFA) (19). Culture of all stool specimens was not performed in other studies evaluating the sensitivity and specificity of the Illumigene assay, and when culture was carried out, CCFA medium was generally used without any indication of prior alcohol shock treatment (10, 11, 17). Alternatively, the relatively low sensitivity could relate to low pathogen burden in the clinical specimens evaluated. For 17 specimens that were positive by toxigenic culture but negative by the Illumigene assay, only 7 (41.2%) were positive by the cell cytotoxicity assay and only 1 (5.9%) was positive by toxin A/B EIA, supporting the idea that organism load may indeed be important. Further, in a recent study, specimens that tested negative for C. difficile by the Illumigene but positive by another molecular platform (the Xpert C. difficile/Epi real-time PCR assay [Cepheid]) were found to have C. difficile DNA concentrations below or just above the limit of detection for the Illumigene instrument (20). Finally, the Illumigene assay works by detection of the toxin A gene by loop-mediated isothermal amplification. It cannot be excluded that the suboptimal sensitivity reported here may have been affected by mutations in the primer binding region. However, repeat testing with the Illumigene was performed in 11 of the 17 negative specimens that were positive by toxigenic culture, and of these, 5 (45.5%) were positive. This suggests that a significant proportion of the Illumigene false negatives were not related to primer binding site mutations.
In general, the sensitivities reported in this study for the cell cytotoxicity assay, the Tox A/B Quik Chek test, and the C. Diff Quik Chek test were slightly lower than in other publications in the literature (5, 6). This may in part be related to the choice of C. difficile culture method (alcohol shock of stools followed by culture on CDMN agar) with molecular detection of the toxin A and/or B gene as the microbiologic reference standard.
This study has several strengths relative to others published in the literature. These include the large number of specimens evaluated; the multicenter design, which, in turn, increases the applicability of the results to other institutions; and the clinical chart review for discordant specimens. However, there are several limitations which deserve attention. Not all specimens that were positive by toxigenic culture and negative by the Illumigene had repeat testing performed. This hindered further explanation of the suboptimal sensitivity of the Illumigene assay in the current study. Additionally, the chart reviews carried out were limited to patients with discordant test results. Ideally, a chart review should have been performed for all patients who tested positive by toxigenic culture, even if all other test results were concordant, in order to truly evaluate the significance of a positive test. This was not done due to limited resources. Further, related to the retrospective nature of the chart reviews, the clinical data available in patient charts were often incomplete, making it difficult to accurately determine if a positive laboratory test for C. difficile was in fact clinically significant. Analysis of the data using toxigenic culture, an objective laboratory reference standard, was performed to account for this limitation, but toxigenic culture may itself lack specificity in the detection of clinically significant disease.
In summary, the Illumigene assay was found be marginally more sensitive than an algorithmic approach for C. difficile detection in comparison with toxigenic culture as the reference standard. While the sensitivities of both the Illumigene assay and algorithms involving GDH followed by confirmatory or toxin testing were suboptimal, the predictive value of a positive and negative test exceeded 94%, suggesting that either approach may be acceptable for routine use in a clinical microbiology laboratory depending on the prevalence of CDI. The Illumigene assay as part of an algorithm or as a stand-alone test may offer some advantage in terms of reducing turnaround time to reporting of results, with the trade-off being a small reduction in specificity for diagnosis of clinical disease. Based on the results of this study, DSM laboratories have decided to adopt an algorithmic approach to C. difficile diagnosis using the GDH antigen as a screening test and the Illumigene as the follow-up confirmatory test (algorithm 2). This decision was based on the reduction in turnaround time in comparison with the algorithm currently in place, comparable and acceptable sensitivity and specificity in comparison with other algorithms, and slightly lower cost relative to Illumigene as a stand-alone test.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Bartlett JG, Gerding DN. 2008. Clinical recognition and diagnosis of Clostridium difficile infection. Clin. Infect. Dis. 46:S12–S18 [DOI] [PubMed] [Google Scholar]
- 2. Kelly CP, LaMont JT. 2008. Clostridium difficile—more difficult than ever. N. Engl. J. Med. 359:2932–2940 [DOI] [PubMed] [Google Scholar]
- 3. Poutanen SM, Simor AE. 2004. Clostridium difficile-associated diarrhea in adults. Can. Med. Assoc. J. 171:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gravel D, Miller M, Simor A, Taylor G, Gardam M, McGeer A, Hutchinson J, Moore D, Kelly S, Boyd D, Mulvey M, the Canadian Nosocomial Infection Surveillance Program 2009. Heath care-associated Clostridium difficile infection in adults admitted to acute care hospitals in Canada: a Canadian Nosocomial Infection Surveillance Program study. Clin. Infect. Dis. 48:568–576 [DOI] [PubMed] [Google Scholar]
- 5. Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455 [DOI] [PubMed] [Google Scholar]
- 6. Crobach MJT, Dekkers OM, Wilcox MH, Kuijper EJ. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin. Microbiol. Infect. 15:1053–1066 [DOI] [PubMed] [Google Scholar]
- 7. Larson AM, Fung AM, Fang FC. 2010. Evaluation of tcdB real-time PCR in a three-step diagnostic algorithm for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 48:124–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swindells J, Brenwald N, Reading N, Oppenheim B. 2010. Evaluation of diagnostic tests for Clostridium difficile infection. J. Clin. Microbiol. 48:606–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CAD, Dunne WM., Jr 2011. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J. Clin. Microbiol. 49:2887–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lalande V, Barrault L, Wadel S, Eckert C, Petit JC, Barbut F. 2011. Evaluation of a loop-mediated isothermal amplification assay for diagnosis of Clostridium difficile infections. J. Clin. Microbiol. 49:2714–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Norén T, Alriksson I, Andersson J, Akerlund T, Unemo M. 2011. Rapid and sensitive loop-mediated isothermal amplification test for Clostridium difficile detection challenges cytotoxin B cell test and culture as gold standard. J. Clin. Microbiol. 49:710–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Techlab Inc 2009. C. Diff Quik Chek package insert. Techlab Inc., Blacksburg, VA. [Google Scholar]
- 13. Techlab Inc 2009. Tox A/B Quik Chek package insert. Techlab Inc., Blacksburg, VA [Google Scholar]
- 14. Trinity Biotech Company 2011. Bartels cytotoxicity assay for Clostridium difficile, toxin kit insert. Trinity Biotech Company, Carlsbad, CA [Google Scholar]
- 15. Meridian Bioscience 2011. Illumigene package insert. Meridian Bioscience, Cincinnati, OH [Google Scholar]
- 16. Alfa M, Swan B, VanDekerkhove B, Pang P, Harding GKM. 2002. The diagnosis of Clostridium difficile-associated diarrhea: comparison of Triage® C. difficile panel, EIA for tox A/B and cytotoxin assays. Diagn. Microbiol. Infect. Dis. 43:257–263 [DOI] [PubMed] [Google Scholar]
- 17. Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM. 2012. Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J. Clin. Microbiol. 50:1331–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boyanton BL, Jr, Sural P, Loomis CR, Pesta C, Gonzalez-Krellwitz L, Robinson-Dunn B, Riska P. 2012. Loop-mediated isothermal amplification compared to real-time PCR and enzyme immunoassay for toxigenic Clostridium difficile detection. J. Clin. Microbiol. 50:640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aspinall ST, Hutchisnson DN. 1992. New selective medium for isolating Clostridium difficile from faeces. J. Clin. Pathol. 45:812–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gyorke CE, Wang S, Leslie JL, Cohen SH, Solnick JV, Polage CR. 2013. Evaluation of Clostridium difficile fecal load and limit of detection during a prospective comparison of two molecular tests, the illumigene C. difficile and Xpert C. difficile/Epi tests. J. Clin. Microbiol. 51:278–280 [DOI] [PMC free article] [PubMed] [Google Scholar]