Abstract
The clinical picture of Ochrobactrum anthropi infection is not well described because the infection is rare in humans and identification of the pathogen is difficult. We present a case of O. anthropi bacteremia that was initially misidentified as Ralstonia paucula and later identified by 16S rRNA sequencing and recA analysis.
CASE REPORT
An 85-year-old man was admitted to our hospital to receive transcatheter arterial chemoembolization (TACE). He had liver cirrhosis (Child-Pugh score A) and hepatocellular carcinoma (HCC) caused by hepatitis C virus infection. His medical history included repeated radio frequency ablation and a total of nine TACE procedures for the treatment of HCC. He had also experienced urinary bladder and ureteral carcinoma.
The patient was free from any complications; however, he suddenly developed high fever (39°C), chills, and rigors 12 days after TACE. Physical examination did not reveal any significant findings, but laboratory testing showed a highly inflammatory state: his white blood cell count was 17,700/mm3, and his C-reactive protein level was 4.5 mg/dl. Urine examination was normal, and whole-body computed tomography revealed no particular pathogenic lesion. An echocardiogram was not performed. Oral cefcapene pivoxil was prescribed for 1 week based on the suspicion of some forms of infection relating to TACE.
Two sets of blood cultures were obtained at the time of high fever. Of these four bottles, two different aerobic bottles became positive for Gram-negative rods after an incubation period of 36 h. However, the patient's condition and laboratory measurements improved promptly, and he was discharged on day 16 without distinct diagnosis. Later, the organism was identified as Ralstonia paucula by the Microscan walkaway system (Siemens) with a concordance rate of 99.9%. Susceptibility testing showed the organism to be sensitive to imipenem, meropenem, amikacin, minocycline, and colistin but resistant to β-lactams such as piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, aztreonam, levofloxacin, ciprofloxacin, gentamicin, and trimethoprim-sulfamethoxazole. Although the pathogen was resistant to the antibiotic prescribed, he responded well to the treatment and the antibiotic did not need to be changed.
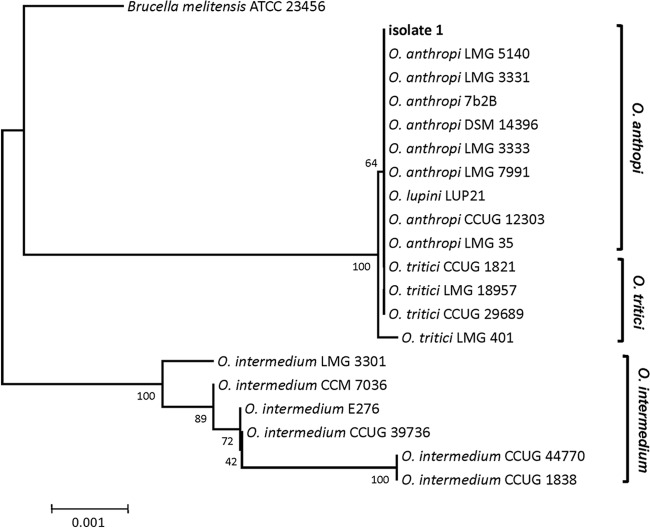
The organism had been rarely encountered at our hospital, and therefore the sample was sent to Kochi University for further identification. The two samples derived from different blood cultures were examined separately. Blood samples were inoculated onto agar media and incubated at 28°C. Pale yellow colonies grew in culture and released a popcorn-like smell. Almost the entire region of the 16S rRNA gene was amplified by colony PCR using a pair of primers (pr0R2, 5′-AGAGTTTGATCMTGGCTCAG-3′; 9Rev, 5′-AAGGAGGTGATCCAGCC-3′) and sequenced with the BigDye Terminator v3.1 cycle sequencing kit. The resulting sequences from the two samples were identical. BLAST analysis against the NCBI database revealed the organism not to be R. paucula but rather to be of the Ochrobactrum anthropi cluster, which is highly homologous to Brucella spp. (Fig. 1). The 16S rRNA partial sequences of isolated organisms are 100% identical to those of the O. anthropi cluster (Fig. 1). Subsequently, almost the entire region of the recA gene was amplified by colony PCR with a pair of primers (1) (recA-BrucOchro-f, 5′-ATGTCTCAAAATTCATTGCGAC-3′; recA-BrucOchro-r, AGCATCTTCTTCCGGTCCGC-3′) and sequenced. Again, two colonies had the same sequences and were 100% identical to recA of several O. anthropi strains (Fig. 2), confirming that the organism is O. anthropi.
Fig 1.
Phylogenetic tree of partial 16S rRNA nucleotide sequences (1,430 nucleotides) using neighbor-joining analysis.
Fig 2.
Phylogenetic tree of partial recA nucleotide sequences (897 nucleotides) using neighbor-joining analysis.
Ochrobactrum spp. belongs to the Brucellaceae and is known to be isolated from Leguminosae nodules (2, 3). Its name is derived from the Greek ochros, meaning pale yellow; this is the characteristic color of Ochrobactrum colonies. The organism has the potential to colonize an exceptionally wide variety of habitats (4–8). The genus Ochrobactrum currently comprises 9 species; to date, only 3 species, O. anthropi, O. intermedium, and O. pseudintermedium, have been reported to occur in clinical samples (9).
Of the clinically relevant species, O. anthropi is becoming increasingly recognized as a potentially problematic, opportunistic, and nosocomial pathogen (10, 11). O. anthropi is an aerobic, oxidase-positive, urease-positive, Gram-negative, motile, non-lactose-fermenting bacillus previously known as “Achromobacter group Vd” (12). Most cases of human disease due to this pathogen that have been reported have been associated with central venous catheter line infection (13–18). However, this organism has also been reported as a cause of infective endocarditis (10, 19, 20), pancreatic abscess (21), puncture wound osteochondritis (22), endophthalmitis (23), urinary tract infection (24), meningitis (25), pelvic abscess (26), and osteomyelitis (27). Infection is most commonly, although not exclusively, seen in immunocompromised patients, such as those with debilitating illnesses or malignancy (13, 14, 19, 26, 28–30). Opportunistic infections and nosocomial outbreaks of O. anthropi are being increasingly reported.
Yu et al. reported the clinical characteristics of 15 cases of O. anthropi bacteremia (31). They stated that all patients had severe underlying disease and manifested primary O. anthropi bacteremia without obvious focus; however, none of the patients died directly from the infection. Thus, although O. anthropi can be pathogenic in critically ill or immunocompromised patients, the organism is considered to be of relatively low virulence. However, the clinical picture of O. anthropi infection, especially bacteremia, has still not been well described. The reason is the difficulty of differentiating Ochrobactrum spp. from other organisms by physiological tests because of their high phenotypic similarity (32). To gain a better understanding of the clinical manifestations of O. anthropi infection, we need to be able to clearly identify the pathogen.
Commercially available test systems used in routine diagnosis are not suitable for species discrimination within the genus Ochrobactrum. In fact, the pathogen in the present case was first misidentified as R. paucula. Although the biochemical properties of O. anthropi and R. paucula are similar, they have distinct differences, especially regarding malonic acid (MAL), the oxidation-fermentation test with glucose (OF/G), tobramycin (TO4), and nitrate reduction (NIT) reactions (Table 1). Of these, a positive reaction with MAL and a negative result with NIT were assumed to be the major reasons for misidentification in this case, while the results of OF/G and TO4 were rather consistent with those expected for O. anthropi.
Table 1.
Comparison of the biochemical properties of Ochrobactrum anthropi, Ralstonia paucula, and the isolate in the present case
| Propertya | Resultb for |
||
|---|---|---|---|
| Ochrobactrum anthropi | Ralstonia paucula | Isolate in the present case | |
| URE | 10 | 5 | (+) |
| H2S | 1 | 1 | (−) |
| IND | 1 | 1 | (−) |
| LYS | 1 | 1 | (−) |
| ARG | 5 | 1 | (−) |
| ORN | 1 | 1 | (−) |
| TDA | 25 | 1 | (−) |
| ESC | 1 | 1 | (−) |
| VP | 1 | 1 | (−) |
| CIT | 75 | 75 | (−) |
| MAL | 10 | 75 | (+) |
| ONPG | 1 | 1 | (−) |
| TAR | 5 | 50 | (−) |
| ACE | 1 | 25 | (+) |
| CET | 1 | 10 | (−) |
| OF/G | 99 | 10 | (+) |
| P4 | 99 | 90 | (+) |
| K4 | 99 | 90 | (+) |
| CL4 | 50 | 25 | (−) |
| FD64 | 95 | 90 | (+) |
| TO4 | 25 | 75 | (−) |
| NIT | 95 | 10 | (−) |
| OXI | 99 | 99 | (+) |
All biochemical properties were derived from the MicroScan system (Siemens). URE, urea; H2S, hydrogen sulfide; IND, indole; LYS, lysine; ARG, arginine; ORN, ornithine; TDA, tryptophan deaminase; ESC, esculin; VP, Voges-Proskauer reaction; CIT, citric acid; MAL, malonic acid; ONPG, o-nitrophenyl-β-d-galactopyranoside; TAR, tartaric acid; ACE, acetamide; CET, cetrimide; OF/G, oxidation-fermentation test/glucose; P4, resistance rate to penicillin G 4 μg/ml; K4, resistance rate to kanamycin 4 μg/ml; CL4, resistance rate to colistin 4 μg/ml; FD64, resistance rate to nitrofurantoin 64 μg/ml; TO4, resistance rate to tobramycin 4 μg/ml; NIT, nitrate reduction; OXI, oxidase.
The numbers represents the estimated percentages of the positive rates. 1, very rare for positive reaction; 5, rare for positive reaction; 10, usually not a positive reaction; 25, mostly negative reaction; 50, positive or negative reaction; 75, usually positive reaction; 90, mostly positive reaction; 95, almost all are positive reaction; 99, positive reaction.
Misidentification of O. anthropi as other members of the Brucellaceae has already been reported (33). At present, 16S rRNA gene sequencing is used for the identification and differentiation of O. anthropi and Brucella spp. However, this approach, in particular partial sequencing, is prone to misidentifying these pathogens because of their high sequence similarities (9). Meanwhile, recA analysis provides more accurate identification and differentiation (9, 34).
In the present case, the pathogen was first identified as a member of the O. anthropi cluster by 16S rRNA analysis (Fig. 1). At that point, the pathogen could not be completely differentiated from other Ochrobactrum spp. such as O. lupini (1, 35). Since there are few clinical reports concerning infections with other Ochrobactrum spp., further investigation was required for identification. Eventually, the pathogen was successfully matched and confirmed as O. anthropi subclade II by recA analysis (34) (Fig. 2).
As discussed, catheter line infection is the most common way by which O. anthropi causes infection in humans (13–18). However, our patient had not received a central venous line or an arterial line during his hospitalization. Physical examination and systemic investigation did not reveal any symptoms or findings, and considering his clinical course, it was quite unlikely that infective endocarditis existed although an echocardiogram was not performed. The possibility of blood culture contamination was extremely low since genetically identical organisms were detected from separately obtained blood cultures. The only possible focus of infection was assumed to be a peripheral intravenous line, i.e., peripheral line-associated bloodstream infection; however, it could be a primary bacteremia as reported elsewhere (31).
Most O. anthropi isolates have been proven to be widely resistant to chloramphenicol and all β-lactams (except imipenem) via production of the AmpC β-lactamase OCH-1. This β-lactamase is chromosomal, inducible, and resistant to inhibition by clavulanic acid (14, 36). Generally, the organism is considered susceptible to gentamicin, fluoroquinolones, sulfamethoxazole-trimethoprim, and colistin (23). The pathogen in the present case was resistant to various β-lactams except carbapenems, gentamicin, ciprofloxacin, levofloxacin, and sulfamethoxazole-trimethoprim. The patient was treated with an oral cephalosporin, to which the pathogen was later shown to be resistant, but he recovered well without any complications, presumably because of the low virulence of the organism. In fact, according to previous studies, immunocompetent patients with O. anthropi infection survived and experienced clinical cure without any long-term sequelae (26, 31), and 4 of 5 immunosuppressed patients (organ transplant recipients) with O. anthropi bacteremia experienced resolution of bacteremia without antibiotic administration (37).
We performed an environmental survey for O. anthropi in the ward to which the patient had been admitted; however, no environmental isolates matched the patient isolates. Therefore, we conclude that the patient had been colonized with the organism somewhere in his body, such as his throat or intestine, and his bacteremia resulted from bacterial translocation across the mucous membrane (38).
Nucleotide sequence accession numbers.
Nucleotide sequences of 16S rRNA and recA have been deposited in DDBJ/EMBL/GenBank under the accession numbers AB778289 and AB778290.
ACKNOWLEDGMENT
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Scholz HC, Al Dahouk S, Tomaso H, Neubauer H, Witte A, Scholter M, Kämpfer P, Falsen E, Pfeffer M, Engel M. 2008. Genetic diversity and phylogenetic relationships of bacteria belonging to the Ochrobactrum-Brucella group by recA and 16S rRNA gene-based comparative sequence analysis. Syst. Appl. Microbiol. 31:1–16 [DOI] [PubMed] [Google Scholar]
- 2. Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludena D, Mateos PF, Martinez-Molina E, Velazquez E. 2005. Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl. Environ. Microbiol. 71:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zurdo-Piñero JL, Rivas R, Trujillo ME, Vizcaíno Carrasco JA, Chamber Palomares A, Mateos PF, Martínez-Molina E, Velázquez E. 2007. Ochrobactrum cytisi sp. nov. isolated from nodules of Cytisus scoparius in Spain. Int. J. Syst. Evol. Microbiol. 57:784–788 [DOI] [PubMed] [Google Scholar]
- 4. Chang BV, Chiang BW, Yuan SY. 2007. Biodegradation of nonylphenol in soil. Chemosphere 66:1857–1862 [DOI] [PubMed] [Google Scholar]
- 5. Abou-Shanab RA, Angle JS, van Berkum P. 2007. Chromate-tolerant bacteria for enhanced metal uptake by Eichhornia crassipes (Mart.). Int. J. Phytoremediation 9:91–105 [DOI] [PubMed] [Google Scholar]
- 6. Babic I, Fisher-Le Saux M, Giraud E, Boemare N. 2000. Occurrence of natural dixenic association between the symbiont Photorhabdus luminescens and bacteria related to Ochrobactrum spp. in tropical entomopathogenic Heterorhabditis spp. (Nematoda, Rhabditida). Microbiology 146:709–718 [DOI] [PubMed] [Google Scholar]
- 7. Zurek L, Schal C, Watson DW. 2000. Diversity and contribution of the intestinal bacterial community to the development of Musca domestica (Diptera: Muscidae) larvae. J. Med. Entomol. 37:924–928 [DOI] [PubMed] [Google Scholar]
- 8. Shilton CM, Brown GP, Benedict S, Shine R. 2008. Spinal arthropathy associated with Ochrobactrum anthropi in free-ranging cane toads (Chaunus [Bufo] marinus) in Australia. Vet. Pathol. 45:85–94 [DOI] [PubMed] [Google Scholar]
- 9. Kämpfer P, Citron DM, Goldstein EJ, Scholz HC. 2007. Difficulty in the identification and differentiation of clinically relevant Ochrobactrum species. J. Med. Microbiol. 56:1571–1573 [DOI] [PubMed] [Google Scholar]
- 10. Romero Gómez MP, Peinado Esteban AM, Sobrino Daza JA, Sáez Nieto JA, Alvarez D, Peña García P. 2004. Prosthetic mitral valve endocarditis due to Ochrobactrum anthropi: case report. J. Clin. Microbiol. 42:3371–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chain PS, Lang DM, Comerci DJ, Malfatti SA, Vergez LM, Shin M, Ugalde RA, Garcia E, Tolmasky ME. 2011. Genome of Ochrobactrum anthropi ATCC 49188T, a versatile opportunistic pathogen and symbiont of several eukaryotic hosts. J. Bacteriol. 193:4274–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruckner DA, Colonna P. 1993. Nomenclature for aerobic and facultative bacteria. Clin. Infect. Dis. 16:598–605 [DOI] [PubMed] [Google Scholar]
- 13. Daxboeck F, Zitta S, Assadian O, Krause R, Wenisch C, Kovarik J. 2002. Ochrobactrum anthropi bloodstream infection complicating hemodialysis. Am. J. Kidney Dis. 40:E17. [DOI] [PubMed] [Google Scholar]
- 14. Cieslak TJ, Robb ML, Drabick CJ, Fischer GW. 1992. Catheter-associated sepsis caused by Ochrobactrum anthropi: report of a case and review of related nonfermentative bacteria. Clin. Infect. Dis. 14:902–907 [DOI] [PubMed] [Google Scholar]
- 15. Gill MV, Ly H, Mueenuddin M, Schoch PE, Cunha BA. 1997. Intravenous line infection due to Ochrobactrum anthropi (CDC group Vd) in a normal host. Heart Lung 26:335–336 [DOI] [PubMed] [Google Scholar]
- 16. Kern WV, Oethinger M, Kaufhold A, Rozdzinski E, Marre R. 1993. Ochrobactrum anthropi bacteremia: report of four cases and short review. Infection 21:306–310 [DOI] [PubMed] [Google Scholar]
- 17. Gransden WR, Eykyn SJ. 1992. Seven cases of bacteremia due to Ochrobactrum anthropi. Clin. Infect. Dis. 15:1068–1069 [DOI] [PubMed] [Google Scholar]
- 18. Stiakaki E, Galanakis E, Samonis G, Christidou A, Maraka S, Tselentis Y, Kalmanti M. 2002. Ochrobactrum anthropi bacteremia in pediatric oncology patients. Pediatr. Infect. Dis. J. 21:72–74 [DOI] [PubMed] [Google Scholar]
- 19. Pérez-Blanco V, García-Caballero J, Domínguez-Melcón FJ, Gómez-Limón IM. 2005. Ochrobactrum anthropi infectious endocarditis in an immunocompetent patient. Enferm. Infecc. Microbiol. Clin. 23:111–112 (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 20. Mahmood MS, Sarwari AR, Khan MA, Sophie Z, Khan E, Sami S. 2000. Infective endocarditis and septic embolization with Ochrobactrum anthropi: case report and review of literature. J. Infect. 40:287–290 [DOI] [PubMed] [Google Scholar]
- 21. Appelbaum PC, Campbell DB. 1980. Pancreatic abscess associated with Achromobacter group Vd biovar 1. J. Clin. Microbiol. 12:282–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barson WJ, Cromer BA, Marcon MJ. 1987. Puncture wound osteochondritis of the foot caused by CDC group Vd. J. Clin. Microbiol. 25:2014–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braun M, Jonas JB, Schönherr U, Naumann GO. 1996. Ochrobactrum anthropi endophthalmitis after uncomplicated cataract surgery. Am. J. Ophthalmol. 122:272–273 [DOI] [PubMed] [Google Scholar]
- 24. Chang HJ, Christenson JC, Pavia AT, Bobrin BD, Bland LA, Carson LA, Arduino MJ, Verma P, Aguero SM, Carroll K, Jenkins E, Daly JA, Woods ML, Jarvis WR. 1996. Ochrobactrum anthropi meningitis in pediatric pericardial allograft transplant recipients. J. Infect. Dis. 173:656–660 [DOI] [PubMed] [Google Scholar]
- 25. Van Horn KG, Gedris CA, Ahmed T, Wormser GP. 1989. Bacteremia and urinary tract infection associated with CDC group Vd biovar 2. J. Clin. Microbiol. 27:201–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vaidya SA, Citron DM, Fine MB, Murakami G, Goldstein EJ. 2006. Pelvic abscess due to Ochrobactrum intermedium in an immunocompetent host: case report and review of the literature. J. Clin. Microbiol. 44:1184–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jelveh N, Cunha BA. 1999. Ochrobactrum anthropi bacteremia. Heart Lung 28:145–146 [DOI] [PubMed] [Google Scholar]
- 28. Kish MA, Buggy BP, Forbes BA. 1984. Bacteremia caused by Achromobacter species in an immunocompromised host. J. Clin. Microbiol. 19:947–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brivet F, Guibert M, Kiredjian M, Dormont J. 1993. Necrotizing fasciitis, bacteremia, and multiorgan failure caused by Ochrobactrum anthropi. Clin. Infect. Dis. 17:516–518 [DOI] [PubMed] [Google Scholar]
- 30. Haditsch M, Binder L, Tschurtschenthaler G, Watschinger R, Zauner G, Mittermayer H. 1994. Bacteremia caused by Ochrobactrum anthropi in an immunocompromised child. Infection 22:291–292 [DOI] [PubMed] [Google Scholar]
- 31. Yu WL, Lin CW, Wang DY. 1998. Clinical and microbiologic characteristics of Ochrobactrum anthropi bacteremia. J. Formos. Med. Assoc. 97:106–112 [PubMed] [Google Scholar]
- 32. Romano S, Aujoulat F, Jumas-Bilak E, Masnou A, Jeannot JL, Falsen E, Marchandin H, Teyssier C. 2009. Multilocus sequence typing supports the hypothesis that Ochrobactrum anthropi displays a human-associated subpopulation. BMC Microbiol. 18:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Elsaghir AA, James EA. 2003. Misidentification of Brucella melitensis as Ochrobactrum anthropi by API 20NE. J. Med. Microbiol. 52:441–442 [DOI] [PubMed] [Google Scholar]
- 34. Scholz HC, Tomaso H, Dahouk SA, Witte A, Schloter M, Kämpfer P, Falsen E, Neubauer H. 2006. Genotyping of Ochrobactrum anthropi by recA-based comparative sequence, PCR-RFLP, and 16S rRNA gene analysis. FEMS Microbiol. Lett. 257:7–16 [DOI] [PubMed] [Google Scholar]
- 35. Teyssier C, Marchandin H, Jean-Pierre H, Masnou A, Dusart G, Jumas-Bilak E. 2007. Ochrobactrum pseudintermedium sp. nov., a novel member of the family Brucellaceae, isolated from human clinical samples. Int. J. Syst. Evol. Microbiol. 57:1007–1013 [DOI] [PubMed] [Google Scholar]
- 36. Nadjar D, Labia R, Cerceau C, Bizet C, Philippon A, Arlet G. 2001. Molecular characterization of chromosomal class Cbeta-lactamase and its regulatory gene in Ochrobactrum anthropi. Antimicrob. Agents Chemother. 45:2324–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ezzedine H, Mourad M, Van Ossel C, Logghe C, Squifflet JP, Renault F, Wauters G, Gigi J, Wilmotte L, Haxhe JJ. 1994. An outbreak of Ochrobactrum anthropi bacteraemia in five organ transplant patients. J. Hosp. Infect. 27:35–42 [DOI] [PubMed] [Google Scholar]
- 38. Holmes B, Popoff M, Kiredjian M, Kersters K. 1988. Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int. J. Syst. Bacteriol. 38:406–416 [Google Scholar]