Abstract
In order to switch from IS6110 and polymorphic GC-rich repetitive sequence (PGRS) restriction fragment length polymorphism (RFLP) to 24-locus variable-number tandem-repeat (VNTR) typing of Mycobacterium tuberculosis complex isolates in the national tuberculosis control program in The Netherlands, a detailed evaluation on discriminatory power and agreement with findings in a cluster investigation was performed on 3,975 tuberculosis cases during the period of 2004 to 2008. The level of discrimination of the two typing methods did not differ substantially: RFLP typing yielded 2,733 distinct patterns compared to 2,607 in VNTR typing. The global concordance, defined as isolates labeled unique or identically distributed in clusters by both methods, amounted to 78.5% (n = 3,123). Of the remaining 855 cases, 12% (n = 479) of the cases were clustered only by VNTR, 7.7% (n = 305) only by RFLP typing, and 1.8% (n = 71) revealed different cluster compositions in the two approaches. A cluster investigation was performed for 87% (n = 1,462) of the cases clustered by RFLP. For the 740 cases with confirmed or presumed epidemiological links, 92% were concordant with VNTR typing. In contrast, only 64% of the 722 cases without an epidemiological link but clustered by RFLP typing were also clustered by VNTR typing. We conclude that VNTR typing has a discriminatory power equal to IS6110 RFLP typing but is in better agreement with findings in a cluster investigation performed on an RFLP-clustering-based cluster investigation. Both aspects make VNTR typing a suitable method for tuberculosis surveillance systems.
INTRODUCTION
DNA fingerprinting of Mycobacterium tuberculosis isolates has been applied for the investigation of epidemiological links between tuberculosis (TB) cases in several countries since DNA typing techniques were developed in the early 1990s (1–4). In The Netherlands, IS6110 restriction fragment length polymorphism (RFLP) typing was used nationwide from 1993 until the end of 2008. Although RFLP typing revolutionized studies on the transmission of M. tuberculosis, the method remained technically demanding and time consuming. The analysis of the complex IS6110 RFLP banding pattern requires a sophisticated computer application and a high degree of expertise (5). Moreover, in the Netherlands, it took on average 44 days to the deliver the RFLP typing result to the Municipal Health Services, after the isolate had reached the laboratory (D. van Soolingen, personal communication). This major drawback significantly limited the usefulness of DNA fingerprinting in routine examination of transmission, as it in fact only offered retrospective confirmation of suspected epidemiological links. In 2006, 24-locus variable-number tandem-repeat (VNTR) typing was favorably evaluated and proposed as the new gold standard for typing of M. tuberculosis (6). In order to switch from RFLP to VNTR typing in 2008 in the Netherlands, it was considered necessary to establish a retrospective VNTR pattern database of possible sources of infection for a preceding period of 5 years. We therefore retyped isolates of all 3,975 culture-confirmed TB cases diagnosed during the period of 2004 to 2008. Since cluster investigation was routinely applied in The Netherlands on the basis of RFLP cluster results, this provided the unique opportunity to compare, on a large population-based scale, the validity of the clustering of cases on the basis of RFLP and VNTR typing and to evaluate the latter, faster method to trace epidemiological links.
MATERIALS AND METHODS
The cohort.
The National Institute for Public Health and the Environment (RIVM) in Bilthoven, The Netherlands, serves as a reference laboratory for the secondary laboratory diagnosis of all TB cases in The Netherlands, offering identification, drug susceptibility testing, and molecular typing. During the period of 2004 to 2008, 3,975 M. tuberculosis isolates of culture-confirmed TB cases were subjected to RFLP typing at the RIVM, using a previously described protocol (1). For isolates with ≤5 copies of IS6110, additional polymorphic GC-rich repetitive sequence (PGRS) RFLP typing (7) was performed. The additional PGRS typing results were combined with the IS6110 RFLP typing result to determine the clustering of cases and to initiate a cluster investigation. Municipal health services in the Netherlands conduct contact tracing and source case finding, according to the “stone in the pond” principle (8), and also cluster investigations; findings are reported to the national surveillance unit.
The lineages of the samples included were determined based on spoligotyping or by the use of the MIRU-VNTRplus online database (www.miru-vntrplus.org). Overall, 65% (n = 2,589) of the isolates belonged to the Euro-American lineage, 9% (n = 373) to the East African-Indian (EAI) lineage, 8% (n = 335) to the Central Asian strain (CAS) lineage, and 7% (n = 296) to the Beijing lineage of M. tuberculosis; 2% (n = 66) represented Mycobacterium africanum and 2% (n = 65) M. bovis. The final 6% (n = 251) of the strains were assigned to the (sub)species Mycobacterium canetti or M. caprae or remained unknown or unidentifiable to the genotype family level.
VNTR typing.
Purified DNA extracts of M. tuberculosis isolates were subjected to VNTR typing, partly by GenoScreen (Lille, France) and partly by the RIVM. Both entities performed 24-locus VNTR typing according to the international standard (6). To monitor and ensure the quality and mutual reproducibility, one negative control, DNA from two positive controls with known VNTR profiles (M. tuberculosis strain H37Rv and M. bovis BCG strain P3) and three blinded duplicates were included on each 96-well plate.
Computer and statistical analysis.
BioNumerics software version 6.6.4 (Applied Maths, Belgium) was used for the analysis of molecular typing results. IS6110 RFLP patterns were recorded as the fingerprint type, and VNTR typing results as the character type.
The statistical significance was measured for the proportion of confirmed or presumed epidemiological links in the subgroup of isolates that were clustered on RFLP and VNTR typing compared to the subgroup of isolates clustered by RFLP but not by VNTR typing. A logistic regression analysis was conducted to model the probability to find an epidemiological link whether an isolate clustered by RFLP typing was confirmed or not by VNTR typing. The relation between the epidemiological status and reconfirmed status by VNTR typing within the same cluster was adjusted for the cluster size and correlation of samples within the same cluster. The models were fitted by using package Ime4 of the statistical software package R.
Clustering on the basis of RFLP and VNTR typing.
For the comparison of the two typing methods, differences in cluster distribution were investigated, as well as the correlation of clustering with findings in a cluster investigation. Because, until 2009, RFLP typing was applied in The Netherlands, a cluster investigation was conducted only on the basis of RFLP clustering of cases.
Two percent (85/3,975) of the isolates with a double allele in one of the 24 VNTR loci were treated as follows. For cases where separate consideration of each of the two alleles resulted in one clustered and one unique VNTR pattern or two clustered patterns, the clustered pattern(s) were included (as two separated cases in the case of two clustered patterns) in the total database for analysis of the correlation with the contact tracing data. Isolates with >1 locus with double alleles were excluded from the analysis.
A total of 52 isolates for which one or two VNTR loci could not be amplified even after repeating the amplification in a single locus PCR were included in the database and all analyses, with the missing loci indicated with an “x” and treated as a specific allele in the pattern.
The concordance in clustering of cases between the two typing methods was calculated on the basis of (i) identical cluster compositions and shared unique RFLP and VNTR profiles and (ii) concordance in labeling as a “clustered” or “unique” isolate, regardless of the cluster composition. For comparisons of deviating VNTR patterns of isolates within RFLP clusters the VNTR pattern with the highest average similarity in the specific RFLP cluster was considered the reference pattern.
RESULTS
During the period of 2004 to 2008, a total of 3,975 culture-confirmed TB cases were diagnosed in The Netherlands and had their isolates subjected to genotyping analysis. Over the 5-year period, 2.1% (n = 85) of the samples displayed a double allele in one of the 24 VNTR loci. When the results of these 85 isolates were analyzed as two separate alleles and genotypes, 53 were unique in the database by either of both alleles/genotypes. For 32 of the 85 cases, one of the VNTR patterns found was clustered with at least one other case, and in three cases, both VNTR patterns were clustered. As for each of three latter cases, the two VNTR patterns were included in the database, and the total number of VNTR patterns of isolates for further analysis amounted to 3,978. Incomplete patterns, lacking one or two alleles among the 24 loci, were detected in 52 (1.3%) of the cases in our database.
In total, for the 3,978 VNTR typing results, 2,607 different VNTR types were detected compared to 2,733 distinct RFLP types. In this data set, 47% (n = 1,857) of the isolates belonged to one of the 486 VNTR clusters and 42% (n = 1,683) belonged to one of the 438 clusters identified by RFLP typing. The distributions of the cluster sizes on the basis of the two typing techniques are highly similar, as depicted in Fig. 1.
Fig 1.
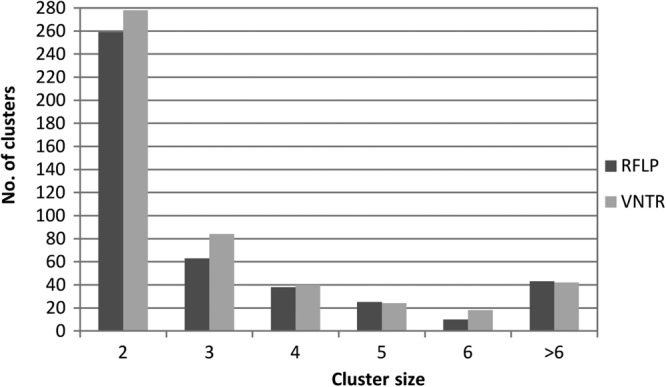
Distribution of cluster sizes obtained on the basis of RFLP (gray bars) and VNTR (black bars) typing.
Comparison of RFLP and VNTR typing results.
The concordance between the two typing techniques was 78.5% (n = 3,123), as defined by 1,307 isolates identically clustered (i.e., with a same cluster composition) and 1,816 unique isolates in both methods (Table 1). For the remaining 855 isolates, 12% (n = 479) were clustered only by VNTR, 7.7% (n = 305) only by RFLP typing, and 1.8% (n = 71) by both typing methods but in a different cluster composition.
Table 1.
Comparison of clustering on the basis of RFLP and VNTR typing results of 3,978 M. tuberculosis isolates in The Netherlands, 2004–2008
| VNTR |
Total | ||
|---|---|---|---|
| Cluster | Unique | ||
| RFLP | |||
| Cluster | 1,378a | 305 | 1,683 |
| Unique | 479 | 1,816 | 2,295 |
| Total | 1,857 | 2,121 | 3,978 |
Seventy-one RFLP- and VNTR-clustered isolates showed different cluster compositions in RFLP and VNTR typing.
Cluster composition on the basis of RFLP and VNTR.
Among the 438 RFLP clusters, comprising 1,683 isolates, 58% (256 clusters, including a total of 784 isolates) (Fig. 2) displayed identical cluster sizes and composition by VNTR typing. An additional 47 RFLP clusters (11%) showed almost identical sizes and compositions, with only one or more single cases of these clusters split off by VNTR typing.
Fig 2.
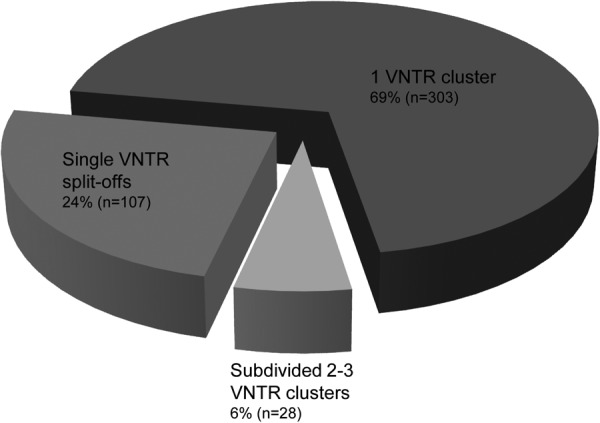
Composition of RFLP clusters by VNTR typing. In total, 69% of the RFLP clusters showed a completely identical (n = 256) or similar (n = 47; with only one or more single cases split off by a distinct VNTR pattern) composition by VNTR typing. For 24% (n = 107) of the RFLP clusters, VNTR typing resulted in sets of only unique patterns, whereas 6% (n = 28) of the RFLP clusters were subdivided into 2 or 3 VNTR clusters.
Twenty-eight (6%) of the 438 RFLP clusters, involving 302 cases, were subdivided by VNTR typing into two or three clusters each, with sizes ranging from 2 to 48 cases. One hundred seven (24%) of the 438 RFLP clusters, comprising 241 cases, were subdivided into single VNTR patterns. Ninety (84%) of these 107 RFLP clusters consisted of only two cases.
Four hundred seventy-nine isolates, which exhibited unique profiles in RFLP typing, were clustered by VNTR typing into 226 clusters. Of these, 65% (n = 148) represented “new” VNTR clusters, as they consisted of isolates (n = 307) with unique RFLP profiles only. The other isolates (n = 172) were grouped within 78 existing VNTR clusters.
The aforementioned scenarios also occurred vice versa. Out of the 486 VNTR clusters, including 1,857 isolates, 51% (n = 241) consisted of identically sized and composed RFLP clusters, 15% (n = 71) consisted of one defined RFLP cluster enlarged by unique RFLP cases, 4% (n = 21) were subdivided by RFLP typing into “subclusters” in two to four cases, and 31% (n = 152), mostly (n = 118) including two isolates only, consisted of cases with unique RFLP patterns.
Typing results in relation to cluster investigation.
Information on cluster investigation was available for 87% (n = 1,462) of the isolates in RFLP clusters. Of these, the epidemiological link with another cluster member was confirmed in 35% of the cases (n = 514), there was a presumed link in 16% (n = 226) of the cases, and no link was found in 49% (n = 722) of the cases. Ninety-four percent (n = 484) of the isolates clustered by RFLP and with a confirmed epidemiological link were also clustered in VNTR typing. Among the isolates clustered by RFLP but with only a presumed link, 88% (n = 200) proved clustered by VNTR typing. In contrast, only 64% (n = 462) of the isolates clustered by RFLP but devoid of epidemiological links in cluster investigation were clustered by VNTR typing (Fig. 3, top).
Fig 3.
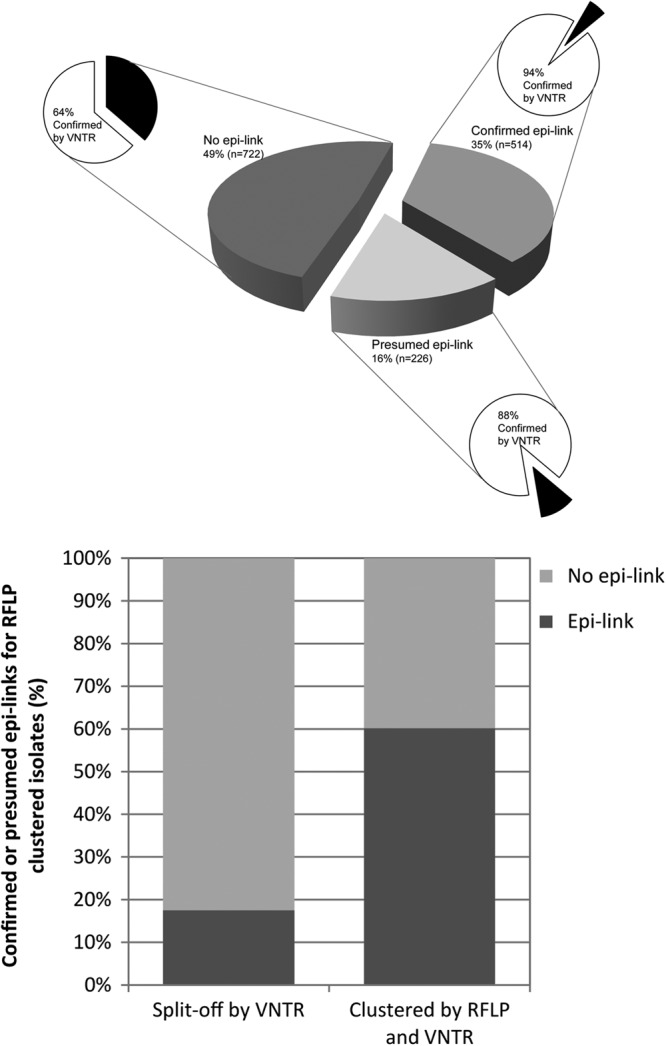
(Top) RFLP-clustered isolates (n = 1,462) divided by cluster investigation results, classified as confirmed, presumed, and no epidemiological link. For each segment, the distribution of confirmed clustering by VNTR typing is shown. (Bottom) Percentages of confirmed or presumed epidemiological links for RFLP-clustered cases. Isolates with and without VNTR-supported clustering.
Among the 1,462 isolates in RFLP clusters, an epidemiological link with another member of the cluster was confirmed or presumed in 51% (n = 740) of the cases. In the subgroup of 1,146 isolates that were clustered on the basis of both RFLP and VNTR typing, the proportion of confirmed (n = 484) or presumed epidemiological links (n = 200) rose to 60% (Fig. 3, bottom). In contrast, among the 316 cases clustered by RFLP but distinguished from the cluster by a VNTR typing result, only 56 (18%) showed a confirmed (n = 30) or presumed (n = 26) epidemiological link in contact tracing. This difference in percentage is statistically highly significant (P < 0.001).
VNTR locus differences among RFLP clustered cases.
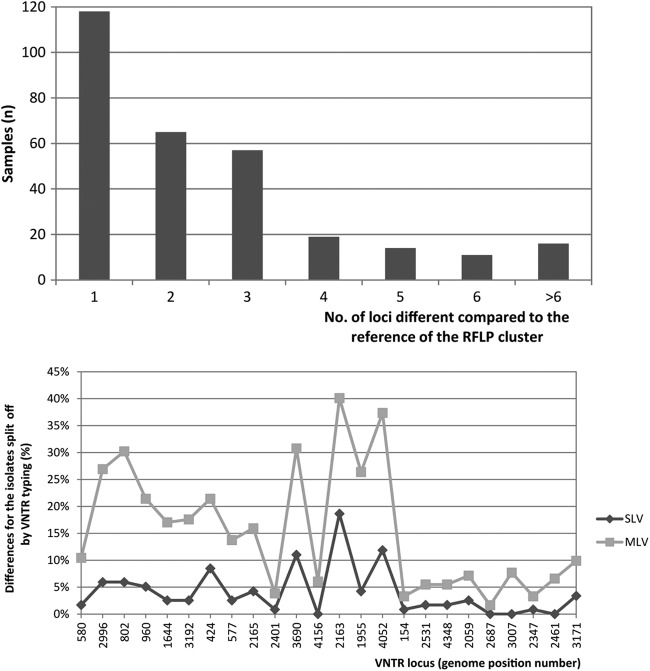
In total, 21% (n = 359) of all 1,683 isolates clustered by RFLP were split off from their respective cluster on the basis of the VNTR typing result, and this involved 300 different VNTR patterns. The distribution of the number of loci by which the pattern deviated from the reference pattern in the cluster is shown in Fig. 4 (top). Single-locus variations were detected for 118 VNTR patterns. The patterns that diverged by more than two loci were not related to isolates of specific genotypes or phylogenetic groups on the basis of their RFLP patterns.
Fig 4.
(Top) Number of loci that differ in VNTR typing patterns (n = 300) split off from their RFLP cluster. (Bottom) Distribution of detected differences by locus in 24-locus VNTR genotypes of samples clustered by RFLP typing and split off by VNTR typing, separated by single-locus variation (SLV) cases and multilocus variation cases (MLV).
For the single-locus variant (SLV) and the multilocus variant (MLV) patterns, the loci that were mostly responsible for isolates being split off their RFLP clusters on the basis of the VNTR pattern difference are the loci with genome position numbers 2163, 4052, and 3690. For MLV patterns, changes were also frequently seen in locus 802 and 2996 (30% and 17% of the patterns, respectively) (Fig. 4, bottom).
Interestingly, the number of loci that differed in isolates split off from their RFLP cluster by a unique VNTR pattern was associated with the strength of the epidemiological links observed for the respective patients. Epidemiological link data were available for 87% (n = 311) of the isolates split off their RFLP clusters by VNTR typing. In isolates of patients with proven epidemiological links that were split off by VNTR typing (n = 19), we found a single-locus variation in 74% (n = 14) of the isolates and >1 locus variation in the remainder (26%; n = 5). In contrast, the percentage of >1 locus variation rose to 61% (n = 11) among isolates of patients with presumed links only and 71% (n = 103) of isolates from patients without epidemiological links (Fig. 5).
Fig 5.
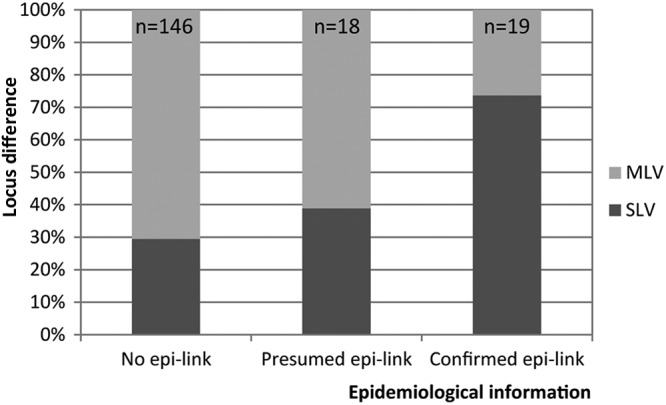
Percentages of single-locus variants (SLV) and multilocus variants (MLV) for the isolates clustered by RFLP and split off by VNTR typing in relation to the epidemiological information.
DISCUSSION
This report represents the first nationwide, surveillance program-based comparative study of RFLP and VNTR typing, integrating full population coverage over a 5-year time period and extensive epidemiological data. The comparison of the typing results of 3,978 M. tuberculosis isolates showed an overall concordance in clustering of 79% and highly similar discriminatory power in the two methods (2,607 versus 2,733 distinct patterns, respectively). Such a concordance is consistent with linkage between markers expected from the clonal evolution of Mycobacterium tuberculosis (9, 10). The expectable remaining 21% reflect the stochastic component in the respective evolution of the independent molecular markers used.
We detected a slightly higher level of discriminatory power of RFLP compared to VNTR typing (i.e., more distinct RFLP types), and hence, more clusters were identified by VNTR typing. However, this result does not reflect a comparison between only IS6110 RFLP and VNTR typing, because additional PGRS RFLP typing was used for strains with ≤5 copies of IS6110 (n = 358). Nevertheless, it is consistent with those of previous comparative studies that were performed at regional (11), population-based scales (12) or at a national level (13), which showed slightly higher to slightly lower discriminatory power of VNTR versus IS6110 RFLP typing.
Importantly, cases with isolates that were clustered by both VNTR and RFLP typing had significantly more confirmed or presumed epidemiological links than cases with isolates clustered by RFLP only. Most (82%) split-offs by VNTR of isolates previously clustered by RFLP occurred in clusters without an epidemiological link or without epidemiological information, and this finding was in agreement with findings in a previous regional study in The Netherlands (14). Of note, 71 isolates (2% of the data set) were split off from RFLP clusters by VNTR typing and formed new VNTR-based clusters or expanded existing VNTR clusters. For 82% (n = 58) of the isolates in this subset, no epidemiological link could be detected on the basis of the originally identified RFLP clusters. Taken together, these findings suggest that VNTR typing results could, in general, be in better agreement with the true chain of transmission than the RFLP typing results were. Yet, this can only be confirmed by a renewed source case investigation on the basis of the VNTR cluster information.
In contrast, we found no clustering by VNTR typing in only 6% of the cases with proven epidemiological links. This is in line with the rate of change (4.9%) among VNTR patterns found among sets of epidemiologically linked or serial isolates (6). Most (74%; n = 14) of the changes seen between isolates with proven epidemiological links were consistently restricted to single-locus variations, again in general agreement with previous findings (15). These changes most likely reflect limited genetic drift among these markers in clonal populations during disease progression and transmission. Consistently, this drift was observed most frequently in VNTR loci that are in general the most variable in various strain populations, such as loci with genome positions 2163b and 4052 (6).
As the cluster investigation was performed only on the basis of RFLP clustering, we cannot formally evaluate the epidemiological significance of the newly formed clusters on the basis of VNTR typing that were not found as such by RFLP typing. However, we can estimate the impact of the slight overclustering by VNTR typing. We found that 479/3,978 (12%) isolates were clustered on the basis of VNTR typing, where they earlier revealed unique RFLP patterns, but also found that 305 (8%) isolates that were clustered in RFLP typing had unique VNTR patterns. This will predictably result in a 12% (174 [479 − 305] of the total 1,462 RFLP-clustered cases for which a cluster investigation has been performed) increase in source case-finding activities. This may be worth executing because clustering on the basis of VNTR typing seems in better agreement with findings in conventional contact tracing. This will most likely optimize the yield of confirmed epidemiological links in the future. From a TB control perspective, the slightly lower discriminatory power in VNTR typing (i.e., bias toward clustering) could therefore be considered preferable over a bias to nonclustering in RFLP typing. Failure to recognize TB transmission, on the basis of typing results, may result in less source case finding and contact tracing and may enable further transmission of M. tuberculosis.
To improve the discriminatory power of molecular typing of M. tuberculosis isolates by VNTR typing, spoligotyping could be used in combination with this, as it has been shown to result in slightly increased discriminatory power in regional studies (11, 12, 16). Additional hypervariable VNTR loci have also been investigated and were recommended especially for high-incidence settings where the Beijing genotype is prevalent (15, 17). However, Beijing genotype strains only make up about 7% of the current data set and thus are not the full explanation for the extent of discrepancies observed between RFLP and VNTR typing.
The switch to VNTR typing also has several technical advantages. First, VNTR typing is less time-consuming and provides a faster time to result, because the technique is based on DNA amplification and thus requires far less DNA (i.e., less biomass) and requires hardly any culture delay, in contrast to RFLP typing. Second, the results of VNTR typing are in a simple format, which facilitates a simple exchange of typing information and interlaboratory comparison. Third, VNTR typing is much easier to perform and can be implemented more efficiently (13). These advantages are in agreement with the implementation of VNTR typing for the outbreak detection of methicillin-resistant Staphylococcus aureus (MRSA) (18) and Salmonella enterica (19).
The switch to VNTR typing also had down sides. The first round of worldwide proficiency testing initiated by our laboratory within the framework of an ECDC project revealed that both the intralaboratory and interlaboratory reproducibilities are to be improved in a number of laboratories, indicating the need for careful implementation (20). Moreover, new patient cluster identification numbers and compositions resulting from the discordances with RFLP typing introduced a major challenge for the TB units of the Municipal Health Services.
However, with a concordance of 78.5%, a strong suggestion that VNTR typing results are more in line with the true chains of transmission than the RFLP typing and the technical advantages of VNTR typing, we conclude that VNTR typing is more suitable than RFLP typing of M. tuberculosis in national tuberculosis surveillance programs, such as that in The Netherlands. We expect that the use of this method will ensure the transition until whole-genome sequencing, which provides the ultimate level of discrimination and is under evaluation for its feasibility and impact (21, 22), will become cost effective in routine TB molecular-guided control and surveillance.
ACKNOWLEDGMENTS
We thank all the municipal health services that reported their epidemiological results to the national surveillance unit and the microbiological laboratories in The Netherlands for their efforts to send Mycobacterium tuberculosis isolates to the RIVM. Maarten Schipper is acknowledged for statistical analysis of the data.
P. Supply is a consultant for GenoScreen, Lille, France. All other authors have declared that no competing interests exist.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM, Small PM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prodinger WM. 2007. Molecular epidemiology of tuberculosis: toy or tool? A review of the literature and examples from Central Europe. Wien Klin. Wochenschr. 119:80–89 [DOI] [PubMed] [Google Scholar]
- 3. van Soolingen D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1–26 [DOI] [PubMed] [Google Scholar]
- 4. Garcia de Viedma D, Mokrousov I, Rastogi N. 2011. Innovations in the molecular epidemiology of tuberculosis. Enferm. Infecc. Microbiol. Clin. 29(Suppl 1):8–13 [DOI] [PubMed] [Google Scholar]
- 5. van Soolingen D, Arbeit RD. 2001. Dealing with variation in molecular typing of Mycobacterium tuberculosis: low-intensity bands and other challenges. J. Med. Microbiol. 50:749–751 [DOI] [PubMed] [Google Scholar]
- 6. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rhee JT, Tanaka MM, Behr MA, Agasino CB, Paz EA, Hopewell PC, Small PM. 2000. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int. J. Tuberc. Lung Dis. 4:1111–1119 [PubMed] [Google Scholar]
- 8. Veen J. 1992. Microepidemics of tuberculosis: the stone-in-the-pond principle. Tuber. Lung Dis. 73:73–76 [DOI] [PubMed] [Google Scholar]
- 9. Supply P, Warren RM, Banuls AL, Lesjean S, Van Der Spuy GD, Lewis LA, Tibayrenc M, Van Helden PD, Locht C. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529–538 [DOI] [PubMed] [Google Scholar]
- 10. Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. U. S. A. 101:4871–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roetzer A, Schuback S, Diel R, Gasau F, Ubben T, di Nauta A, Richter E, Rusch-Gerdes S, Niemann S. 2011. Evaluation of Mycobacterium tuberculosis typing methods in a 4-year study in Schleswig-Holstein, Northern Germany. J. Clin. Microbiol. 49:4173–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allix-Beguec C, Fauville-Dufaux M, Supply P. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 46:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bidovec-Stojkovic U, Zolnir-Dovc M, Supply P. 2011. One year nationwide evaluation of 24-locus MIRU-VNTR genotyping on Slovenian Mycobacterium tuberculosis isolates. Respir. Med. 105(Suppl 1):S67–S73 [DOI] [PubMed] [Google Scholar]
- 14. van Deutekom H, Supply P, de Haas PE, Willery E, Hoijng SP, Locht C, Coutinho RA, van Soolingen D. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 43:4473–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yokoyama E, Kishida K, Uchimura M, Ichinohe S. 2007. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect. Genet. Evol. 7:499–508 [DOI] [PubMed] [Google Scholar]
- 16. Oelemann MC, Diel R, Vatin V, Haas W, Rusch-Gerdes S, Locht C, Niemann S, Supply P. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45:691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Hoek KG, Ndabambi SL, Jordaan AM, Victor TC, van Helden PD, Warren RM. 2008. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J. Clin. Microbiol. 46:3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes A, Edwards GF, Girvan EK, Hannant W, Danial J, Fitzgerald JR, Templeton KE. 2010. Comparison of two multilocus variable-number tandem-repeat methods and pulsed-field gel electrophoresis for differentiating highly clonal methicillin-resistant Staphylococcus aureus isolates. J. Clin. Microbiol. 48:3600–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sintchenko V, Wang Q, Howard P, Ha CW, Kardamanidis K, Musto J, Gilbert GL. 2012. Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA). BMC Infect. Dis. 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Beer JL, Kremer K, Kodmon C, Supply P, van Soolingen D. 2011. First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 50:662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJ, Brinkman FS, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364:730–739 [DOI] [PubMed] [Google Scholar]
- 22. Schurch AC, Kremer K, Daviena O, Kiers A, Boeree MJ, Siezen RJ, van Soolingen D. 2010. High-resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J. Clin. Microbiol. 48:3403–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]