Abstract
We describe the development of a multiplex reverse transcription-PCR (RT-PCR) with Luminex microarray hybridization for detection of influenza virus subtypes (FLULUM). Performance of FLULUM was evaluated by comparing it to our real-time RT-PCR influenza virus assay on samples collected during two influenza seasons. Both assays targeted the matrix genes of influenza virus A (FluA M) and influenza virus B (FluB M) and the hemagglutinin genes of seasonal H3N2 (H3) and H1N1 (H1) and 2009 pandemic H1N1 (2009 H1). We evaluated FLULUM on both the Luminex LX200 and the Luminex MagPix instruments. Compared to real-time PCR, FLULUM tested on 259 specimens submitted in the 2010-2011 season showed sensitivities of 97.3% for FluA M, 90.5% for 2009 H1, 96.9% for H3, and 88.9% for FluB M. No specimens were positive for seasonal H1. FLULUM tested on 806 specimens submitted in the 2011-2012 season showed a sensitivity of 100% for FluA M, 89.9% for 2009 H1, 96.4% for H3, and 95.6% for FluB M. No cross-reactivity was observed for other respiratory viruses. Analytical sensitivity was assessed by testing dilutions of specimens with high viral loads. The limits of detection of FLULUM were comparable to those of the real-time PCR assay for FluA M, FluB M, and H3. The limits of detection for seasonal H1 and 2009 H1 were 10-fold higher for the FLULUM assay compared to real-time PCR. The FLULUM is an economic assay with high clinical sensitivity and specificity. It is particularly suited to high-volume detection of influenza viruses.
INTRODUCTION
The impact of seasonal and pandemic influenza includes significant morbidity and mortality, as well as a socio-economic burden in medical care costs and loss of productivity (1–3). During the influenza A 2009 H1N1 virus pandemic, the detection of influenza virus types A and B and differentiation between influenza virus A subtypes became important for monitoring the outbreak. In addition, specific and accurate diagnosis can improve patient management, since antiviral treatment is effective if the disease is identified early in the course of illness (4). Moreover, the antiviral susceptibility of influenza virus A subtypes can differ. CDC data from a limited number of states in late 2008 indicated that the prevalence of influenza A H1N1 (seasonal) virus strains resistant to the antiviral oseltamivir was high. Influenza types A H3N2, A 2009 H1N1 pandemic, and B viruses are generally susceptible to oseltamivir. The Centers for Disease Control and Prevention (CDC) recommends when influenza A H1N1 (seasonal) virus infection or exposure is suspected, zanamivir or a combination of oseltamivir and rimantadine are more appropriate options than oseltamivir alone (5).
Traditionally, respiratory viral infections have been diagnosed by culture, rapid antigen test, or DFA (6). However, many studies have demonstrated that molecular diagnostic assays show superior sensitivity compared to that of conventional assays and are now becoming acceptable as the new gold standard (7–12). Real-time PCR in particular offers significant advantages due to its high sensitivity and rapid turnaround time (13–16).
The University of Washington Molecular Virology Laboratory has developed a real-time PCR assay to detect influenza virus types and subtypes in samples from patients seen at the university's medical centers, hospitals, and clinics. The TaqMan assay is multiplexed to detect six targets (influenza virus A matrix gene [FluA M], influenza virus B matrix gene [FluB M], the hemagglutinin genes of types H3N2 [H3], H1N1 seasonal [H1], and H1N1 pandemic 2009 [2009 H1], and an extraction control) in three reactions. However, the number of clinical specimens that can be tested on a daily basis may be insufficient due to the limited multiplexing capacity of real-time PCR. In particular, the 2009 influenza pandemic prompted our investigation of multiplexing technologies that would provide influenza virus typing and subtyping in a single reaction to increase throughput capacity and potentially reduce the overall cost of testing.
The Luminex xMap technology is a bead-based array platform that can detect up to 100 DNA targets simultaneously (17). We developed an assay combining one-tube asymmetric multiplex reverse transcription-PCR (RT-PCR) with xMap bead hybridization and detection (FLULUM) and compared its performance with our real-time RT-PCR assay as the gold standard. The FLULUM assay was evaluated on both the Luminex LX200 and the MagPix instrument. In addition, we compared the turnaround time, direct costs, and throughput of FLULUM to real-time RT-PCR.
MATERIALS AND METHODS
Clinical specimens.
A total of 1,065 respiratory specimens submitted to the University of Washington Molecular Virology Laboratory from 2010 to 2012 for influenza diagnosis by real-time RT-PCR were used in the present study. The specimens included nasal washes in saline, nasal or nasopharyngeal swabs in viral transport medium, bronchoalveolar lavage, throat or oropharyngeal swabs, tracheal aspirates, or sputum.
Nucleic acid extraction.
Total nucleic acid was extracted with a MagNA Pure LC using the total nucleic acid extraction kit (Roche Diagnostics, Indianapolis, IN). The sample and elution volumes were 200 μl. In order to monitor RNA extraction efficiency and amplification inhibition, a 262-base RNA transcript derived from jellyfish DNA (EXO) was spiked into the lysis buffer at a final concentration of 1,000 copies/PCR (18). A mixed positive control containing 200 to 1,000 copies/PCR of each influenza virus harvested from cell culture and diluted in minimal essential medium and one negative control consisting of cultured, uninfected human epithelial cells were processed with each batch of clinical specimens. All of the samples were tested by real-time RT-PCR the same day. Samples extracted between September 2010 and April 2011 were stored at −80°C and tested by FLULUM in May 2011. For samples extracted between September 2011 and May 2012, some (n = 63) were tested the same day as the RT-PCR test, whereas most were stored at −80°C for retrospective FLULUM testing in May and June 2012.
Laboratory-developed influenza virus real-time RT-PCR.
The influenza virus RT-PCR TaqMan assay detects six targets: FluA M, FluB M, H3, H1, 2009 H1, and EXO. The assay consists of three duplex reactions (FAM/VIC or HEX/VIC). RT-PCR was performed in 40-μl reaction volumes containing 10 μl of nucleic acid template, 1× buffer, 1× enzyme mix, and 1× ROX dye (RNA UltraSense One-Step qRT-PCR System; Invitrogen, Carlsbad, CA). The target genes, amplicon sizes, final reaction concentrations, 5′ and 3′ probe labels, and sequences of the primers and probes used in the laboratory-developed real-time RT-PCR assay are shown in Table 1. Each of the three reaction mixes in each PCR run included nucleic acid extracted from the positive mix and negative controls. Amplification and real-time fluorescence detections were performed on a 7500 Real-Time PCR system (Applied Biosystems) using the following conditions: 15 min at 50°C and 2 min at 95°C, 45 cycles of 15 s at 95°C, and then 1 min at 60°C for 40 cycles. Specimens with PCR threshold cycle (CT) values of <40 were considered positive. The assay limit of detection was 1,000 viral RNA copies/ml of specimen.
Table 1.
Target genes, amplicon sizes, final reaction concentrations, labels, and sequences of primers and TaqMan probes used in a panel of three laboratory-developed real-time RT-PCR assays for the detection of influenza viruses
| Target gene | Amplicon size (bp) | Reaction concn (nM) | Functiona | Sequence (5′-3′)b |
|---|---|---|---|---|
| FluA matrix | 82 | 150 | Forward 1 | TCATGGAGTGGCTAAAGACAAGAC |
| 150 | Forward 2 | TCATGGAATGGCTAAAGACAAGAC | ||
| 250 | Reverse | GGCACGGTGAGCGTGAA | ||
| 100 | Probe 1* | TCACCTCTSACTAAGGG | ||
| 50 | Probe 2* | TCACCTCTAATTAAGGG | ||
| FluA hemagglutinin H3 | 68 | 250 | Forward | GACCTTTTTGTTGAACGMAGMA |
| 250 | Reverse | GAGRCATAATCYKGCACATC | ||
| 100 | Probe 1* | CAGCAAYTGTTACCCTTA | ||
| 100 | Probe 2* | CAGCAGYGTTTACCCTTA | ||
| FluA hemagglutinin H1 | 76 | 250 | Forward | CGAAATATTCCCCAAAGARAGCT |
| 250 | Reverse | CCCRTTATGGGAGCATGATG | ||
| 100 | Probe† | TGGCCCAACCACACCGTAACCG | ||
| FluA hemagglutinin 2009 H1 | 64 | 250 | Forward | AAGACCCAAAGTGAGGGATCAA |
| 250 | Reverse | TTGTCTCCCGGCTCTACTAGTGT | ||
| 100 | Probe* | AAGGGAGAATGAACTATTAC | ||
| FluB matrix | 76 | 250 | Forward | CACAATTGCCTACCTGCTTTCA |
| 250 | Reverse | CCAACAGTGTAATTTTTCTGCTAGTTCT | ||
| 100 | Probe‡ | CTTTGCCTTCTCCATCTT | ||
| Jellyfish (EXO) | 64 | 100 | Forward | GGCGGAAGAACAGCTATTGC |
| 200 | Reverse | GGAACCTAAGACAAGTGTGTTTATGG | ||
| 100 | Probe‡ | AACGCCATCGCACAAT |
*, Probes labeled on the 5′ end with FAM (6-carboxyfluorescein) and on the 3′ end with a minor groove binder nonfluorescent quencher; †, probe labeled on the 5′ end with HEX (5′-hexachlorofluorescein) and on the 3′ end with Black Hole Quencher 1; ‡, probes labeled on the 5′ end with VIC and on the 3′ end with a minor groove binder nonfluorescent quencher.
Degenerate bases: K, G or T; M, A or C; R, A or G, S,G or C; and Y, C or T.
The FluA M/Flu B M assay demonstrated excellent performance compared to the results of 1,180 nasal wash specimens tested by using fluorescent antibodies (9). The H3/H1 assay was validated by testing a panel of FluA-positive and -negative specimens by an Food and Drug Administration (FDA)-approved CDC influenza virus A subtyping assay performed at the Washington State Public Health Laboratory. The 2009 H1 assay was validated by testing a panel of FluA-positive and -negative specimens by the CDC-approved influenza virus A H1 novel 2009 assay performed at the Washington State Public Health Laboratory. The results of both tests agreed for all samples tested.
One-tube asymmetric multiplex RT-PCR.
The design of the primers and probes (Table 2) were based on those used in the real-time RT-PCR assay, but with some modifications. Single-stranded target was produced by adjusting the melting temperatures of the forward and reverse primers to approximately 52 and 68°C, respectively. The temperature differential between primer sets allows for a 50°C RT step, followed by exponential PCR amplification with an annealing temperature at 52°C. Increasing the annealing temperature of the reaction to 68°C for additional nonexponential amplification favors extension by the biotin-labeled reverse primer. RT and two-stage PCR are performed in a single reaction vial using an Invitrogen UltraSense One-Step RT-PCR System (Invitrogen). The multiplex reaction was carried out in a 40-μl volume containing 10 pmol of each forward and reverse primer, 1 μl of enzyme mix, and 10 μl of nucleic acid template under the following conditions: 50°C for 20 min; 95°C for 2 min; 40 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 45 s; followed by 20 cycles of 95°C for 30 s and 68°C for 30 s. The positive and negative controls were the same as those used for the real-time RT-PCR assay.
Table 2.
Target genes, amplicon sizes, labels, and sequences of primers and probes used in the Luminex assay for detection of influenza viruses (FLULUM)
| Target gene | Amplicon size (bp) | Functiona | Sequence (5′-3′)b |
|---|---|---|---|
| FluA matrix | 83 | Forward | TGGARTGGCTAAAGACAAGAC |
| Reverse* | CACTGGGCACGGTGAGCGTGAACAC | ||
| Probe† | CACCTCTGACTAAGGGRATTT | ||
| FluA hemagglutinin H3 | 98 | Forward | CTGGAGAACCAACATACAATTG |
| Reverse* | GCCCATATCCTCAGCATTTTCCCTCAGTTG | ||
| Probe† | CTGACTCAGAAATGAACAAACTGTT | ||
| FluA hemagglutinin H1 | 122 | Forward | GAAATATTCCCCAAAGARAGCT |
| Reverse* | ACCATTCTTCCCCGTCAGCCATAGCAAA | ||
| Probe† | TGGCCCAACCACACCGTAACCG | ||
| FluA hemagglutinin 2009 H1 | 115 | Forward | GGTGCTATAAACACCAGCCT |
| Reverse* | GGGAYATTCCTCAATCCTGTGGCCAGTCTC | ||
| Probe† | CCCATTTCAGAATATACATCCGAT | ||
| FluB matrix | 90 | Forward | GAAGATGGAGAAGGCAAAG |
| Reverse* | TCCATTCCAAGGCAGAGTCTAGGTCA | ||
| Probe† | GCAGAAAAATTACACTGTTGGTT | ||
| Jellyfish (EXO) | 84 | Forward* | CAAATTGAACGGTCAATTGGAAGTGG |
| Reverse | CCTAAGACAAGTGTGTTTATGG | ||
| Probe† | CGTTTGCAATAGCTGTTCTTC |
*, Primer labeled with 5′ biotin; †, probe modified with 5′ amino-C12 linker.
Degenerate bases: R, A or G.
Probe coupling to beads.
Capture probes modified with an amino-C12 linker at the 5′ end were coupled to Luminex carboxylated microspheres (“beads”) by a carbodiimide-based procedure according to the manufacturer's recommended protocol. Although the sequences of the probes were the same, MicroPlex carboxylated beads were used for the LX200 assay and MagPlex magnetic carboxylated beads were used for the MagPix assay (Luminex Corp., Austin, TX). For each combination of probe and bead set, 5.0 million beads were resuspended in 0.1 M MES (morpholineethanesulfonic acid) buffer (pH 4.5), with 0.2 nmol probe, and treated twice with 25 μg of 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride (EDC; Thermo Scientific/Pierce, Rockford, IL) at room temperature for 30 min, rinsed in 0.02% Tween 20, rinsed in 0.1% sodium dodecyl sulfate, and resuspended in Tris-EDTA buffer (pH 8.0) to 5.0 × 104 beads/μl.
Hybridization and Luminex analysis.
After PCR amplification, 1 μl of each reaction was transferred to 40 μl of tetramethyl ammonium chloride (TMAC) hybridization buffer (3 M TMAC, 50 mM Tris-HCl, 4 mM EDTA, 0.1% Sarkosyl) containing 3,000 probe-coupled beads for each target. The mixture was denatured at 95°C for 5 min and hybridized at 52°C. After 30 min, 35 μl of reporter solution (TMAC hybridization buffer containing 10 μg of phycoerythrin-conjugated streptavidin/ml) was added, followed by incubation at 52°C for 5 min. The beads were then analyzed on a Luminex LX200 or MagPix instrument at 52°C. The median fluorescence intensity (MFI) of at least 100 beads was reported for each bead set. An MFI value above the threshold level determined empirically for each target indicated a positive result for that target.
RESULTS
Clinical performance of the Luminex assay.
To assess the clinical performance of the FLULUM, we compared the results to those of the real-time RT-PCR assay for the detection of influenza viruses in 259 specimens (159 positive, 100 negative) submitted in the 2010-2011 season (LX200 instrument), and 841 specimens (236 positive, 605 negative) submitted in the 2011-2012 season (MagPix instrument).
The results of the 2010-2011 specimens performed on the LX200 are shown in Table 3. The sensitivities of FLULUM for each viral target as determined using the results of our laboratory-developed real-time RT-PCR assays as the comparator were as follows: FluA M, 97.3% (n = 150); 2009 H1, 90.5% (n = 85); H3, 96.9% (n = 65); and FluB M, 88.9% (n = 9). No data were available for seasonal H1 due to the lack of positive specimens.
Table 3.
Clinical sensitivity and specificity of the FLULUM assay for 259 specimens submitted in the 2010-2011 seasona
| Target | Sensitivity |
Specificity |
||
|---|---|---|---|---|
| TP/(TP+FN) | % | TN/(TN+FP) | % | |
| FluA matrix | 146/150 | 97.3 | 109/109 | 100.0 |
| 2009 H1 | 77/85 | 90.5 | 174/174 | 100.0 |
| H3 | 63/65 | 96.9 | 194/194 | 100.0 |
| FluB matrix | 8/9 | 88.9 | 250/250 | 100.0 |
The assay was performed on the Luminex LX200 instrument. Abbreviations: TP, true positive; FN, false negative; TN, true negative; FP,false positive.
The results of the 2011-2012 specimens performed on the MagPix are shown in Table 4. The sensitivities for each target were as follows: FluA M, 100% (n = 89); 2009 H1, 89.9% (n = 69); H3, 96.4% (n = 28); and FluB M, 95.6% (n = 112).
Table 4.
Clinical sensitivity and specificity of the FLULUM assay for 806 specimens submitted in the 2010-2011 seasona
| Target | Sensitivity |
Specificity |
||
|---|---|---|---|---|
| TP/(TP+FN) | % | TN/(TN+FP) | % | |
| FluA matrix | 89/89 | 100.0 | 717/717 | 100.0 |
| 2009 H1 | 62/69 | 89.9 | 743/743 | 100.0 |
| H3 | 27/28 | 96.4 | 784/784 | 100.0 |
| FluB matrix | 107/112 | 95.6 | 694/694 | 100.0 |
The assay was performed on the Luminex MagPix instrument. Abbreviations: TP, true positive; FN, false negative; TN, true negative; FP, false positive.
All samples with FluB M false-negative results determined by FLULUM had real-time PCR results of >37, which was close to the limit of detection (>40), suggesting the lower limit of detection for FLULUM. The CT values of samples with FluA M false-negative results by FLULUM ranged from 19.6 to 24.4; for 2009 H1, the values ranged from 30.9 to 39.1, and for H3, the values ranged from 33.8 to 36.2. We observed no correlation of failure in FLULUM with specimen type.
The analytical specificity of FLULUM was assessed on the MagPix by testing clinical specimens determined to be positive by RT-PCR for noninfluenza respiratory viruses, including respiratory syncytial virus, rhinovirus, adenovirus, coronavirus, parainfluenza virus types 1 to 3, metapneumovirus, and bocavirus. None of these samples gave positive signals for any FLULUM target. All clinical samples determined to be negative by real-time PCR were also negative by FLULUM.
Analytical sensitivity of the Luminex assay.
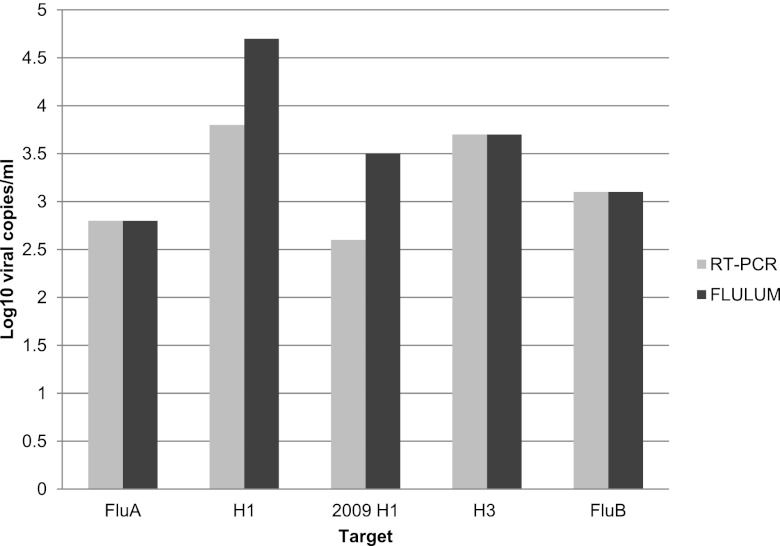
We assessed the relative analytical sensitivity (limit of detection) of FLULUM by testing in duplicate 10-fold serial dilutions of specimens with high viral loads (a positive culture harvest was used for testing the H1 target). Influenza virus A and B copy numbers in the samples were determined by using the real-time FluA M and FluB M RT-PCR assays, respectively, by comparing the CT values of the samples to standard curves generated by amplification of 10-fold serial dilutions of 107 copies of RNA transcripts/reaction containing the sequences of the FluA M and FluB M amplicons. The mean numbers of viruses (expressed as log10 copies/ml) in the lowest dilutions that were positive by each assay are shown in Fig. 1. The limit of detection of the FLULUM matched that of the real-time PCR assay for FluA M, FluB M, and the H3 targets. The limits of detection for seasonal H1 and 2009 H1 were 10-fold higher for the FLULUM assay compared to real-time PCR.
Fig 1.
Comparison of the analytical sensitivity of the FLULUM assay versus the real-time RT-PCR assay. The relative limit of detection in is expressed in log10 copies/ml.
Cost, turnaround time, and throughput comparison.
Table 5 presents a cost comparison of the reagents used in the FLULUM and real-time RT-PCR assays. Although the cost of the Luminex beads is significant, the higher-throughput capability of the assay (93 samples per plate versus 30 samples per plate for real-time RT-PCR) reduces the reagent cost from $9.42 per test for real-time RT-PCR to $3.90 for FLULUM.
Table 5.
Cost comparison of reagents used in the FLULUM and real-time RT-PCR assaya
| Method (details) | Cost/96-well plate | Cost/test |
|---|---|---|
| FLULUM (3 controls, 93 tests/plate) | ||
| RT-PCR reagents | $270.00 | $2.90 |
| Primers (forward and biotin-reverse) | $2.63 | $0.03 |
| MagPlex beads | $60.48 | $0.65 |
| Streptavidin-phycoerythrin | $9.15 | $0.10 |
| Probes/coupling reagents | $3.49 | $0.04 |
| Instrument reagents | $16.58 | $0.18 |
| Total | $362.33 | $3.90 |
| Real-time RT-PCR (2 controls, 30 tests/plate) | ||
| RT-PCR reagents | $270.00 | $9.00 |
| Primers | $0.64 | $0.02 |
| Fluorescent probes | $11.92 | $0.40 |
| Total | $282.56 | $9.42 |
Based on use of the Luminex MagPix Instrument. These values do not include costs for extraction, disposables, common lab reagents, labor, instrument(s), quality control, and development.
The real-time RT-PCR assay, with the advantage of simultaneous target amplification and detection, required ∼2 h to test one plate (30 samples) and ∼5.5 h for 90 samples when only one real-time instrument was available. FLULUM took up to 2 h longer to test one plate (93 samples) due to a longer amplification procedure and the subsequent hybridization and detection steps. However, the turnaround time of the assay from extraction to result is still within an 8-h clinical shift. If additional samples needed to be tested in the same shift, two FLULUM assays can be set up by one technician to run 186 samples with a second standard thermocycler. This is an easier and more cost-effective alternative to purchasing and running several real-time instruments in parallel.
DISCUSSION
We compared the performance of two laboratory-developed tests (LDT) for the detection of influenza virus subtypes in clinical samples: real-time TaqMan RT-PCR and asymmetric multiplex RT-PCR with Luminex bead-based amplicon detection performed on two instruments. To our knowledge, this is the first report to describe a Luminex-based LDT designed specifically for influenza virus subtyping. Commercial Luminex assays that detect influenza virus include the xTag respiratory virus panel (RVP) and the xTag RVP Fast (Luminex Corp). The xTag RVP detects 12 viral targets, including influenza viruses A and B, with additional identification of virus A subtypes H1, H3, and H5, but not the 2009 pandemic H1 subtype (19). Due to its lengthy protocol and multiple manipulations of amplified product, the RVP classic assay was streamlined to have a shorter protocol, with the addition of the 2009 H1 target, and is now marketed as the xTag RVP Fast assay (20).
Our clinical laboratory regularly performs a real-time RT-PCR LDT to diagnose virally caused respiratory tract illnesses, including those caused by respiratory syncytial virus, metapneumovirus, influenza virus types A and B, parainfluenza virus types 1 to 4, coronavirus, rhinovirus, bocavirus, and adenovirus. This well-characterized and validated LDT has been in use for more than 5 years (9, 21–23). The influenza virus real-time RT-PCR panel used as the comparator assay to assess the performance of FLULUM was developed using the same FluA M and FluB M primers and probes as used in the previously validated respiratory virus panel and newly designed primer-probe sets to detect the hemagglutinin genes of FluA virus subtypes seasonal H3 and H1 and 2009 pandemic H1. The five influenza virus targets are detected in three duplex reactions. Quantitative RT-PCR using standard curves generated from RNA transcripts has shown a sensitivity of 1,000 viral copies/ml of specimen.
Multiplexing technologies are growing increasingly popular since they offer increased test throughput capacity and can reduce overall cost. We developed an influenza virus diagnostic test for the Luminex multiplex platform as an alternative to our real-time RT-PCR test to accommodate high demand during influenza season. The disadvantage to multiplex molecular assays is often decreased sensitivity and specificity. In particular, hybridization of labeled target DNA can be less than optimal due to the steric hindrance of microarray surfaces, which leads to preferential binding of the target to the nonlabeled antisense strand (24). During development, the sensitivity of our assay was greatly improved by adapting a form of asymmetric PCR based on temperature-differential primer design (25, 26). The increased rounds of amplification add to the overall turnaround time of the assay; however, the one-tube protocol minimizes handling.
The subsequent hybridization and analysis steps require the manipulation of amplified product, which can be a source of contamination, producing false-positive results in subsequent reactions. In order to prevent carryover contamination, physical separation between pre- and postamplification steps is necessary. All postamplification steps were performed in a separate, dedicated room, with separate pipettes and other lab equipment, and personnel don clean lab coats and gloves.
Compared to our real-time RT-PCR assay, our Luminex assay developed for the LX200 and the MagPix performed well for the detection of FluA and FluB in clinical specimens. The FLULUM showed a clinical sensitivity of >95% for all targets except for FluB in the 2010-2011 season and 2009 H1 in both seasons. The very low number of FluB-positive specimens (n = 9) available for the 2010-2011 season accounts for the discrepancy between seasons. The discordant results between FLULUM and real-time RT-PCR for the FluB target were associated with CT values close to the limit of detection by real-time RT-PCR (CT > 40), indicating failure of the FLULUM for low viral loads for influenza virus type B. The detection of the 2009 H1 target by FLULUM was both clinically and analytically less sensitive compared to real-time RT-PCR. The CT values for positive specimens missed by FLULUM had a wide range, indicating that factors other than viral load, such as suboptimal primer binding, may be affecting the sensitivity. The detection of the seasonal H1 target by FLULUM was analytically less sensitive compared to the real-time RT-PCR, but we were unable to determine the clinical performance for this target because the virus was not detected in our laboratory during the study period.
The LX200 uses a system of lasers and photo-multiplying tubes for fluorescent detection of the bead and hybridization signals and can detect 100 targets. The newer MagPix utilizes light-emitting diodes (LED) and a charge-coupled device camera and can detect 50 targets. Both are compatible with the same xMap technology, although the MagPix requires the use of magnetic beads. We adapted our FLULUM assay to use magnetic beads in order to evaluate the newer instrument in a clinical setting. The MagPix has several advantages over the LX200 in a clinical setting. It is a less expensive instrument to purchase and operate, and it takes up less bench space. The LED detection system significantly reduces the warm-up times and simplifies the laborious probe adjustment and calibration protocols required by the LX200. In addition, the MagPix is a much more robust instrument, since it is not sensitive to motion and changes in temperature and requires less maintenance.
Although real-time PCR and multiplexed assays such as FLULUM have high sensitivity and specificity, these tests are complex and must be performed by highly trained medical technologists, usually in specialized molecular diagnostic laboratories with limited hours of operation. Thus, the turnaround times of these tests can still be too long for optimal clinical management of patients, especially when considering targeted antiviral treatment, minimizing unnecessary antibacterial treatment, and timely cohorting to control infection (27). Recently, several diagnostic companies have developed FDA-approved rapid sample-to-result systems that automate all steps of viral diagnosis, including nucleic acid extraction, amplification, and detection. Systems that test for influenza virus include the Xpert Flu assay (Cepheid, Sunnyvale, CA), Liat influenza A/B assay (IQuum, Marlborough, MA), Simplexa Flu A/B & RSV (Focus Diagnostics, Cypress, CA), and FilmArray RVP (BioFire Diagnostics, Salt Lake City, UT). The characteristics of these systems include ease of use, minimal hands-on time, rapid assay time (≤1 h), and a sensitivity and specificity comparable to real-time PCR (28, 29). Thus, use of these systems can be preferable for on-site testing at emergency department and urgent care centers (27). However, the limited throughput (one sample for Xpert, Liat, and FilmArray; eight samples for Simplexa) can be a significant drawback, especially in high influenza season. Even with the purchase of several instruments to run in parallel, the extraordinarily high demand for molecular influenza diagnosis such as that experienced during the 2009 pandemic could easily overwhelm medical centers equipped with these types of platforms alone. Given the much higher throughput potential of our FLULUM assay, it is an important component of our overall influenza virus diagnostic capability.
In conclusion, when tested on clinical specimens, our laboratory-developed Luminex influenza virus assay showed clinical sensitivity and specificity comparable to real-time RT-PCR. The assay can process large numbers of specimens at the same time, while significantly reducing the cost. Furthermore, the platform's flexibility allows for the possibility of adding more targets and of easily changing primer sequences in order to adapt to seasonal changes in circulating influenza virus types and sequences.
ACKNOWLEDGMENT
We thank the respiratory virus clinical technologists at the UW Molecular Virology Laboratory for specimen processing and testing.
Footnotes
Published ahead of print 23 January 2013
REFERENCES
- 1. Neuzil KM, Mellen BG, Wright PF, Mitchel EF, Jr, Griffin MR. 2000. The effect of influenza on hospitalizations, outpatient visits, and courses of antibiotics in children. N. Engl. J. Med. 342:225–231 [DOI] [PubMed] [Google Scholar]
- 2. Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096 [DOI] [PubMed] [Google Scholar]
- 3. Thompson WW, Comanor L, Shay DK. 2006. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J. Infect. Dis. 194(Suppl 2):S82–S91 [DOI] [PubMed] [Google Scholar]
- 4. Lee N, Chan PK, Hui DS, Rainer TH, Wong E, Choi KW, Lui GC, Wong BC, Wong RY, Lam WY, Chu IM, Lai RW, Cockram CS, Sung JJ. 2009. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J. Infect. Dis. 200:492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM. 2011. Antiviral agents for the treatment and chemoprophylaxis of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 60:1–24 [PubMed] [Google Scholar]
- 6. Uyeki TM. 2003. Influenza diagnosis and treatment in children: a review of studies on clinically useful tests and antiviral treatment for influenza. Pediatr. Infect. Dis. J 22:164–177 [DOI] [PubMed] [Google Scholar]
- 7. Letant SE, Ortiz JI, Bentley Tammero LF, Birch JM, Derlet RW, Cohen S, Manning D, McBride MT. 2007. Multiplexed reverse transcriptase PCR assay for identification of viral respiratory pathogens at the point of care. J. Clin. Microbiol. 45:3498–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuypers J, Campbell AP, Cent A, Corey L, Boeckh M. 2009. Comparison of conventional and molecular detection of respiratory viruses in hematopoietic cell transplant recipients. Transpl. Infect. Dis. 11:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuypers J, Wright N, Ferrenberg J, Huang ML, Cent A, Corey L, Morrow R. 2006. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J. Clin. Microbiol. 44:2382–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Templeton KE, Scheltinga SA, Beersma MF, Kroes AC, Claas EC. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 42:1564–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinberg GA, Erdman DD, Edwards KM, Hall CB, Walker FJ, Griffin MR, Schwartz B. 2004. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J. Infect. Dis. 189:706–710 [DOI] [PubMed] [Google Scholar]
- 12. Hodinka RL, Kaiser L. 2013. Is the era of viral culture over in the clinical microbiology laboratory? J. Clin. Microbiol. 51:2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ellis JS, Zambon MC. 2002. Molecular diagnosis of influenza. Rev. Med. Virol. 12:375–389 [DOI] [PubMed] [Google Scholar]
- 14. Harnden A, Brueggemann A, Shepperd S, White J, Hayward AC, Zambon M, Crook D, Mant D. 2003. Near patient testing for influenza in children in primary care: comparison with laboratory test. BMJ 326:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kok J, Blyth CC, Foo H, Patterson J, Taylor J, McPhie K, Ratnamohan VM, Iredell JR, Dwyer DE. 2010. Comparison of a rapid antigen test with nucleic acid testing during cocirculation of pandemic influenza A/H1N1 2009 and seasonal influenza A/H3N2. J. Clin. Microbiol. 48:290–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Szewczuk E, Thapa K, Anninos T, McPhie K, Higgins G, Dwyer DE, Stanley KK, Iredell JR. 2010. Rapid semi-automated quantitative multiplex tandem PCR (MT-PCR) assays for the differential diagnosis of influenza-like illness. BMC Infect. Dis. 10:113 doi:10.1186/1471-2334-10-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dunbar SA. 2006. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Limaye AP, Huang ML, Leisenring W, Stensland L, Corey L, Boeckh M. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377–382 [DOI] [PubMed] [Google Scholar]
- 19. Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45:2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gadsby NJ, Hardie A, Claas EC, Templeton KE. 2010. Comparison of the Luminex Respiratory Virus Panel fast assay with in-house real-time PCR for respiratory viral infection diagnosis. J. Clin. Microbiol. 48:2213–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuypers J, Wright N, Corey L, Morrow R. 2005. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J. Clin. Virol. 33:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. 2007. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics 119:e70–e76 [DOI] [PubMed] [Google Scholar]
- 23. Renaud C, Kuypers J, Ficken E, Cent A, Corey L, Englund JA. 2011. Introduction of a novel parechovirus RT-PCR clinical test in a regional medical center. J. Clin. Virol. 51:50–53 [DOI] [PubMed] [Google Scholar]
- 24. Shchepinov MS, Case-Green SC, Southern EM. 1997. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 25:1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lodes MJ, Suciu D, Wilmoth JL, Ross M, Munro S, Dix K, Bernards K, Stover AG, Quintana M, Iihoshi N, Lyon WJ, Danley DL, McShea A. 2007. Identification of upper respiratory tract pathogens using electrochemical detection on an oligonucleotide microarray. PLoS One 2:e924 doi:10.1371/journal.pone.0000924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanchez JA, Pierce KE, Rice JE, Wangh LJ. 2004. Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc. Natl. Acad. Sci. U. S. A. 101:1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu M, Qin X, Astion ML, Rutledge JC, Simpson J, Jerome KR, Englund JA, Zerr DM, Migita RT, Rich S, Childs JC, Cent A, Del Beccaro MA. 2012. Implementation of FilmArray respiratory viral panel in a core laboratory improves testing turn-around-time and patient care. Am. J. Clin. Pathol. 139:118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Novak-Weekley SM, Marlowe EM, Poulter M, Dwyer D, Speers D, Rawlinson W, Baleriola C, Robinson CC. 2012. Evaluation of the Cepheid Xpert Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses. J. Clin. Microbiol. 50:1704–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Renaud C, Crowley J, Jerome KR, Kuypers J. 2012. Comparison of FilmArray respiratory panel and laboratory-developed real-time reverse transcription-polymerase chain reaction assays for respiratory virus detection. Diagn. Microbiol. Infect. Dis. 74:379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]