Abstract
Microbiology laboratories continually strive to streamline and improve their urine culture algorithms because of the high volumes of urine specimens they receive and the modest numbers of those specimens that are ultimately considered clinically significant. In the current study, we quantitatively measured the impact of the introduction of CHROMagar Orientation (CO) medium into routine use in two hospital laboratories and compared it to conventional culture on blood and MacConkey agars. Based on data extracted from our Laboratory Information System from 2006 to 2011, the use of CO medium resulted in a 28% reduction in workload for additional procedures such as Gram stains, subcultures, identification panels, agglutination tests, and biochemical tests. The average number of workload units (one workload unit equals 1 min of hands-on labor) per urine specimen was significantly reduced (P < 0.0001; 95% confidence interval [CI], 0.5326 to 1.047) from 2.67 in 2006 (preimplementation of CO medium) to 1.88 in 2011 (postimplementation of CO medium). We conclude that the use of CO medium streamlined the urine culture process and increased bench throughput by reducing both workload and turnaround time in our laboratories.
INTRODUCTION
Urinary tract infections are one of the most common infectious diseases for which patients seek medical attention, and although many of these infections are treated empirically, urine cultures account for a significant portion of every clinical microbiology laboratory's daily workload (1, 2, 3, 4). The laboratory diagnosis of urinary tract infection requires quantitative urine culture on standard agar media. Because only 20 to 30% of urine samples result in significant growth, a considerable amount of time is expended evaluating samples that do not have clinical utility (5). Therefore, any new medium or method with the ability to streamline urine culture processing in a meaningful way, such as reducing technologist workload, improving result turnaround times (TATs), or reducing laboratory costs, would be welcomed and has the potential to have considerable laboratory impact.
Urine cultures have traditionally been performed using sheep blood agar (BA), a nonselective medium, and a selective medium such as MacConkey (MAC) agar, cysteine lactose electrolyte-deficient (CLED) agar, or eosin methylene blue (EMB) agar (6). Chromogenic media applicable to urine culture processing and reporting have been commercially available for more than 10 years and offer another option for diagnostic laboratories. Chromogenic media are intended to correctly identify more-frequently occurring bacteria and yeasts or organism groups on primary culture with no further testing or a minimum number of confirmatory tests. Substrates present in chromogenic media target specific classes of enzymes produced by certain bacteria and yeasts (7). Target enzymes hydrolyze chromogenic substrates generating colored products which allow for easy identification of specific organisms (7, 8). Chromogenic media have been reported to be an acceptable alternative to traditional media for the isolation of urinary pathogens (1, 9, 10, 11, 12, 13, 14). Chromogenic media may facilitate improved sensitivity of identification of some Gram-positive cocci (e.g., enterococci) in mixed cultures with Enterobacteriaceae and may promote more uniform interpretation of urine culture plates by less experienced bench technologists (1, 10, 13, 15). Chromogenic media may also promote more rapid identification of the etiological agent(s) of infection and may provide clinicians with relevant information regarding their choice of empirical antimicrobial therapy for their patients. Consequently, this may decrease inappropriate use of antibacterial and antifungal agents (1, 4, 16, 17). Published data support performing antimicrobial susceptibility testing on colonies taken directly from CHROMagar Orientation (CO) medium (Becton, Dickinson, Cockeysville, MD) (1, 14).
Only two previous reports have been published describing an assessment of workload and/or cost savings associated with chromogenic media used for urine cultures (1, 13). D'Souza et al. reported that the use of CO resulted in a >50% reduction in inoculation time and a >20% reduction in workup time (1). Ohkusu (13) reported that the cost associated with identification of Gram-negative bacilli using CO medium was reduced 70% compared to that of identifications generated using the Crystal E/NF system (Becton, Dickinson).
The purpose of the current study was to add to the limited literature in this area by determining if a reduction in additional test workload, TAT, and/or labor costs could be realized by implementing CO medium as the primary medium for urine culture. The use of CO medium was compared to the traditional method of using BA and MAC agar for routine urine cultures in two hospital microbiology laboratories.
MATERIALS AND METHODS
Study sites and timeline.
The study was conducted at Diagnostic Services of Manitoba's two largest clinical microbiology laboratories (Health Sciences Centre and St. Boniface Hospital), which collectively process approximately 65,000 urine specimens per year. Between 2006 and 2011, a number of protocol changes were implemented at both sites to streamline urine culture workup. In 2006 and 2007, urine cultures were performed using BA and MAC agar split plates. In 2007, the Clinical and Laboratory Standards Institute (CLSI) methods for the abbreviated identification of bacterial organisms (18) were implemented. In 2008, the BA and MAC agar split plates were replaced by CO medium as the primary culture medium following in-house validation by both laboratories.
Workload and turnaround time assessment following CO implementation.
Workload was evaluated at both laboratory sites using their laboratory information systems (LIS) (Delphic LIS; Sysmex, Auckland, New Zealand) which track workload units (WLUs) automatically. The Standards for Management Information Systems (MIS) have defined one WLU as representing 1 min of hands-on labor. Data on WLUs related to urine cultures were assessed for a 6-month period each year from 2006 to 2011. Extra WLUs per urine specimen were assessed prior to and after implementation of CO medium and followed until the end of 2011. WLUs associated with extra plate reading (i.e., urine plates read on two separate days when split plates were used versus a single reading for CO) and antimicrobial susceptibility testing were not included in the calculation. WLU calculations assessed only the extra tests done to identify microorganisms.
To determine average TAT, 240 positive urine specimens (120 urine samples that grew Escherichia coli and 120 urine samples that grew Enterococcus species) were randomly selected from the two laboratory sites in 2007 (preimplementation of CO medium) and 2009/2011 (postimplementation of CO medium). The TAT for each sample was calculated from time of receipt by the microbiology laboratory to the time identification results were released to the treating physician.
Urine culture protocol.
Urine samples were inoculated onto BA/MAC agar split plates using a calibrated 0.001-ml loop and streaked manually. CO medium was also inoculated with a calibrated 0.001-ml loop, but the plates were streaked using an automated streaking device (ISOplater; Vista Technology, Edmonton, Canada). All culture plates (BA/MAC agar or CO medium) were incubated at 35 to 37°C for 24 h before the first reading. As per the manufacturer's instructions, CO plates were read only once at 24 h, in contrast to the BA/MAC agar split plates, which were read at 24 and 48 h. The number and identification of different organisms grown on CO medium were primarily assessed by colony color and morphology, whereas when using BA/MAC agar split plates other tests and procedures were often required to differentiate between organisms. The additional tests included rapid tests such as Gram stain, rapid biochemical tests, and latex and slide agglutination tests. In addition, tests requiring overnight incubation, such as organism subcultures, tube biochemical tests, and commercial Vitek or API identification panels (bioMérieux, Marcy l' Etoile, France), were also required. Culture interpretation was based on the number of different organisms, quantitation of organisms, and identification of uropathogens (Table 1).
Table 1.
Criteria used to interpret urine culturesa
| Culture interpretationb | Culture results |
|---|---|
| Significant growth | Growth at ≥104 CFU/ml of 1 or 2 uropathogens |
| Growth at ≥104 CFU/ml of 1 or 2 uropathogens at a 10-fold higher level than nonuropathogens | |
| Growth at ≥105 CFU/ml of a pure culture of any organism | |
| Insignificant growth | Growth at <104 CFU/ml of any organism(s) |
| Growth at ≥104 CFU/ml of ≥2 nonuropathogens | |
| No growth | <103 CFU/ml |
| Possible contamination | Growth at ≥104 CFU/ml of ≥3 organisms |
Adapted from reference 19 with permission from Elsevier.
Interpretation of culture results was based on the routine urine culture protocol used by Diagnostic Services of Manitoba (DSM) clinical microbiology laboratories.
Survey of technologists.
One month after CO medium was implemented in the two laboratories, a brief, anonymous survey regarding CO medium was performed at one site (St. Boniface Hospital). Those technologists who had experience on the urine bench and had been using CO medium regularly participated in the survey. In 2012, the same survey was performed at both sites to determine if the results were consistent after CO medium had been used as the primary urine culture medium for 4 years. The questions and breakdown of answers from the survey are shown in Table 2.
Table 2.
Survey of technologists who routinely used CHROMagar Orientation (CO) mediuma
| Question | No. of responses (% of total) |
|||
|---|---|---|---|---|
| 2008 data |
2012 data |
|||
| Yes | No | Yes | No | |
| 1. Do you routinely (>80% of the time) have sufficient growth on CO medium for susceptibility testing on day 1 (i.e., subculture not required before testing) for | ||||
| E. coli | 18/18 (100%) | 0 | 34/34 (100%) | 0 |
| Enterococcus | 11/18 (61%) | 7/18 (39%) | 16/34 (47%) | 18/34 (53%) |
| Klebsiella Enterobacter Serratia group | 18/18 (100%) | 0 | 34/34 (100%) | 0 |
| Other Gram-negative bacilli | 18/18 (100%) | 0 | 34/34 (100%) | 0 |
| Other Gram-positive cocci | 16/18 (89%) | 2/18 (11%) | 18/34 (53%) | 16/18 (47%) |
| 2. Do you encounter difficulties with any spot/rapid tests that are performed from CO medium? | 1/18 (6%) | 17/18 (94%) | 5/34 (15%) | 29/34 (85%) |
| 3. Does CO medium produce consistent color for E. coli and Enterococcus to allow you to report the organism without additional tests? | 17/18 (94%) | 1/18 (6%) | 30/34 (88%) | 4/34 (12%) |
| 4. Do you feel CO medium aids in distinguishing mixed cultures? | 17/18 (94%) | 1/18 (6%) | 32/34 (94%) | 2/34 (6%) |
| 5. Do you find colors on CO medium are easy to distinguish for the majority of isolates? | 17/18 (94%) | 1/18 (6%) | 34/34 (100%) | 0 |
| 6. Do you feel CO medium has improved efficiency on the bench? | 18/18 (100%) | 0 | 33/34 (97%) | 1/34 (3%) |
| 7. Do you feel confident in reporting E. coli and Enterococcus without additional identification tests? | 18/18 (100%) | 0 | 32/34 (94%) | 2/34 (6%) |
| 8. What percentage of the time do you need to subculture organisms from CO medium before starting workup? | ||||
| ≤10% | 17/18 (94%) | 31/34 (91%) | ||
| 11–50% | 1/18 (6%) | 3/34 (9%) | ||
| >50% | 0 | 0 | ||
| 9. To help identify organisms that will be “listed” in a final report, what percentage of the time do you need to do a Gram stain? | ||||
| ≤10% | 6/18 (34%) | 7/34 (21%) | ||
| 11–50% | 8/18 (44%) | 15/34 (44%) | ||
| >50% | 4/18 (22%) | 12/34 (35%) | ||
The survey was of technologists who routinely used CHROMagar Orientation (CO) medium at the urine culture bench 1 month after and 4 years after it was implemented as the primary medium in the clinical microbiology laboratories.
Statistical analysis.
The significance of the differences observed in extra WLUs per urine specimen between pre- and postimplementation of CO medium were determined by two-tailed, unpaired t tests. All statistical analyses were done using GraphPad Prism software, version 5.02 (San Diego, CA).
RESULTS
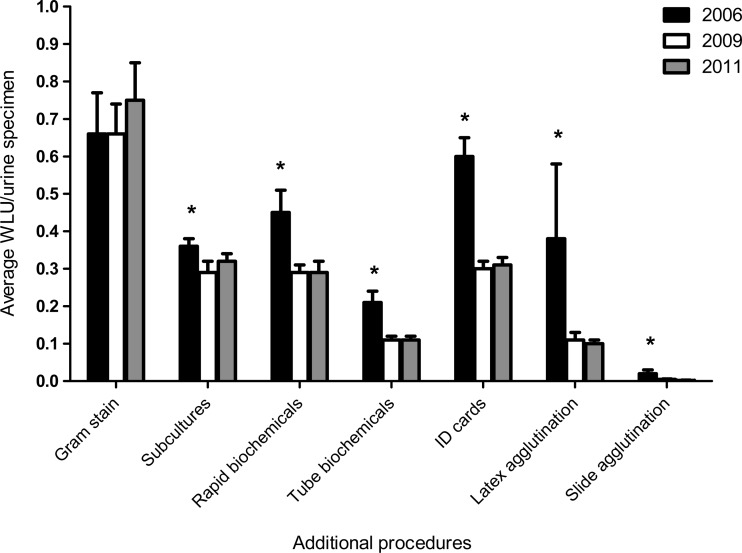
During the 6-month assessment periods from 2006 to 2011, 187,018 urine samples were analyzed at the two hospital laboratories; in 2006, 2009, and 2011, there were 28,251, 33,750, and 32,125 urine samples, respectively. Overall, 28% fewer additional identification procedures were required when using CO medium compared to BA/MAC agar split plates. The breakdown of specific tests and the impact of CO medium implementation are shown in Fig. 1. The Gram stain was the only procedure that was not affected by the implementation of CO medium. For all tests combined, the average ± standard deviation WLU counts per specimen were 2.67 ± 0.24 in 2006, 1.94 ± 0.37 in 2009, and 1.88 ± 0.15 in 2011. There was a statistically significant reduction from 2006 to 2009 (P = 0.0023; 95% confidence interval [CI], 0.3298 to 1.131) that was maintained in 2011 (P = <0.0001, 95% CI, 0.5326 to 1.047). The average workload reduction after CO medium was introduced was 1.04 WLU per specimen.
Fig 1.
Impact of CHROMagar Orientation (CO) medium on workload for identification tests for preimplementation (2006) and postimplementation (2009, 2011) periods. CO medium was implemented in 2008. *, statistically significant (P < 0.05) reduction in average WLU/urine specimen observed when 2006 was compared to either 2009 or 2011. Bars indicate standard deviations. The WLUs per procedure were 3.0 for Gram stain, 1.5 for subcultures, 1.0 for rapid biochemicals, 1.5 for tube biochemicals, 5.0 for Vitek ID panels, 6.0 for API ID panels, 4.0 for latex agglutination, and 1.0 for slide agglutination.
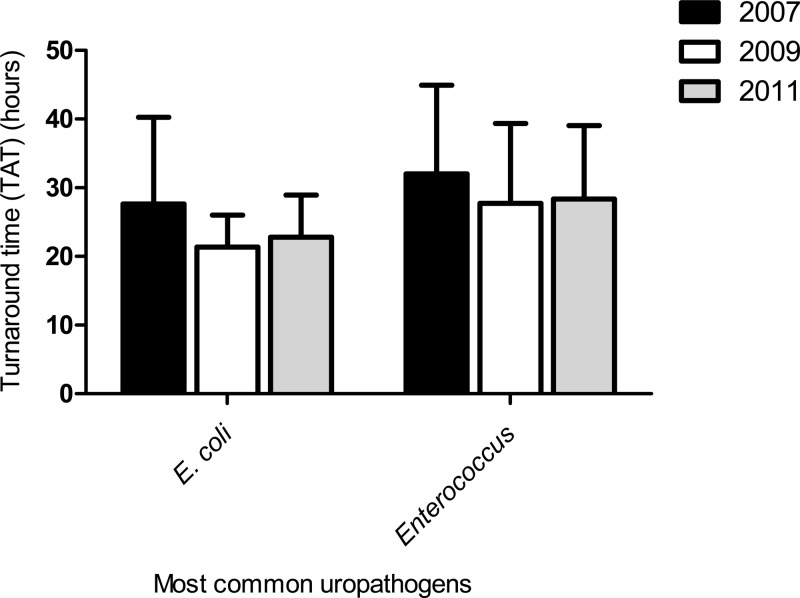
In addition, a reduction in TAT for reporting the identification of the most common Gram-negative (E. coli) and Gram-positive (Enterococcus species) uropathogens was observed (Fig. 2). Following the implementation of CO medium, the TAT for reporting E. coli and Enterococcus isolates decreased by approximately 6 h (20% reduction) and 4 h (12.5% reduction), respectively (Fig. 2); however, this reduction was not statistically significant (P = >0.5) for either uropathogen.
Fig 2.
Turnaround time (TAT) for identification of E. coli and Enterococcus species preimplementation and postimplementation for CHROMagar Orientation (CO) medium. P values were not significant for either uropathogen. Bars indicate standard deviations.
Improved throughput on the urine bench was also documented at both hospital laboratory sites. Historically, there were two technologists scheduled daily to complete work at the urine bench in each laboratory. However, after implementation of CO medium, the workload could be completed by a single technologist in each laboratory. The survey completed by the bench technologists supported this finding, as 97% of technologists felt that CO medium improved efficiency on the urine bench (Table 2).
DISCUSSION
Chromogenic media may be used by clinical laboratories for more rapid identification of several common pathogens causing urinary tract infections (1, 7, 8, 9, 10, 11, 12, 14). Our study demonstrated that CO medium as the primary culture medium is advantageous in the microbiology laboratory to streamline identification and reduce workload of urine cultures. Bench throughput was improved because of easier detection of polymicrobial cultures, and the workload was reduced, as many of the extra tests associated with conventional culture methods were no longer required (Fig. 1). A similar finding was reported by Sharmin et al. (15), who performed medium comparison studies on 400 urine samples and plated to BA and MAC agar, CLED agar, and a chromogenic medium. They concluded that the chromogenic medium considerably reduced workload and minimized the use of conventional identification tests. Unlike the results in our study, the reductions in workload that they reported were based on subjective assessment rather than quantitative data taken from an LIS to objectively evaluate workload and TAT as we did. Perry et al. (7) and Ciragil et al. (16) also suggested that chromogenic medium reduces the need for extra reagents and decreases the labor associated with processing urine cultures. Our data support the findings of these investigators.
D'Souza et al. (1) tested a total of 1,023 urine samples sent for routine culture by plating them on BA and MAC agar as well as on CO medium. Results from all media were compared, and a reduction in workup time of >20% was reported when using CO medium. Further, Sekikawa et al. (4) stated that the initial assessment of colony morphology and enumeration of growth on a chromogenic medium plate was easier, faster, and more reliable than conventional culture methods. Data collected from our staff survey (Table 2) found that 94% to 100% of technologists surveyed felt that CO medium was reliable at distinguishing mixed cultures and improving efficiency on the urine bench. In addition, the majority of technologists surveyed indicated that they were confident in reporting the most common uropathogens (i.e., E. coli and Enterococcus species) from CO medium without the use of additional identification tests.
We observed a reduction in TAT for the identification and reporting of results for E. coli and Enterococcus species, but the reduction failed to achieve statistical significance (Fig. 2). The lack of statistical significance is likely a reflection of the small sample size (120 isolates of each species in both study periods) used to assess differences in TAT. These data were obtained by performing a manual search. To do this for all urine specimens was beyond the capabilities of the LIS. A reduction in TAT was expected, as it has been reported by de Vasconcelos et al. that using chromogenic media for rapid identification of Candida species in urine reduced TAT by between 24 and 48 h (17).
Since the use of CO medium reduces the need for further subculture and extra confirmatory tests, there is an overall reduction in reagent use, suggesting that CO medium is a cost-effective replacement for conventional urine culture methods (3). Ohkusu (13) estimated $3,850 in savings per year when using a chromogenic agar system in their laboratory. Results of our study extend Ohkusu's findings, as we found there was an average reduction of 1.04 WLU per specimen after CO medium was implemented. Therefore, if a laboratory processed 100,000 urine specimens per year, the use of CO medium would result in an approximate reduction of 104,000 WLUs, which represents approximately one equivalent full-time bench technologist. Thus, the use of CO medium not only decreases supply costs but also reduces labor costs in the microbiology laboratory.
In conclusion, our quantitative data demonstrated that CO medium significantly reduced workload in the microbiology laboratory compared to that for BA/MAC agar split plates and should be considered as an alternative to conventional culture methods for detecting and reporting uropathogens.
ACKNOWLEDGMENTS
We thank Becton, Dickinson Microbiology for providing the laboratories with CHROMagar Orientation medium during the preliminary studies.
We are grateful to the clinical microbiology technologists from Health Sciences Centre and St. Boniface General Hospital in Winnipeg, MB, Canada, who performed the work for this study and provided the feedback regarding the use of CHROMagar Orientation medium.
In 2009, P. R. S. Lagacé-Wiens accepted a travel grant from CHROMagar, Paris, France, to present data on a chromogenic extended-spectrum β-lactamase (ESBL) medium at an international conference. He has had no financial dealings with them since. The remaining authors report no conflicts of interest relevant to this article.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. D'Souza HA, Campbell M, Baron EJ. 2004. Practical bench comparison of BBL CHROMagar Orientation and standard two-plate media for urine cultures. J. Clin. Microbiol. 42:60–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fallon D, Ackland G, Andrews N, Frodsham D, Howe S, Howells K, Nye KJ, Warren RE. 2003. A comparison of the performance of commercially available chromogenic agars for the isolation and presumptive identification of organisms from urine. J. Clin. Pathol. 56:608–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qaiser S, Zeeshan M, Jabeen K, Ahsan T, Zafar A. 2011. Comparison of chromogenic urinary tract infection medium with cysteine lactose electrolyte deficient media in a resource limited setting. J. Pak. Med. Assoc. 61:632–635 [PubMed] [Google Scholar]
- 4. Sekikawa E, Vidicki M, Bilkey M. 2011. Comparison of two chromogenic media and conventional media in the primary isolation and identification of urinary tract pathogens. N. Z. J. Med. Lab. Sci. 65:77–82 [Google Scholar]
- 5. Scarparo C, Piccoli P, Ricordi P, Scagnelli M. 2002. Comparative evaluation of two commercial chromogenic media for detection and presumptive identification of urinary tract pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 21:283–289 doi:10.1007/s10096-002-0718-0 [DOI] [PubMed] [Google Scholar]
- 6. McCarter Y, Burd EM, Hall GS, Zervos M. 2009. Cumitech 2C: laboratory diagnosis of urinary tract infections. ASM Press, Washington, DC [Google Scholar]
- 7. Perry JD, Freydiere AM. 2007. The application of chromogenic media in clinical microbiology. J. Appl. Microbiol. 103:2046–2055 doi:10.1111/j.1365-2672.2007.03442.x [DOI] [PubMed] [Google Scholar]
- 8. Rank EL. 2012. Chromogenic agar media in the clinical, food, and environmental testing arenas, part I. Clin. Microbiol. Newsl. 34:43–47 [Google Scholar]
- 9. Aspevall O, Osterman B, Dittmer R, Sten L, Lindback E, Forsum U. 2002. Performance of four chromogenic urine culture media after one or two days of incubation compared with reference media. J. Clin. Microbiol. 40:1500–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang JC, Chien ML, Chen HM, Yan JJ, Wu JJ. 2008. Comparison of CPS ID 3 and CHROMagar Orientation chromogenic agars with standard biplate technique for culture of clinical urine samples. J. Microbiol. Immunol. Infect. 41:422–427 [PubMed] [Google Scholar]
- 11. Hengstler KA, Hammann R, Fahr AM. 1997. Evaluation of BBL CHROMagar orientation medium for detection and presumptive identification of urinary tract pathogens. J. Clin. Microbiol. 35:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. 1996. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J. Clin. Microbiol. 34:1788–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohkusu K. 2000. Cost-effective and rapid presumptive identification of gram-negative bacilli in routine urine, pus, and stool cultures: evaluation of the use of CHROMagar orientation medium in conjunction with simple biochemical tests. J. Clin. Microbiol. 38:4586–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samra Z, Heifetz M, Talmor J, Bain E, Bahar J. 1998. Evaluation of use of a new chromogenic agar in detection of urinary tract pathogens. J. Clin. Microbiol. 36:990–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sharmin S, Alamgir F, Begum F, Jaigirdar Q. 2010. Use of chromogenic agar media for identification of uropathogen. Bangladesh J. Med. Microbiol. 4:18–23 [Google Scholar]
- 16. Ciragil P, Gul M, Aral M, Ekerbicer H. 2006. Evaluation of a new chromogenic medium for isolation and identification of common urinary tract pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 25:108–111 doi:10.1007/s10096-006-0103-5 [DOI] [PubMed] [Google Scholar]
- 17. de Vasconcelos AA, Jr, Menezes EA, Cunha FA. 2011. Chromogenic medium for direct susceptibility testing of Candida spp. isolated from urine. Mycopathologia 172:125–130 doi:10.1007/s11046-011-9407-9 [DOI] [PubMed] [Google Scholar]
- 18. Clinical and Laboratory Standards Institute 2008. Abbreviated identification of bacteria and yeast; approved guideline–second ed M35-A2 CLSI, Wayne, PA [Google Scholar]
- 19. Kadkhoda K, Manickam K, Degagne P, Sokolowski P, Pang P, Kontzie N, Alfa M. 2011. UF-1000i flow cytometry is an effective screening method for urine specimens. Diagn. Microbiol. Infect. Dis. 69:130–136 doi:10.1016/j.diagmicrobio.2010.09.013 [DOI] [PubMed] [Google Scholar]