Abstract
We report two cases of bacteremia with the anaerobic bacterium Ruminococcus gnavus. In both cases, the bacteremia was associated with diverticular disease. Preliminary conventional identification suggested peptostreptococci, and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis did not produce scores high enough for species identification. Finally, the bacteria were identified by 16S rRNA gene sequencing.
CASE REPORTS
Patient number I was a 67-year-old man admitted to Odense University Hospital, Odense, Denmark, in February 2012 because of abdominal pain. He was receiving chemotherapy for disseminated small cell lung carcinoma. His temperature was 37.0°C; however, the patient was receiving acetaminophen. The patient had neutropenia, with a white blood cell count of 0.8 × 109/liter (normal value range, 3.5 × 109 to 8.8 × 109/liter). The C-reactive protein level was 362 mg/liter (normal value, <0.6 mg/liter). Two sets of blood cultures (40 ml total), each including an aerobic bottle and an anaerobic bottle, were obtained (BacT/Alert; bioMérieux, Marcy l'Etoile, France), and treatment with piperacillin-tazobactam (4 g) and metronidazole (500 mg) intravenously three times a day was initiated. An X-ray demonstrated large amounts of free intra-abdominal air, and an exploratory laparotomy revealed fecal peritonitis caused by perforated diverticulitis. After the laparotomy, the patient's condition slowly deteriorated and he died 1 week later.
Growth was detected in one of the anaerobic bottles after 2 days of incubation. Gram staining showed large Gram-positive diplococci (Fig. 1). Twenty-four hours later, growth on solid medium was observed under anaerobic conditions only. Escherichia coli, fully susceptible to piperacillin-tazobactam, was recovered in one of the aerobic blood culture bottles after 2 days of incubation.
Fig 1.
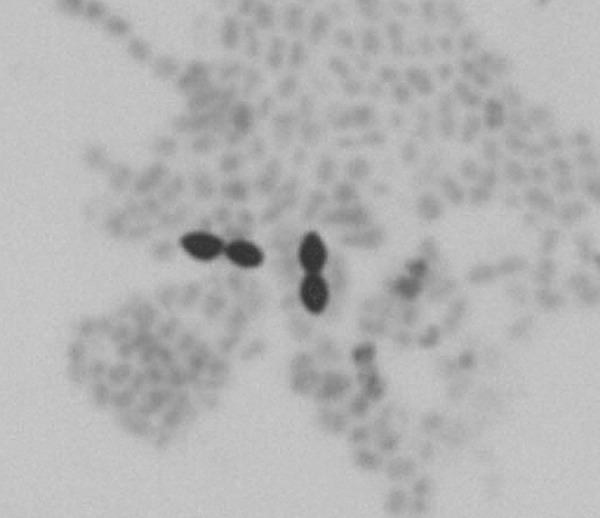
Black and white photomicrograph of methylene blue staining of the Lillebaelt Ruminococcus gnavus isolate.
Patient number II was a 90-year-old man admitted to Lillebaelt Hospital, Vejle, Denmark, in July 2012. He had had abdominal pain for the past 8 days, and the pain had been increasing in the lower left side of the abdomen for the last 9 h before admission. He had also started vomiting. He had a temperature of 39.7°C. The white blood cell-count was 11.8 × 109/liter, and the C-reactive protein level was 67 mg/liter. Two sets of blood cultures (40 ml total), each including an aerobic bottle and an anaerobic bottle, were obtained (BacT/Alert). Because the patient was known to have diverticular disease, a computerized axial tomography (CAT) scan was performed, showing widespread diverticulosis in the colon sigmoideum, but without any signs of perforation. He was treated with a combination of cefuroxime (1.5 g) and metronidazole (500 mg) intravenously three times a day. The patient improved and was discharged 3 days after admission, without fever and with a white blood cell count of 5.8 × 109/liter and a C-reactive protein level of 22 mg/liter. He was discharged without any antibiotics and before any results from blood cultures existed. After growth was detected in the blood cultures, the ward was informed and advised to evaluate the patient for further treatment.
Three days after incubation of the blood cultures, growth was detected in one of the anaerobic blood culture bottles. Primary Gram staining showed Gram-positive diplococci only. Surprisingly, growth of Pseudomonas aeruginosa was discovered on the aerobic plates from the anaerobic bottle the following day. However, after one more day of culturing, an anaerobic bacterium was found on the plates incubated in the anaerobic atmosphere and Gram staining showed large Gram-positive diplococci with tapered ends.
Both anaerobic isolates were catalase negative, and growth was inhibited by metronidazole, bile, and vancomycin but not by kanamycin or colistin.
The isolates were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Biotyper, software v.3.0) with and without formic acid. The sample was inoculated onto a ground steel MALDI target plate followed by 1 μl of matrix (1). When formic acid was used, the sample was also inoculated onto the plate, and after drying, 1 μl of formic acid was applied directly on the sample. After drying once more, 1 μl of matrix was applied. Ruminococcus gnavus was suggested as a possibility for both isolates, but the scores of 1.723 and 1.675, which were the highest scores, were below the cutoff for species identification (≥2.0). Several other aerobic and anaerobic species were also suggested, such as different Clostridium spp., Staphylococcus spp., Weissella viridescens, and Blautia coccoides. The Biotyper database contained only one isolate of the genus Ruminococcus, which was R. gnavus.
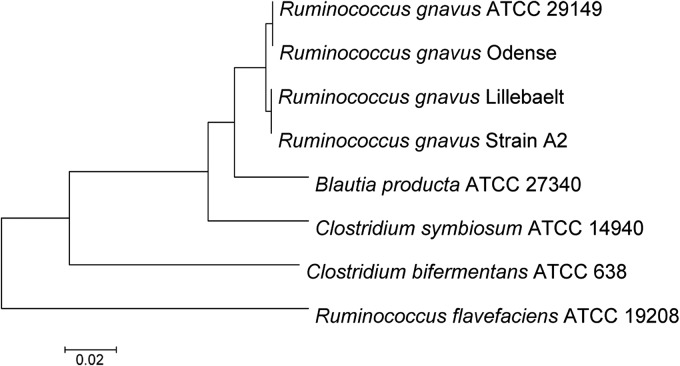
Finally, both isolates were identified by partial 16S rRNA gene sequencing (MicroSeq 500 system; Perkin-Elmer, Applied Biosystems Division, Foster City, CA). The Odense isolate consensus sequence (479 bp) had the highest similarity (99.6% match) to Ruminococcus gnavus ATCC 29149 (GenBank accession no. L76597), and the Lillebaelt isolate consensus sequence (477 bp) had the highest similarity (99.2% match) to Ruminocoocus gnavus strain A2 (GenBank accession no. EU139255) (Fig. 2).
Fig 2.
Phylogenetic analyses showing the relationship between Ruminococcus, Blautia, and Clostridium on the basis of the sequences of the 16S region for each species were performed by the neighbor-joining method in the MEGA 5.0 program package (http://www.megasoftware.net). Sequences were aligned by using the ClustalW program built into the MEGA 5.0 program package.
In the subsequent Biotyper version 3.1, the database still contained only R. gnavus, but one more strain had been entered into the database. Identification of the isolates was repeated with the new database, version 3.1, and R. gnavus was suggested, with maximum scores of 2.181 and 2.124. The best scores were obtained when formic acid was used, and R. gnavus was suggested as the two highest ranking out of ten proposed.
Antimicrobial susceptibility of the two isolates was determined by the Etest gradient method (bioMérieux, Lyon, France) on Brucella blood agar supplemented with hemin and vitamin K (Becton, Dickinson GmbH, BD Diagnostics, Heidelberg, Germany) according to the manufacturer's instructions (Table 1). Tests with a nitrocefin disc (Cefinase; Becton, Dickinson, Sparks, MD) for the detection of β-lactamase production were negative for both isolates after 30 min.
Table 1.
Antimicrobial susceptibility testing results from the two Ruminococcus gnavus isolates in this study
| Antimicrobial agent(s) | MIC (mg/liter) for R. gnavus isolate: |
|
|---|---|---|
| Odense | Lillebaelt | |
| Benzylpenicillin | 2 | 0.125 |
| Piperacillin-tazobactam | 4 | 0.5 |
| Meropenem | 1 | 0.125 |
| Clindamycin | >256 | 0.25 |
| Metronidazole | 0.125 | 0.25 |
| Moxifloxacin | 4 | 8 |
| Vancomycin | 1 | 1 |
| Tigecycline | 0.125 | 0.125 |
New technologies, such as MALDI-TOF MS, have revolutionized the identification of anaerobic bacteria. MALDI-TOF MS is fast, reliable, and inexpensive, and the majority of clinically relevant anaerobic bacteria can be identified to the species level (1). However, the method is limited by the species and number of entries of each species included in the database, as demonstrated in our cases. In both of our cases, the identification of the bacteria caused problems, since the routine identification system in our laboratories is MALDI-TOF MS supplemented with a few biochemical tests (e.g., catalase and a few diagnostic tablets or disks as described). The preliminary identification of our isolates would suggest some kind of Peptostreptococcus spp. according to the Wadsworth-KTL Anaerobic Bacteriology Manual (2). This result is potentially misleading, as Peptostreptococcus spp. are often associated with skin, soft tissue, and bone infections. In light of the close relationship between the species in the order Clostridiales, the varied findings by MALDI-TOF MS are not surprising and can probably be resolved by adding more entries of the different species. The number of species entries in the Biotyper database is of high importance, as illustrated by our two cases, as the presence of only one more isolate resulted in identification scores above the cutoff.
If MALDI-TOF MS does not provide identification, and species identification is warranted, as in cases of bacteremia, we use 16S rRNA gene sequencing. In these situations, with rare and difficult to identify bacteria, 16S rRNA gene sequencing has been shown to be very useful, although it is considerably more expensive (3). Sequencing of anaerobic bacteria from blood cultures has demonstrated the pathogenic potential of many new anaerobic species that were originally found in the intestinal flora and considered nonpathogenic (3, 4).
The susceptibilities of our two R. gnavus isolates varied considerably (Table 1), but both were susceptible to metronidazole, which was part of the treatment in both cases. Empirical treatment of bacteremia with Gram-positive anaerobic cocci is often with metronidazole, but resistance to metronidazole, as well as significant differences in susceptibilities to antibiotics between the species, has been described (5).
Taxonomically, Ruminococcus gnavus originally belonged to the genus Ruminococcus in the family Ruminococcaceae. The Ruminococcus type species is Ruminococcus flavefaciens, which was described in 1948 by Sijpesteijn (6, 7). The genus contained 18 species, but on the basis of 16S rRNA gene sequencing, some of the species have been reassigned to the new genus Blautia within the family Lachnospiraceae, which, like Ruminococcaceae, is a part of the large order Clostridiales (8). R. gnavus had earlier been designated Ruminococcus AB and was in 1976 thoroughly described by Moore et al. (9). Later the name was changed to R. gnavus due to its active fermentative ability, and the type strain was designated ATCC 29149 (6, 9). R. gnavus belongs to a group of reclassified Ruminococcus spp. that have been placed in the genus Blautia, but R. gnavus has retained the Ruminococcus genus name. In Fig. 2, the close relationship between Ruminococcus, Blautia, and Clostridium is shown.
R. gnavus cells are described as anaerobic Gram-positive cocci that may be nonmotile or motile with one to three flagella. The cells are slightly elongated and often have tapered ends. R. gnavus is strictly anaerobic and does not produce spores. Ruminococcus spp. are found to be a part of the intestinal flora in humans and to colonize the rumen of animals such as cattle, sheep, and goats (7, 9, 10). Blautia producta comb. nov. (previously Ruminococcus productus) is one of the most common organisms in the gastrointestinal tract in humans and has been isolated in cases of necrotizing fasciitis and liver and epidural abscesses (8).
Bacteremia in humans with Ruminococcus gnavus has been reported once before, but without any clinical details. The isolate was reported in a study by Simmon et al. describing the genotypic diversity of anaerobic isolates from bloodstream infections (4). We have not been able to obtain any clinical details from this case. Both of our cases were associated with diverticular disease. It is not clear and still needs to be investigated whether R. gnavus possesses specific pathogenic properties or if the bacteremia in our cases is merely a result of translocation from its normal habitat to the blood.
The presented cases remind us of the difficulties of identifying anaerobic bacteria by conventional methods. As the MALDI-TOF MS system databases are expanded, more and more species will probably be identified by this technique, and for the remaining unidentified species, 16S RNA gene sequencing is an excellent tool if species identification is warranted. Species identification is important because unknown associations between anaerobic species and specific clinical syndromes (e.g., diverticulitis) can be revealed and misleading conclusions can be avoided.
Footnotes
Published ahead of print 30 January 2013
REFERENCES
- 1. Justesen US, Holm A, Knudsen E, Andersen LB, Jensen TG, Kemp M, Skov MN, Gahrn-Hansen B, Møller JK. 2011. Species identification of clinical isolates of anaerobic bacteria: a comparison of two matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol. 49:4314–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Josimies-Sommer HR, Summanen P, Citron DM, Baron EJ, Wexler HM, Finegold SM. 2002. Wadsworth-KTL anaerobic bacteriology manual, 6th ed Star Publishing Co, Belmont, CA [Google Scholar]
- 3. Justesen US, Skov MN, Knudsen E, Holt HM, Søgaard P, Justesen T. 2010. 16S rRNA gene sequencing in routine identification of anaerobic bacteria isolated from blood cultures. J. Clin. Microbiol. 48:946–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simmon K, Mirrett S, Reller LB, Petti CA. 2008. Genotypic diversity of anaerobic isolates from bloodstream infections. J. Clin. Microbiol. 46:1596–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Veloo ACM, Welling GW, Degener JE. 2011. Antimicrobial susceptibility of clinically relevant Gram-positive anaerobic cocci collected over a three-year period in The Netherlands. Antimicrob. Agents Chemother. 55:1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skerman VBD, McGowan V, Sneath PHA. 1980. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30:225–420 [PubMed] [Google Scholar]
- 7. Sijpesteijn AK. 1948. Cellulose-decomposing bacteria from the rumen of cattle, p 152 Thesis University of Leiden, The Netherlands [Google Scholar]
- 8. Liu C, Finegold SM, Song Y, Lawson PA. 2008. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int. J. Syst. Bacteriol. 58:1896–1902 [DOI] [PubMed] [Google Scholar]
- 9. Moore WE, Johnson JL, Holdeman LV. 1976. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int. J. Syst. Bacteriol. 26:238–252 [Google Scholar]
- 10. Moore WE, Moore LH. 1995. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 61:3202–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]