Abstract
Multiyear molecular epidemiological surveillance of multidrug-resistant Pseudomonas aeruginosa (MRPA) in a pediatric cystic fibrosis care center identified an endemic MRPA strain (Houston-1). Recent hospitalization was found to be a statistically significant risk factor for acquisition of the endemic strain. Multiple infection control improvements led to the reduced incidence of the Houston-1 strain in the CF population.
TEXT
Multidrug-resistant Pseudomonas aeruginosa (MRPA) is a common respiratory pathogen found in patients with cystic fibrosis (CF) that routinely leads to chronic pulmonary infections (1, 2). While the national annual average of MRPA infection in patients with CF in 2006 was 16%, the average annual percentage of MRPA infection for patients at the CF Care Center at Texas Children's Hospital (TCH) in Houston, Texas, was 30.1% (1). This trend toward an increased prevalence of patients with a MRPA infection at TCH remained present through the latest CF center-specific data for 2010 with a center-specific MRPA infection prevalence of almost twice the national average (3). Infection with MRPA, rather than a susceptible strain, has been associated with high risk of death or lung transplantation. Molecular typing of P. aeruginosa strains in CF populations has repeatedly revealed endemic or epidemic (clonally related) strains (4–10), and infection with specific P. aeruginosa epidemic strains has been implicated in increased patient morbidity (5, 8, 11–13). Identification of risk factors for acquisition of a specific MRPA clone or information regarding outcomes for patients infected with a specific MRPA clone would be valuable tools for health care providers and infection control practitioners.
In 2004, initial rep-PCR-based molecular typing pilot project studies of MRPA in the CF population at TCH identified an endemic strain (Houston-1) present in half of the MRPA isolates tested. The Houston-1 strain persists in the patient population, but several improvements in infection control practices have led to decreased incidence of this endemic MRPA strain. Multiyear molecular epidemiological surveillance has allowed for the monitoring of infection patterns within the patient population, provided key data for additional studies pertaining to risk factors and patient outcomes, and served as a quality improvement metric for the CF Care Center at TCH.
Respiratory specimens such as sputa, throat/nasal cultures, or bronchoalveolar lavages were collected from a total of 71 patients (out of approximately 325 patients cared for at the center) during visits to the CF clinic and as part of inpatient care from 2004 to 2009 (1, 14). Only those patients with a confirmed MRPA infection and who had previously consented to participate in the national CF Patient Registry were included in the study. Routine culture was performed on multiple media (blood agar, chocolate agar, MacConkey agar, BC [Burkholderia cepacia] agar, CNA agar [Columbia agar with colistin and nalidixic acid], and CHROMagar Staph aureus), and antibiotic susceptibility testing for all P. aeruginosa isolates was performed by disk diffusion on Mueller-Hinton plated medium. MRPA was defined as being resistant to all antibiotics that were routinely evaluated in two or more of the following groups: aminoglycosides (tobramycin, gentamicin, and amikacin), fluoroquinolones (ciprofloxacin), and beta-lactams (ceftazidime, meropenem, piperacillin, ticarcillin-clavulanate, and aztreonam) (15). Within the Houston-1 group, there was variation in the antibiotic susceptibility profiles, with some found to be pan-resistant but most found to be resistant to two of the three groups. Multiple MRPA isolates were collected from each patient over the course of the study, and molecular profiles were compared over time. Patients identified as members of the Houston-1 group consistently produced the same molecular profiles.
Bacterial DNA was extracted from bacterial cells using the Mo Bio UltraClean microbial DNA kit (Mo Bio Laboratories, Inc., Solana Beach, CA). Rep-PCR was performed using the DiversiLab Pseudomonas fingerprinting kit (bioMérieux, Durham, NC) according to the manufacturer's instructions, and the amplified products were loaded into the DNA LabChip and size fractionated by microfluidics and electrophoresis in the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) (16). DiversiLab 3.1 software was utilized for subsequent analysis of molecular profiles by the Pearson correlation coefficient and the unweighted-pair group method with arithmetic mean (UPGMA) to compare the presence or absence of amplified fragments and the intensity of each fragment.
Isolates from 32 patients (45%) were found to be members of the Houston-1 group, and the remaining 39 patients (55%) yielded isolates that were classified as a strain other than Houston-1. For isolates to be considered members of an MRPA clonal group, a threshold of 95% or greater similarity was used (17). The molecular profiles and dendrogram are displayed in Fig. 1 with the scatterplot representation presented in Fig. 2.
Fig 1.
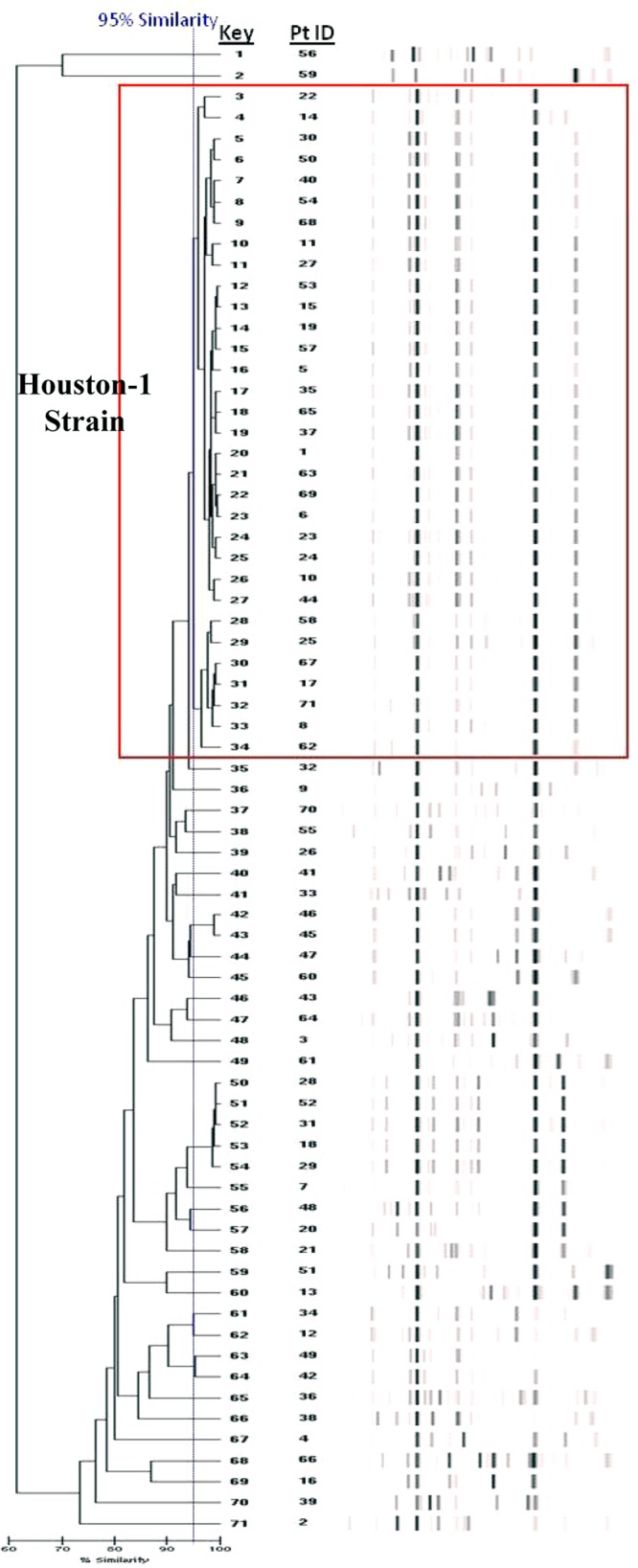
The endemic Houston-1 strain depicted by a dendrogram. The dendrogram is a hierarchical tree representation that uses group averages to display relative similarities or clusters of the isolates. Each patient in the report data set is assigned a key number by the online report generator, and the “Pt ID” column represents the anonymous identifier assigned to each patient for the study. The vertical line delineates the 95% similarity cutoff, and the box identifies the Houston-1 strain. Isolates with a molecular profile that is ≥95% similar to all other isolates within a group indicate a cluster of interest. The Houston-1 group is the largest and most significant cluster identified in the complete data set.
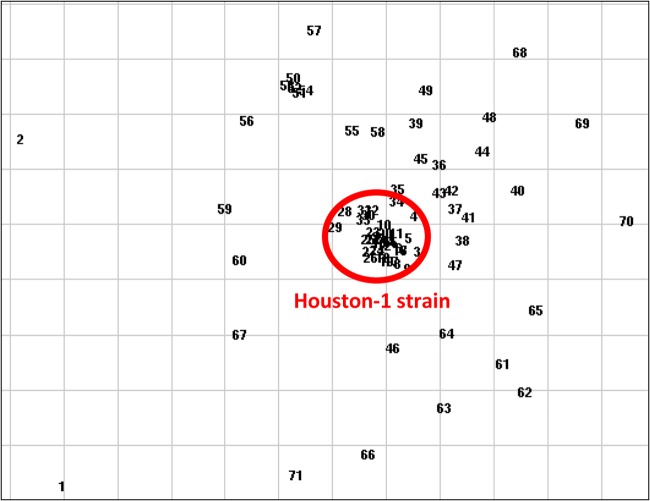
Fig 2.
Scatterplot representation of MRPA clones from patients with CF. The scatterplot is a nonhierarchical data display that uses a two-dimensional spatial representation to display relative similarities of isolates by virtue of relative distances on a grid. The Houston-1 strain is highlighted by the circle in the center of the scatterplot grouping. This graphical view provides a two-dimensional interpretation of groups of related isolates, allowing for clusters and their resulting outliers to be easily detected.
While no differences were observed for most demographic and clinical variables, including outpatient clinic visits, statistical analysis highlighted the differences in timing of hospitalization prior to MRPA infection in the two groups. Logistic regression identified recent hospitalization as a statistically significant risk factor for infection with the Houston-1 strain as opposed to the other strains (P = 0.035), whereby infection with the Houston-1 strain was four times more likely than infection with any of the other strains if the patient was hospitalized ≤90 days prior to infection (Table 1). Patients hospitalized 91 to 180 days prior to infection were three times more likely to belong to the Houston-1 group (P = 0.113). These findings are consistent with prior reports from CF centers in the United Kingdom and Australia, whereby hospitalization was implicated as a risk factor for epidemic strain acquisition (9, 18). Patients in the Houston-1 group also spent an average of 12 more days in the hospital in the year prior to MRPA infection compared to those infected with another strain.
Table 1.
Logistic regression output for variables related to chronology of MRPA infection and comparison of Houston-1 group versus all others
| Predictor | P valuea | Odds ratiob | 95% CIc |
|---|---|---|---|
| Age at MRPA infection | 0.678 | 0.974 | 0.859–1.104 |
| Days from clinic visit to MRPA infection (≤60 days) | 0.329 | ||
| 61–120 days | 0.490 | 0.596 | 0.137–2.588 |
| >120 days | 0.149 | 0.361 | 0.091–1.439 |
| Days from hospitalization to MRPA infection (>365 days) | 0.076 | ||
| ≤90 days | 0.035* | 4.019 | 1.100–14.677 |
| 91–180 days | 0.113 | 2.960 | 0.775–11.314 |
| 181–365 days | 0.509 | 0.453 | 0.043–4.755 |
P value denotes statistical significance (*, P < 0.05) related to infection with the Houston-1 strain as opposed to another strain.
Odds ratios indicate greater likelihood of infection with the Houston-1 strain as opposed to another strain.
CI, confidence interval.
Since no other clone has established a significant cluster of infected patients, the proliferation of the Houston-1 strain during a period of several years supports the potentially increased transmissibility of this endemic strain in this particular patient care environment. Previous analysis of Australian epidemic strains in the CF population suggested that protease activity and differential gene expression may play a role in the increased infectivity of the strains (19, 20). The potentially increased transmissibility of the Houston-1 strain further reinforces the need for rigorous infection control standards. Following the confirmation of the existence of the endemic strain in 2006, our hospital has improved several facets of infection control practices for patients with CF, including contact isolation for all inpatients until final culture results are obtained, immediate placement into an exam room for known contact isolation patients in the outpatient clinic, and greater accessibility to gowns and gloves for all care providers. These changes appear to have diminished the incidence of patients becoming infected with the Houston-1 strain (see the supplemental material).
These results further confirm the clinical utility of molecular typing as a tool for epidemiological surveillance in patients with cystic fibrosis. By identifying an endemic or epidemic MRPA strain in the CF population, molecular typing enables health care providers to investigate possible sources of transmission and evaluate transmissibility of the identified strain. In addition, stratification of patients based on molecular typing results would allow determination of differences in patient morbidity associated with infection with particular strains, which in turn would provide information on potential virulence of the strain. The discovery of an endemic strain in this CF care center resulted in improved infection control practices with respect to both clinic processes and inpatient protocols, and a decrease in the number of patients infected with the endemic strain was realized at the end of the data collection period for this study. Continued monitoring by molecular typing will enable us to determine if this trend continues, and further diligence in infection control based on the data provided by this study should allow for the elimination of common sources of multidrug-resistant Pseudomonas aeruginosa infections in patients with cystic fibrosis.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this work was provided by the Department of Pathology at Texas Children's Hospital.
We acknowledge and appreciate the contributions of the Clinical Microbiology and Molecular Microbiology laboratories of Texas Children's Hospital and the Department of Infection Control and Prevention and CF Care Center of Texas Children's Hospital.
Footnotes
Published ahead of print 9 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02157-12.
REFERENCES
- 1. Cystic Fibrosis Foundation 2008. Patient registry: annual data report 2006. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 2. Falagas ME, Koletsi PK, Bliziotis IA. 2006. The diversity of definitions of multidrug-resistant (MDR) and pandrug-resistant (PDR) Acinetobacter baumannii and Pseudomonas aeruginosa. J. Med. Microbiol. 55:1619–1629 [DOI] [PubMed] [Google Scholar]
- 3. Cystic Fibrosis Foundation 2011. Patient registry: annual data report 2010. Cystic Fibrosis Foundation, Bethesda, MD [Google Scholar]
- 4. de Vrankrijker AM, Brimicombe RW, Wolfs TF, Heijerman HG, van Mansfeld R, van Berkhout FT, Willems RJ, Bonten MJ, van der Ent CK. 2011. Clinical impact of a highly prevalent Pseudomonas aeruginosa clone in Dutch cystic fibrosis patients. Clin. Microbiol. Infect. 17:382–385 [DOI] [PubMed] [Google Scholar]
- 5. Edenborough FP, Stone HR, Kelly SJ, Zadik P, Doherty CJ, Govan JR. 2004. Genotyping of Pseudomonas aeruginosa in cystic fibrosis suggests need for segregation. J. Cyst. Fibros. 3:37–44 [DOI] [PubMed] [Google Scholar]
- 6. Kersulyte D, Struelens MJ, Deplano A, Berg DE. 1995. Comparison of arbitrarily primed PCR and macrorestriction (pulsed-field gel electrophoresis) typing of Pseudomonas aeruginosa strains from cystic fibrosis patients. J. Clin. Microbiol. 33:2216–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Logan C, Habington A, Lennon G, Grogan J, Byrne M, O'Leary J, O'Sullivan N. Genetic relatedness of Pseudomonas aeruginosa isolates among a paediatric cystic fibrosis patient cohort in Ireland. J. Med. Microbiol. 61:64–70 [DOI] [PubMed] [Google Scholar]
- 8. O'Carroll MR, Syrmis MW, Wainwright CE, Greer RM, Mitchell P, Coulter C, Sloots TP, Nissen MD, Bell SC. 2004. Clonal strains of Pseudomonas aeruginosa in paediatric and adult cystic fibrosis units. Eur. Respir. J. 24:101–106 [DOI] [PubMed] [Google Scholar]
- 9. Scott FW, Pitt TL. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53:609–615 [DOI] [PubMed] [Google Scholar]
- 10. Syrmis MW, O'Carroll MR, Sloots TP, Coulter C, Wainwright CE, Bell SC, Nissen MD. 2004. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J. Med. Microbiol. 53:1089–1096 [DOI] [PubMed] [Google Scholar]
- 11. Al-Aloul M, Crawley J, Winstanley C, Hart CA, Ledson MJ, Walshaw MJ. 2004. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax 59:334–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lechtzin N, John M, Irizarry R, Merlo C, Diette GB, Boyle MP. 2006. Outcomes of adults with cystic fibrosis infected with antibiotic-resistant Pseudomonas aeruginosa. Respiration 73:27–33 [DOI] [PubMed] [Google Scholar]
- 13. Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K. 2001. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J. Pediatr. 138:699–704 [DOI] [PubMed] [Google Scholar]
- 14. Saiman L, Siegel J. 2003. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Infect. Control Hosp. Epidemiol. 24:S6–S52 [DOI] [PubMed] [Google Scholar]
- 15. Lang BJ, Aaron SD, Ferris W, Hebert PC, MacDonald NE. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with multiresistant strains of Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 162:2241–2245 [DOI] [PubMed] [Google Scholar]
- 16. Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Schrock R, Manry J, Renwick A, Nieto R, Woods C, Versalovic J, Lupski JR. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doleans-Jordheim A, Cournoyer B, Bergeron E, Croize J, Salord H, Andre J, Mazoyer MA, Renaud FN, Freney J. 2009. Reliability of Pseudomonas aeruginosa semi-automated rep-PCR genotyping in various epidemiological situations. Eur. J. Clin. Microbiol. Infect. Dis. 28:1105–1111 [DOI] [PubMed] [Google Scholar]
- 18. Jones AM, Govan JR, Doherty CJ, Dodd ME, Isalska BJ, Stanbridge TN, Webb AK. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557–558 [DOI] [PubMed] [Google Scholar]
- 19. Manos J, Arthur J, Rose B, Bell S, Tingpej P, Hu H, Webb J, Kjelleberg S, Gorrell MD, Bye P, Harbour C. 2009. Gene expression characteristics of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa during biofilm and planktonic growth. FEMS Microbiol. Lett. 292:107–114 [DOI] [PubMed] [Google Scholar]
- 20. Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Al Nassafi K, Manos J, Elkins M, Bye P, Willcox M, Bell S, Wainwright C, Harbour C. 2007. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J. Clin. Microbiol. 45:1697–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.