Abstract
While the ribosome has evolved to function in complex intracellular environments, these contexts do not easily allow for the study of its inherent capabilities. We have used a synthetic, well-defined, Escherichia coli (E. coli)-based translation system in conjunction with ribosome display, a powerful in vitro selection method, to identify ribosome binding sites (RBSs) that can promote the efficient translation of messenger RNAs (mRNAs) with a leader length representative of natural E. coli mRNAs. In previous work, we used a longer leader sequence and unexpectedly recovered highly efficient cytosine-rich sequences with complementarity to the 16S ribosomal RNA (rRNA) and similarity to eukaryotic RBSs. In the current study, Shine-Dalgarno (SD) sequences were prevalent but non-SD sequences were also heavily enriched and were dominated by novel guanine- and uracil-rich motifs which showed statistically significant complementarity to the 16S rRNA. Additionally, only SD motifs exhibited position-dependent decreases in sequence entropy, indicating that non-SD motifs likely operate by increasing the local concentration of ribosomes in the vicinity of the start codon, rather than by a position-dependent mechanism. These results further support the putative generality of mRNA-rRNA complementarity in facilitating mRNA translation, but also suggest that context (e.g., leader length and composition) dictates the specific subset of possible RBSs that are used for efficient translation of a given transcript.
The 5′ untranslated region (5′ UTR) of messenger RNA (mRNA) is one of the major determinants of translational efficiency. This region often contains ribosome binding sites (RBSs), binding sites for inhibitory or stimulatory trans-acting factors, and secondary structural features that may affect access to the start codon. In prokaryotes, the 5′ UTR frequently contains some variation of the Shine-Dalgarno (SD) consensus sequence, 5′-GGAGGU-3′, which is able to base-pair with the 3′ tail of the 16S ribosomal RNA (rRNA) to promote initiation (1); however, SD-led genes are less common than non-SD-led genes in microbial genomes (2). In vertebrates, the Kozak consensus sequence, GCCGCC(A/G)CCAUGG (start codon underlined) has been reported (3), but only a very small fraction of vertebrate genes (~0.2%) have this exact sequence (4) and the mechanism of action of this sequence is not straightforward. Although there exist fundamental differences in initiation between eukaryotes and bacteria (e.g., a generally more regulated process in eukaryotes involving nuclear export of 5′-capped mRNA prior to ribosomal association vs. simultaneous transcription and translation in bacteria with capless, often polycistronic, mRNA), there are also common themes, including the importance of the initiation region of the mRNA in allowing access to the start codon and in forming appropriate contacts with the ribosome. The ribosome is able to initiate translation on mRNAs with a wide variety of 5′ UTRs and, therefore, is considered a broad-specificity ribozyme (5-7).
In our previous work (8), we asked the following question: what 5′ UTR sequences inherently enable a ribosome to bind mRNA, initiate translation, and proceed to elongation as quickly as possible? To address this question, the 18 bases before the start codon of the ribosome display vector (pRDV) (9) were fully randomized to create a large starting library (~6.9×1010 unique sequences) in a 5′ UTR context of 81 bases. To avoid unnecessary confounding variables that might be present in vivo, ribosome display in a minimal, well-defined, Escherichia coli (E. coli)-based translation system (10-12) was used to select 5′ UTRs that enabled the most efficient translation of a model protein, FLAG-off7. After three rounds of selection, the majority of the sequences were unexpectedly C-rich and exhibited significant complementarity to the E. coli 16S rRNA. Additionally, short motifs within the selected sequences exhibited striking similarities to the Kozak consensus sequence.
The purpose of the present work was to investigate the selection of RBSs in a different context. Forty-two bases were deleted from the constant 5′ UTR used in our previous work (8) to create a construct with a leader length more representative of the leader lengths naturally found in E. coli, typically 20–40 bases (13). The mRNA template had only 21 bases upstream of the 18-base randomized RBS to allow recovery by reverse transcription-polymerase chain reaction (RT-PCR). All other aspects of the ribosome display construct were kept the same. Mostly non-SD motifs were revealed by high-throughput sequencing after three rounds of increasingly stringent selection for translational efficiency. However, the nature of these non-SD motifs differed greatly from the non-SD motifs found in our previous work (8). The shorter leader enabled the selection of G- and U-rich motifs, many of which exhibited unmistakable complementarity to C- and A-rich motifs of E. coli 16S rRNA. Furthermore, the shorter leader allowed for the selection of RBSs with striking similarity to natural E. coli RBSs, and some of these performed extremely well in vivo.
Based on stringent statistical analyses, we conclude that the complementarity of short motifs in the 5′ UTR to short motifs in the 16S rRNA likely facilitates the most efficient translation in a minimal system. Transient contacts between the mRNA and the rRNA of the small ribosomal subunit may allow the mRNA to increase the local concentration of ribosomes in its vicinity and to reposition itself as necessary to form a productive preinitiation complex that is able to join the large ribosomal subunit and begin elongation. Comparing our earlier study (8) with the present work, it is evident that the length and/or composition of the 5′ UTR affects the specific identities of the complementary sequences that are most efficient in translating a particular transcript. However, the experimental and computational results in both studies support mRNA-rRNA complementarity as a general, broad-specificity mechanism for efficient translation.
RESULTS AND DISCUSSION
Enrichment of RBSs in a minimal system
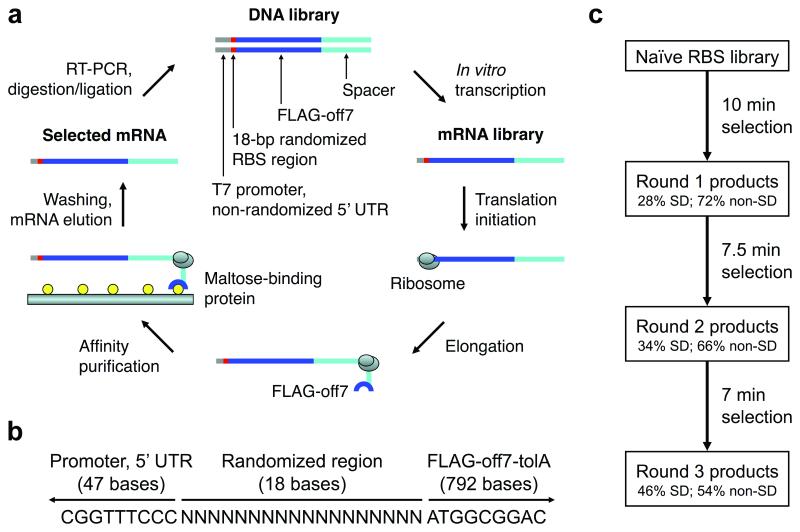
To investigate what upstream sequences promote the most efficient translation in a 5′ UTR context representative of E. coli, we chose a minimal, reconstituted, E. coli-based translation system: PURExpress (New England Biolabs) developed from PURE technology (10, 14, 15). This system has previously been used to investigate what upstream sequences promote fast translation in a longer leader context, ~80 bases (8) vs. ~40 bases in the present study. Briefly, ribosome display is typically used to evolve peptides and proteins with desirable properties, such as improved affinity and stability (16-19). The method involves repeated cycles of generating a DNA library, in vitro transcription of the DNA library, in vitro translation of the resultant mRNA library to generate selection particles, selection of desired library members through binding, and recovery of the mRNA of selected library members. At a minimum, the mRNA transcript contains a 5′ UTR with an RBS followed by a coding region with the gene of interest fused to the gene of an unstructured protein spacer with no stop codon, which allows the ribosome to stall at the end of the mRNA, forming an mRNA-ribosome-polypeptide complex (hereafter called a ribosomal complex). Our adaptation (Figure 1a) shortened the translation time in each round to impart an increasing selection pressure on a randomized 5′ UTR (Figure 1b).
Figure 1.
Ribosome display method, library context, and selection scheme. a) In our adaptation of ribosome display for the selection of efficiently translated sequences, the naïve DNA library contained an 18-bp randomized RBS region before the start codon. Selection pressure was increased over multiple rounds by progressively limiting the time of in vitro translation. b) The DNA context of the randomized RBS region is shown. The T7 promoter and 5′ UTR stem-loop derived from the ribosome display vector, pRDV, are upstream (47 bases in the DNA construct, 21 bases in the mRNA transcript). The coding region (downstream) contains a fusion protein with a FLAG tag, Off7 (a designed ankyrin repeat protein which binds maltose-binding protein), TolA (an unstructured spacer derived from E. coli tolA which allows Off7 to exit the ribosomal tunnel and fold properly), and no stop codon. c) The selection scheme is shown. The naïve RBS library was subjected to three selection rounds of increasing stringency: 10 min, 7.5 min, and 7 min translation. SD sequences were enriched between rounds, but many non-SD sequences remained in the pool after three rounds. Adapted from (8). 5′ UTR, 5′ untranslated region; RBS, ribosome binding site; SD, Shine-Dalgarno.
The 5′ UTR in the present study contains a 21-base 5′ stem-loop derived from the ribosome display vector pRDV (9) to minimize degradation followed by a fully randomized 18-base region with a theoretical diversity of 418 = 6.9×1010 unique sequences, which permitted nearly exhaustive sampling in our in vitro system. In nature, the SD sequence (when present) usually has a context-dependent optimal position within approximately 18 bases before the start codon (20), and approximately 15 bases before the start codon may interact closely with the 30S ribosomal subunit during initiation (21). In total, the 5′ UTR consists of 39 bases, which is representative of that in E. coli (13). The invariant coding region was previously reported (8) and encoded an initiating Met, Ala, FLAG-tag, Gly-Ser (BamHI site), Off7 (9), Lys-Leu (HindIII site), and a modified version of the pRDV TolA spacer with out-of-frame stop codons. Off7, a designed ankyrin repeat protein (DARPin), is a model protein that translates and folds well in vitro. Its nanomolar affinity (~4.4 nM) for maltose-binding protein of E. coli (9) enabled easy affinity purification of only those ribosomal complexes with fully translated protein.
We performed three rounds of selection (10 min, 7.5 min, and 7 min translation at 37°C) and, despite increasingly stringent translation times, the ratio of recovered mRNA molecules on a surface with maltose-binding protein to recovered mRNA molecules on a blocked surface without maltose-binding protein climbed from ~10:1 (round 1) to ~40:1 (round 2) to ~137:1 (round 3) (Supplementary Figure 1). The pool from round 3 was analyzed in depth.
Selected RBSs mostly non-SD and G/U-rich
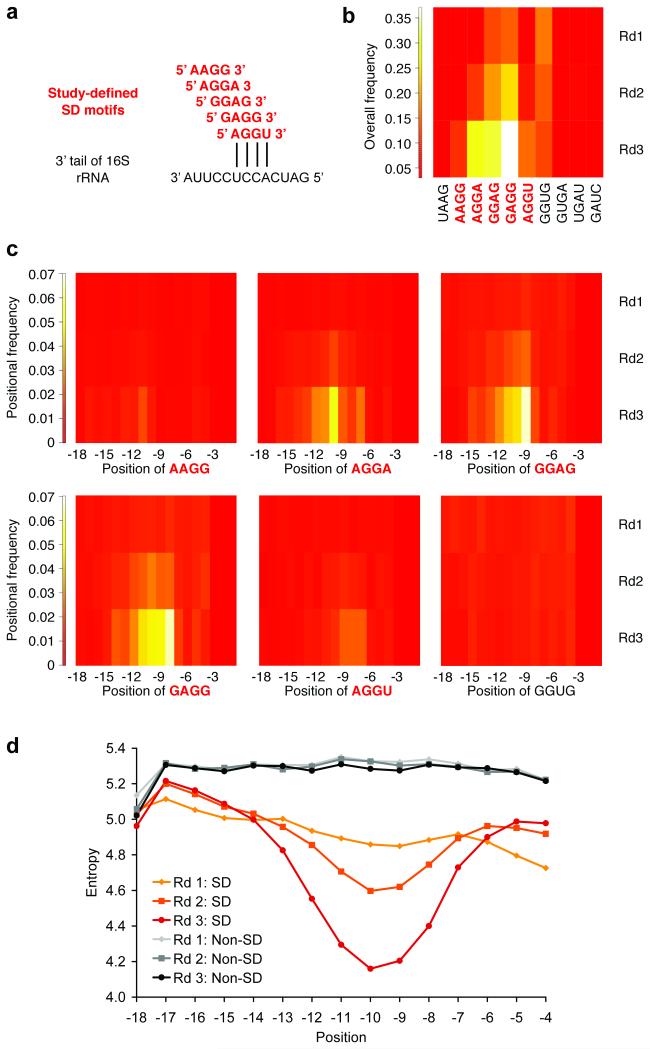
We sequenced the enriched pools from each round using the Roche 454 platform. Approximately 8,000 raw sequences were obtained from each round: 8,782 from round 1; 7,429 from round 2; and 8,889 from round 3. Sequences were excluded from analysis if they did not have exactly 18 bases in the randomized region, if there was an in-frame AUG within the randomized region that could serve as an alternate start site, or if there were errors in the 10 bases upstream or downstream of the randomized region. Approximately 6,000 sequences were analyzed from each round: 6,326 (5,959 unique) from round 1; 5,583 (5,275 unique) from round 2; and 5,542 (5,161 unique) from round 3. SD sequences were broadly defined as any sequence containing at least one of the following four-base motifs that may base-pair to the 3′ tail of the 16S rRNA: AAGG, AGGA, GGAG, GAGG, and AGGU. The overall incidence of SD sequences by round is provided (Figure 1c). The overall and position-dependent enrichment of individual SD sequences and the position-dependent entropy of sequences with or without an SD motif over all three rounds are also presented (Figure 2). In our data, the first G of GGAG is enriched most prevalently around position −9, while the same nucleotide is favored around position −10 in E. coli (22) and position −12 in a longer leader context (8). The optimal position of SD motifs may be affected by mRNA context, as well as different in vitro or in vivo conditions. As in our previous work (8), position-dependent enrichment of SD motifs validated the selection method. Interestingly, the entropy of the pool of SD sequences was highly position-dependent, with the lowest entropy in the vicinity of the strongest enrichment of SD motifs (i.e., low entropy is a direct consequence of high enrichment). In contrast, the entropy of the pool of non-SD sequences was not highly position-dependent and changed very little from round to round. The enrichment of longer (five-, six-, seven-, or eight-base) SD-containing motifs complementary to the 3′ tail of the 16S rRNA is shown (Supplementary Figure 2).
Figure 2.
Enrichment of SD sequences. a) The alignment of study-defined SD motifs (red) with the 3′ tail of the 16S rRNA (black) is shown. b) Overall enrichment of SD sequences over three rounds (Rd1, Rd2, and Rd3) is shown. For comparison, we present all 10 four-base subsets of the reverse complement (5′-UAAGGAGGUGAUC-3′) to the 13 unpaired bases at the 3′ end of the 16S rRNA (5′-GAUCACCUCCUUA-3′) in our selected sequences: UAAG, AAGG, AGGA, GGAG, GAGG, AGGU, GGUG, GUGA, UGAU, and GAUC. c) All study-defined SD motifs (AAGG, AGGA, GGAG, GAGG, and AGGU) exhibited position-dependent enrichment according to their alignment with the 16S rRNA. GGUG (also shown) was enriched overall, but not in a position-dependent manner. d) The position-dependent entropy of four-base windows of 18-base sequences with or without an SD motif over three rounds is shown. Similar plots were observed for five-, six-, seven-, and eight-base windows. Position is specified by the first base of the motif relative to the start codon. SD, Shine-Dalgarno.
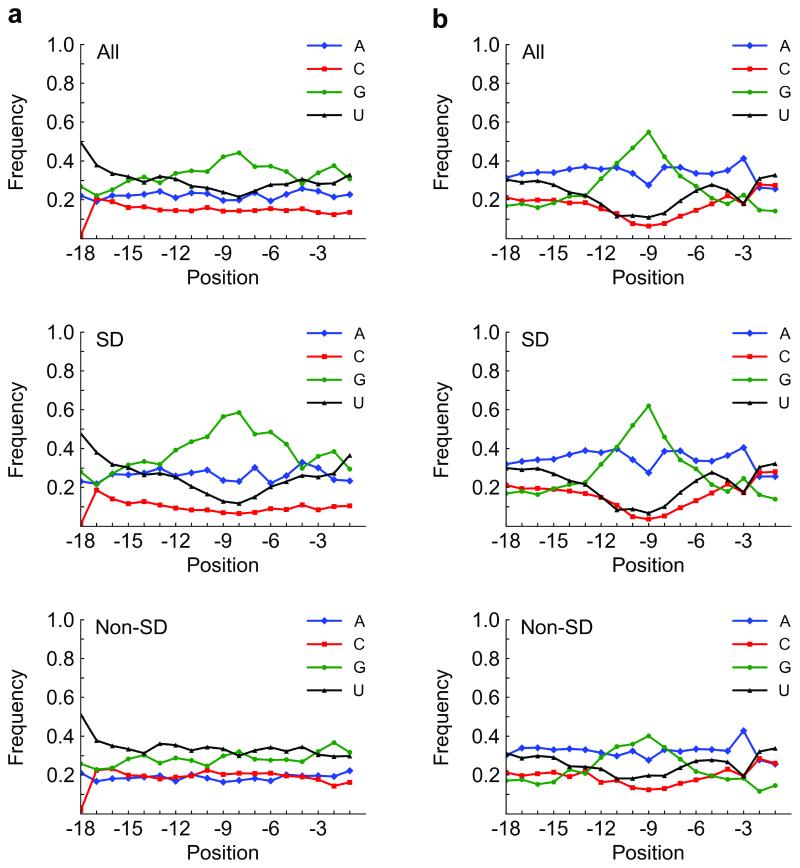
Of the sequences analyzed from the third round, 2,985 (54%) were considered non-SD sequences (2,795 unique). Being highly G/U-rich, these non-SD sequences were remarkably different from the C-rich non-SD sequences selected in the longer leader context (8). The nearly exhaustive sampling of the initial library and the strong first-round signal to background ratio in each study suggest that the different selection outcomes are not the result of a random event. The G/U-richness of the non-SD sequences in the present study appeared to be largely position-independent. Base frequency versus position in all sequences, SD sequences, and non-SD sequences is presented (Figure 3a). These plots closely resemble those of E. coli (Figure 3b), suggesting that the short leader more closely resembles the in vivo context.
Figure 3.
Base content vs. position in our selection and in E. coli. a) Base content vs. position in all, SD (46%), and non-SD (54%) sequences, respectively, in our selection (after third round). Overall (top panel), the nucleotide G is abundant around position −9 or −8, which reflects SD content (middle panel). Interestingly, the non-SD sequences (bottom panel) are mostly G/U-rich. b) Base content vs. position in all, SD (67%), and non-SD (33%) sequences, respectively, from the 18 bases before the start codon in E. coli K12 W3110 (NCBI TaxID 316407; Transterm database (22)). Generally, the base distribution resulting from our selections mimicked that in E. coli. The G peak that appears in our library appears at approximately the same position in E. coli, primarily from the SD sequence. Position is specified by the first base of the motif relative to the start codon. SD, Shine-Dalgarno.
G/U-rich RBSs complementary to 16S rRNA
Based on previous observations of the complementarity of RBSs to the rRNA of the small ribosomal subunit in both prokaryotic (8, 23) and eukaryotic (24) systems, we investigated potential motif complementarity to the 16S rRNA. Overlapping windows of four, five, six, seven, and eight bases from the 18-base randomized region of third-round products were compared to all identically-sized windows of E. coli 16S rRNA. SD and non-SD sequences were considered separately, and native E. coli sequences (18 bases before the start codon) were considered for comparison. We determined the frequency of motifs that were Watson-Crick (A/U or C/G) reverse complements of each window on the 16S rRNA and assigned a p-value to each window based on the probability distribution obtained by analyzing 105 randomly generated libraries equal in size to the dataset (probability of each base = 0.25).
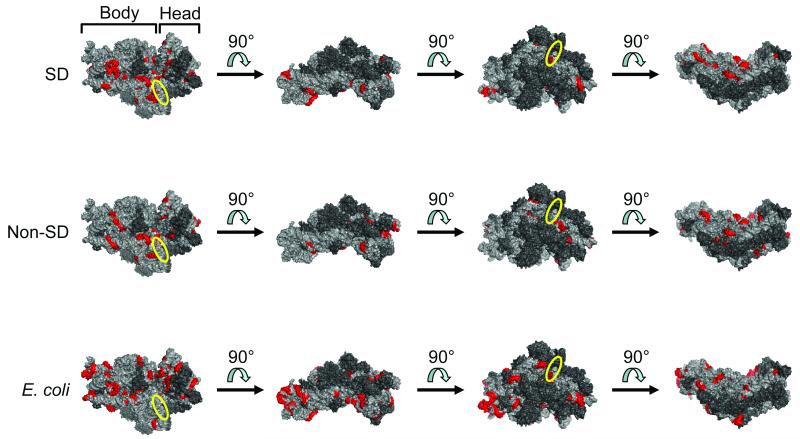
Intriguingly, we found many regions of complementarity despite the lack of C-richness. The number of significant windows on the 16S rRNA was similar for SD and non-SD pools (SD: 42 windows [35 unique sequences], of which 18 windows [12 unique sequences] were not found in the non-SD pool or in E. coli; non-SD: 46 windows [41 unique sequences], of which 37 windows [33 unique sequences] were not found in the SD pool or in E. coli); however, there were many more windows on the 16S rRNA with significant complementarity to native E. coli sequences (82 windows [70 unique sequences], of which 67 windows [55 unique sequences] were not found in the experimentally selected sequences). There were 9 windows (8 unique sequences) with significant complementarity to both SD and non-SD sequences, and there were 15 windows (15 unique sequences) with significant complementarity to both SD and E. coli sequences. There were no windows with significant complementarity to both non-SD and E. coli sequences (Supplementary Figure 3). Potential mRNA-rRNA base-pairing sites on the 30S ribosomal subunit of E. coli (PDB 3DF1; (25)) for the selected library (SD only), the selected library (non-SD only), and native E. coli sequences are shown (Figure 4). To be especially stringent, only significant (p < 0.01; Bonferroni-corrected) six-base windows that shared five bases with at least one neighboring significant window were highlighted. As in our previous study (8), potential mRNA-rRNA base-pairing sites were found primarily on the body of the 30S subunit on the face that becomes buried after assembly with the 50S subunit (Figure 4, leftmost view). The mRNA tunnel is located between the body and head on this face. Complete results from the 16S rRNA comparison are presented (Supplementary Table 1).
Figure 4.
Distribution of potential sites for base-pairing of RBSs (selected library [SD only], top row; selected library [non-SD only], middle row; and E. coli, bottom row) to 16S rRNA. Regions on the E. coli 30S ribosomal subunit with significant complementarity to the RBS population of interest (p-value < 0.01; Bonferroni-corrected) were determined. Significant six-base windows that shared five bases with at least one neighboring significant window are highlighted in red (PyMOL rendering of PDB 3DF1). Four different views convey the general distribution of these potential base-pairing sites over the small ribosomal subunit. The first view in each row shows the face that becomes buried after assembly with the large ribosomal subunit. The approximate position of the anti-SD sequence is indicated by the yellow ellipse. 16S rRNA = light gray; ribosomal proteins = dark gray. RBS, ribosome binding site; SD, Shine-Dalgarno.
The overall propensity of the enriched library to form secondary structure resembled that of the starting library (Supplementary Figure 4), as in our previous study (8), which emphasizes the importance of primary structure (i.e., nucleotide sequence) in ribosome binding. Low secondary structure in the first ~40 nucleotides of the coding region may have compensated for some degree of secondary structure in the RBS region, as such unstructured regions may serve as standby sites for the ribosome (26).
G/U-rich motifs generally prevalent
Based on the observed G/U-rich trend and the complementarity of G/U-rich motifs to C/A-rich regions of 16S rRNA, we decided to perform a naïve motif search to reveal local patterns. The frequency of all possible four-, five-, six-, seven-, and eight-base motifs within the 18 bases was determined, independent of the 16S rRNA, and we checked whether specific motifs were significantly overrepresented compared with what would be expected in the naïve library (i.e., N18). All 5,542 18-base regions from the third-round products, including both SD and non-SD sequences, were included in this analysis. Of the top 20 five-base motifs, 13 contained a four-base SD motif (AAGG, AGGA, GGAG, GAGG, or AGGU) and the other seven were G/U-rich, containing four or five G or U bases. The most frequent motifs in the enriched library exhibited high similarity to the most frequent motifs in the 18 nucleotides before the start codon in E. coli. These results contrast with our previous work that used a longer leader sequence and found that the most frequent motifs in the enriched library were C-rich with high similarity to the most frequent motifs in the 18 nucleotides before the start codon in human (8). In the present study, seven of the top 18 five-base motifs in our selected sequences were also present within the top 15 motifs in E. coli: GGAGG, AGGAG, GAGGA, GAGGU, AAGGA, UAAGG, and UGGAG (Table 1). Complete results from the naive motif search are provided (Supplementary Table 2).
Table 1.
Top 20 motifs from motif search
| 5′ UTR motifs selected for fast translation |
Frequency | 5′ UTR motifs (18b prior to AUG) in E. coli |
Frequency |
|---|---|---|---|
| GGAGG | 0.242 | AGGAG | 0.156 |
| AGGAG | 0.161 | AAGGA | 0.155 |
| GAGGA | 0.119 | GGAGA | 0.116 |
| GAGGU | 0.117 | CAGGA | 0.114 |
| GAGGG | 0.099 | AGGAA | 0.114 |
| GGGAG | 0.078 | GAGGA | 0.090 |
| AAGGA | 0.077 | AAAAA | 0.085 |
| AGGUA | 0.075 | AAAGG | 0.084 |
| UAGGA | 0.061 | GGAGU | 0.084 |
| GGUAU | 0.056 | GAGAA | 0.079 |
| GGUGG | 0.055 | ACAGG | 0.075 |
| UGAGG | 0.054 | GGAGG | 0.075 |
| GUGGG | 0.053 | UGGAG | 0.073 |
| UAAGG | 0.051 | UAAGG | 0.071 |
| UUAUU | 0.050 | GAGGU | 0.069 |
| GGGUG | 0.050 | GGAAA | 0.066 |
| AGGGA | 0.049 | AGAGG | 0.064 |
| UGGAG | 0.049 | GAAAA | 0.063 |
| UAUUU | 0.048 | AGGAU | 0.063 |
| UUAGG | 0.047 | AAAAG | 0.063 |
5′ UTR motifs selected for ability to facilitate translation in an E. coli-based translation system exhibit high similarity to the same region (18 bases prior to AUG) in E. coli. The top 20 five-base motifs from each population are shown. Seven particular motifs (bold and underlined) were present in both sets. Similar results were obtained using other motif lengths. 5′ UTR, 5′ untranslated region.
SD function enhanced by A/U-rich motifs
We further considered the co-occurrence of two significant motifs (false discovery rate [FDR] < 0.01) within the same 18-base randomized RBS region. The number of RBS regions that contained both motif 1 and motif 2 normalized by the number of RBS regions that contained motif 2 only was defined as the co-occurrence metric. Through this measure, we identified A/U-rich motifs co-occurring with canonical SD motifs. A/U-rich motifs are known to improve mRNA expression and stability in E. coli (27). Weak secondary structure and binding to ribosomal protein S1 may contribute to this effect (28). These sequences may also interact transiently through base-pairing to the 16S rRNA because there are at least several A/U-rich potential pairing sites identified in Figure 4 and Supplementary Table 1. The top 10 results from the co-occurrence analysis of five-base motifs are presented (Table 2). Each of the top 10 results includes one five-base SD motif and one A/U-rich motif. Clearly, SD sequences have positional dependence; however, we were also interested in the positional dependence of the 10 A/U-rich motifs most frequently co-occurring with the five-base SD motifs (Supplementary Figure 5). The A/U-rich motifs are most likely to be located upstream or downstream of the optimal SD positions (which are near the middle of the randomized region) in library members containing a five-base SD motif. In library members not containing a five-base SD motif, there is little dependence on position. All pairs of significant four-, five-, and six-base motifs with a non-zero co-occurrence metric are provided (Supplementary Table 3).
Table 2.
Top 10 co-occurrence metrics among significant motifs
| Motifs | Motif 1 | Motif 2 | Coincidence | Motif 1 incidence |
Motif 2 incidence |
Co-occurrence metric |
|---|---|---|---|---|---|---|
| GGAGG|UAUUA | GGAGG | UAUUA | 67 | 1341 | 204 | 0.328 |
| AUUAA|GGAGG | GGAGG | AUUAA | 35 | 1341 | 116 | 0.302 |
| GGAGG|UUAUA | GGAGG | UUAUA | 37 | 1341 | 125 | 0.296 |
| GGAGG|UAUAU | GGAGG | UAUAU | 38 | 1341 | 129 | 0.295 |
| GGAGG|GUUAA | GGAGG | GUUAA | 29 | 1341 | 101 | 0.287 |
| AGGAG|GUAAU | AGGAG | GUAAU | 34 | 893 | 119 | 0.286 |
| AGGAG|GUAUA | AGGAG | GUAUA | 42 | 893 | 157 | 0.268 |
| GGAGG|UUGAA | GGAGG | UUGAA | 28 | 1341 | 106 | 0.264 |
| GAGGU|UUAAG | GAGGU | UUAAG | 38 | 648 | 149 | 0.255 |
| AUAUU|GGAGG | GGAGG | AUAUU | 40 | 1341 | 157 | 0.255 |
The number of sequences that contain both motif 1 and motif 2 is equal to “coincidence.” Co-occurrence metric = coincidence/(motif 2 incidence).
Efficiency of selected RBSs in E. coli
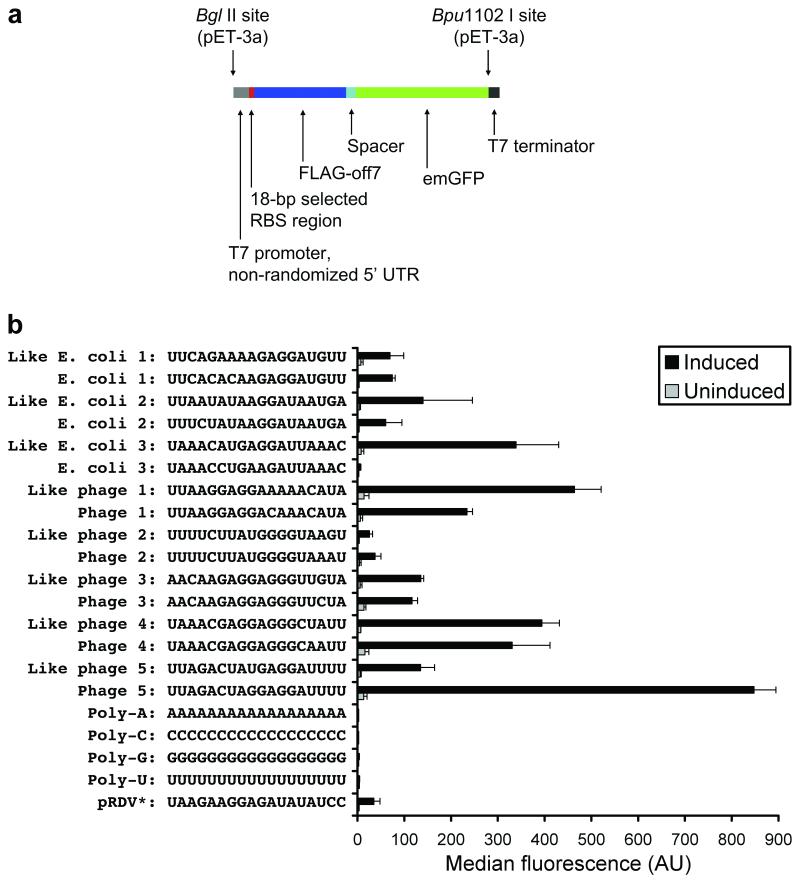
Some of the selected sequences were highly similar to natural E. coli or phage sequences. To evaluate the performance of these clones in vivo, we fused Emerald GFP (emGFP) to Off7 through a short linker and then monitored green fluorescence in E. coli (Figure 5). We investigated the three individual selected library members which exhibited maximal similarity (16 matching positions out of 18) to pre-AUG 18-base regions from E. coli (corresponding genes were rpsP, yjeT, and rpsA) and the five individual selected library members which exhibited maximal similarity (17 matching positions out of 18) to pre-AUG 18-base regions from phage in the EMBL-EBI database (corresponding phages were Lactococcus phage 1706, Salmonella phage ST64B, Pseudomonas phage PaP3, Enterococcus phage phiEF24C, and Staphylococcus phage Twort). Notably, these sequences were not from E. coli phage. Negative controls (poly-A, poly-C, poly-G, and poly-U) and the 18 bases before the start codon in pRDV in the short leader context were also tested in vivo.
Figure 5.
In vivo expression. a) Expression cassettes containing an RBS followed by FLAG-off7-emGFP were cloned into pET-3a and expressed in BL21(DE3)pLysS. b) Average median fluorescence of each clone is shown. Error bars indicate standard deviation of at least three experiments. Library members having maximal similarity to E. coli or phage 5′ UTRs and the corresponding natural sequences are shown, along with the four homopolymers (negative controls) and the 18 bases before the start codon in the ribosome display vector, pRDV (* indicates that these 18 bases are situated in the short leader context). 5′ UTR, 5′ untranslated region; emGFP, Emerald GFP; RBS, ribosome binding site.
These “like E. coli” and “like phage” clones performed extremely well, some even better than their native counterparts. Additionally, the G/U-rich “like phage 2” clone expressed at >8-fold above background despite not having an SD sequence. Expression from homopolymers was low (<2-fold above background) or nonexistent, which partially contrasts with previous work using the longer leader in which poly-A, poly-G, and poly-U facilitated some degree of translation (8); poly-C performed poorly in both contexts. The 18 bases before the start codon in pRDV performed much better in the longer leader context (8) compared with the shorter leader context. The average median fluorescence with the WT pRDV RBS in the longer leader context was 1417±178 AU induced and 15.2±15.6 AU uninduced in similar experiments (8).
Assumptions and justifications
The goal of the present study was to gain insight into the inherent capabilities of the ribosome specifically in an E. coli-like 5′ UTR context. To meet this objective, a large, diverse library of 5′ UTRs was allowed to compete for the use of ribosomes in a minimal, well-defined, E. coli-based translation system. It is possible that some sequences may have been selected for their ability to outcompete other sequences for binding to ribosomes, not necessarily for their efficiency in the overall process, which includes forming the initiation complex, joining with the large ribosomal subunit, and proceeding to elongation. The successful expression of a panel of clones in vivo strongly suggests that these RBSs were independently efficient, although we cannot exclude the possibility that the high protein yield might be at least partially mediated by greater transcript stability. We also expect that the selected RBS regions function primarily in translation initiation, although some effects on elongation may be possible. Together, the 5′ UTR and the beginning of the coding sequence comprise a translation initiation region which affects the efficiency of forming the preinitiation complex and, therefore, overall translational efficiency (29).
The comparison of third-round sequences with the 16S rRNA was performed without knowledge-based bias in regard to intramolecular rRNA base-pairing. While available ribosomal crystal structures have provided evidence that many of the potential pairing sites found in this study may participate in some degree of intramolecular rRNA base-pairing, a large proportion of these intramolecular rRNA base-pairs may be vulnerable to displacement at the translation temperature. Potential pairing sites located near the surface of the ribosome could easily be involved in complementary mRNA-rRNA interactions. This concept has been previously expressed in the form of a competitive displacement model (30).
Insights from experimental system
The combination of a minimal translation system, ribosome display (which allows for very large libraries, up to ~1014 with reasonable scale-up), high-throughput sequencing, and statistical analysis allows for the discovery of new insights related to the biology of the ribosome. The present study complements and greatly expands upon the findings of our previous work (8) that used a 5′ UTR based on pRDV (9). The 5′ UTR in our previous study contained the same 5′ stem-loop used in the present study, but it also contained a translational enhancer and SD RBS derived from enterobacteriophage T7. This leader works very well both in vitro and in vivo to promote expression of the downstream gene.
In this earlier work with the longer leader, we had expected that randomizing the 18 nucleotides before the start codon in pRDV and selecting the fastest-translating sequences would yield predominantly SD sequences; however, 76% of the sequences were non-SD with short, C-rich motifs complementary to the 16S rRNA (8). These C-rich sequences performed extremely well in vitro but did not express well in vivo, likely because of inhibitory nucleic acids, which abolished the activity of C-rich RBSs but not the WT pRDV RBS in vitro. In the present study, we expected to see a similar mixture of sequences, but the selected library contained very different themes. SD sequences were more dominant (46% vs. 24% in the previous study) and the non-SD sequences were predominantly G/U-rich. Because both libraries were constructed with fully randomized oligonucleotides and were nearly exhaustively sampled, our results suggest a clear context-dependent enrichment: C-rich sequences were not most efficient in the shorter leader context, while G/U-rich sequences were not most efficient in the longer leader context. However, based on our previous work, we suspected that short motifs within the 18-base randomized region might still be complementary to certain motifs on the 16S rRNA, even though the base composition of the mRNA-rRNA interaction would be different in the present study. Indeed, we found this to be case. Interestingly, many more significant windows on the 16S rRNA were found when considering natural E. coli mRNAs in comparison with our selected SD and non-SD sequences. Complementarity between the mRNA and rRNA of E. coli has been noted previously (23), but not with the rigorous statistical analyses applied to our data. A more general trend of rRNA-complementary sequences has also been observed in natural mRNAs (31), and we have now tested this hypothesis experimentally with two different 5′ UTR contexts.
Comparing the base frequencies in the selected library with the 18 bases before the start codon in E. coli, the selected library seems to under-represent A and over-represent U. E. coli ribosomes, like the ribosomes of many other organisms (32), are able to translate efficiently using both poly-A and highly U-rich leader sequences, probably at least partially because of their lack of secondary structure. G and C would have the ability to form stronger intramolecular base-pairs and could potentially decrease accessibility of the start codon. Interestingly, the abundance of the various bases in the E. coli 16S rRNA is: 25.2% A; 22.8% C; 31.6% G; and 20.4% U. If mRNA-rRNA complementary were truly a driving force in our selections, it might be expected that U would be more prevalent than A in the selected library, as we have found. Additionally, we selected library sequences which were highly similar to natural E. coli and phage sequences, and these performed very well in vivo.
Most efficient RBSs modulated by 5′ UTR
The length and composition of the 5′ UTR are likely to have a strong influence on how efficiently a downstream coding sequence is translated. For example, longer 5′ UTRs may allow more efficient translation than shorter 5′ UTRs, as long as the start codon remains reasonably accessible (32, 33). The additional length may allow for more interactions (specific or non-specific) with the ribosome, which may increase the local concentration of ribosomes in the vicinity of the start codon and promote the formation of a productive preinitiation complex. A longer 5′ UTR may also be beneficial if particular features (such as the translational enhancer found in pRDV) are introduced. It is interesting that in our minimal, E. coli-based system, a longer 5′ UTR allowed for the selection of C-rich (vertebrate-like) motifs complementary to small subunit rRNA, while a shorter 5′ UTR allowed for the selection of G/U-rich (more E. coli-like) motifs complementary to the small subunit rRNA. The present work provides evidence for non-SD RBSs complementary to E. coli rRNA, which further supports broad-specificity mRNA-rRNA complementarity as a general, broad-specificity mechanism for efficient translation. Our findings also underscore the importance of context-dependent differences in the selection of efficient, rRNA-complementary RBSs, which may help to explain why such sequences can be highly varied across species and even among transcripts within a single organism.
METHODS
Construction of library and clones
Construction of the RBS library and clones used for in vivo expression studies is described in the Supplementary Methods. Oligonucleotide sequences are provided (Supplementary Table 4).
Ribosome display
Ribosome display was performed as described (8) with some modifications. In the first round of selection, 27 μg mRNA (corresponding to ~5.8×1013 molecules) was added to a master translation mix (total volume ~22 μL; RNA:ribosome ratio ~2:1) that was divided into eight 2.5 μL reactions. The translation reactions were incubated at 37°C for 0, 1, 2, 3, 4, 5, 10, and 15 min to allow full translation of any mRNAs that contained an RBS while minimizing degradation. Thin-walled PCR tubes were used for incubation to ensure that the translation temperature was reached quickly. The reactions were stopped using 420 μL cold WBT buffer (50 mM Tris-acetate, pH 7.5 at 4°C, 150 mM NaCl, 50 mM magnesium acetate, 0.05% (w/v) Tween-20; (19)) with RNasin Plus (Promega; 1% v/v). Each stopped translation was mixed and distributed into four wells on a 96-well plate; duplicate wells with or without immobilized maltose-binding protein (100 μL/well) were prepared as described (8). Briefly, NUNC Maxisorp plates (Thermo Fisher Scientific) were prepared by adsorbing NeutrAvidin (Thermo Fisher Scientific), blocking with BlockAce (AbD Serotec), and allowing biotinylated maltose-binding protein of E. coli to bind to the “positive” wells. Binding of the contents of the stopped translation to the prepared wells was performed for 1 h at 4°C with shaking. The plate was then washed three times with WBT and once with WB (same buffer without Tween-20) before reverse transcription. Reverse transcription and subsequent rounds are described in the Supplementary Methods.
Data analysis
Raw sequences obtained from the Roche/454 GS FLX sequencer were filtered to ensure that the randomized region was of the expected length (18 bases) and in the expected context (ACGGTTTCCC upstream and ATGGCGGACT downstream). Sequences that contained an in-frame ATG in the randomized region were excluded from analysis. For entropy calculations, all possible k-base windows of the 18-base randomized region were considered (k = 4–8). For each window, the diversity was quantified by the Shannon entropy (in bits): where i ∈ {all k-base sequences} and p(i) = normalized frequency. The general methodologies for the rRNA comparison, naïve motif search, co-occurrence analysis, and secondary structure analysis have been described (8) and are further detailed in Supplementary Methods.
In vivo experiments
Selected RBSs were cloned into pET-3a (Novagen) with FLAG-off7 and emGFP. Methods for protein expression in E. coli using selected RBSs have been described (8) and are provided in Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Plückthun for providing the original pRDV-off7 plasmid. We also appreciate helpful discussions with S. Jensen and B. Gregory. This work was supported by the National Institutes of Health [T32HG000046 to P.A.B.]; the National Science Foundation [Graduate Research Fellowship to P.A.B. and XSEDE grant MCB110145 to C.A.S.]; and the University of Pennsylvania [C.A.S.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang B, Halgamuge S, Tang S-L. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamoto T. Evolution and the universality of the mechanism of initiation of protein synthesis. Gene. 2009;432:1–6. doi: 10.1016/j.gene.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barendt PA, Shah NA, Barendt GA, Sarkar CA. Broad-specificity mRNA-rRNA complementarity in efficient protein translation. PLoS Genet. 2012;8:e1002598. doi: 10.1371/journal.pgen.1002598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binz HK, Amstutz P, Kohl A, Stumpp MT, Briand C, Forrer P, Grütter MG, Plückthun A. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu Y, Ueda T. PURE technology. Methods Mol. Biol. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- 13.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muñiz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, García-Sotelo JS, López-Fuentes A, Porrón-Sotelo L, Alquicira-Hernández S, Medina-Rivera A, Martínez-Flores I, Alquicira-Hernández K, Martínez-Adame R, Bonavides-Martínez C, Miranda-Ríos J, Huerta AM, Mendoza-Vargas A, Collado-Torres L, Taboada B, Vega-Alvarado L, Olvera M, Olvera L, Grande R, Morett E, Collado-Vides J. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuura T, Yanagida H, Ushioda J, Urabe I, Yomo T. Nascent chain, mRNA, and ribosome complexes generated by a pure translation system. Biochem. Biophys. Res. Commun. 2007;352:372–377. doi: 10.1016/j.bbrc.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi H, Shimizu Y, Ying B-W, Ueda T. Efficient protein selection based on ribosome display system with purified components. Biochem. Biophys. Res. Commun. 2007;352:270–276. doi: 10.1016/j.bbrc.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milovnik P, Ferrari D, Sarkar CA, Plückthun A. Selection and characterization of DARPins specific for the neurotensin receptor 1. Prot. Eng. Des. Sel. 2009;22:357–366. doi: 10.1093/protein/gzp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barendt PA, Sarkar CA. Cell-free display systems for protein engineering. In: Park SJ, Cochran JR, editors. Protein Engineering and Design. 1st ed. CRC Press; Boca Raton, FL: 2009. pp. 51–81. [Google Scholar]
- 19.Dreier B, Plückthun A. Ribosome display: a technology for selecting and evolving proteins from large libraries. Methods Mol. Biol. 2011;687:283–306. doi: 10.1007/978-1-60761-944-4_21. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusupova GZ, Yusupov MM, Cate JHD, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs GH, Chen A, Stevens SG, Stockwell PA, Black MA, Tate WP, Brown CM. Transterm: a database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 2009;37:D72–D76. doi: 10.1093/nar/gkn763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shabalina SA. Regions of intermolecular complementarity in Escherichia coli 16S rRNA, mRNA, and tRNA molecules. Molec. Biol. 2002;36:359–364. [PubMed] [Google Scholar]
- 24.Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 25.Borovinskaya MA, Shoji S, Fredrick K, Cate JHD. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA. 2008;14:1590–1599. doi: 10.1261/rna.1076908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Smit MH, Van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003;331:737–743. doi: 10.1016/s0022-2836(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 27.Komarova AV, Tchufistova LS, Dreyfus M, Boni IV. AU-rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005;187:1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamoto T. Mechanisms of the initiation of protein synthesis: in reading frame binding of ribosomes to mRNA. Mol. Biol. Rep. 2011;38:847–855. doi: 10.1007/s11033-010-0176-1. [DOI] [PubMed] [Google Scholar]
- 30.Sarge KD, Maxwell ES. Evidence for a Competitive-Displacement Model for the initiation of protein synthesis involving the intermolecular hybridization of 5 S rRNA, 18 S rRNA and mRNA. FEBS Lett. 1991;294:234–238. doi: 10.1016/0014-5793(91)81437-d. [DOI] [PubMed] [Google Scholar]
- 31.Tranque P, Hu MC, Edelman GM, Mauro VP. rRNA complementarity within mRNAs: a possible basis for mRNA-ribosome interactions and translational control. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12238–12243. doi: 10.1073/pnas.95.21.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mureev S, Kovtun O, Nguyen UTT, Alexandrov K. Species-independent translational leaders facilitate cell-free expression. Nat. Biotechnol. 2009;27:747–752. doi: 10.1038/nbt.1556. [DOI] [PubMed] [Google Scholar]
- 33.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.