Abstract
Background
The purpose of this study was to evaluate the potential value of circulating miRNA-122a and miRNA-221 in the diagnosis of hepatocellular carcinoma.
Methods
Serum samples were obtained from 85 patients with hepatocellular carcinoma and 85 age-matched and sex-matched healthy volunteers. miRNAs were isolated from the serum samples, and alfa-fetoprotein levels were determined. Expression of miRNA-122a and miRNA-221 in cases and controls was quantified using U6 sn RNA as the internal control. The diagnostic value of miRNA-122a, miRNA-221, and alfa-fetoprotein was compared by receiver operating characteristic analysis.
Results
The serum miRNA-122a level in patients with hepatocellular carcinoma was significantly reduced in comparison with healthy controls and correlated with known risk factors for hepatocellular carcinoma. Circulating miRNA-221 in patients with hepatocellular carcinoma was higher compared with the control group, but the difference was not statistically significant. Receiver operating characteristic analysis revealed that the diagnostic power of miRNA-122a was suboptimal compared with serum alfa-fetoprotein. Further, the serum alfa-fetoprotein and miRNA-122a combined classifier resulted in performance similar to that of alfa-fetoprotein alone.
Conclusion
The serum miRNA-122a level correlates with risk factors for hepatocellular carcinoma. However, use of miRNA-122a as a diagnostic tool for hepatocellular carcinoma is not superior to alfa-fetoprotein. Further analysis is needed to evaluate the diagnostic power of plasma miRNA-122a for hepatocellular carcinoma.
Keywords: circulating miRNA, miRNA-122a, miRNA-221, hepatocellular carcinoma, alpha-fetoprotein, molecular diagnostics
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy and the third leading cause of cancer mortality worldwide. Each year, there is an incidence of more than 500,000 cases and 600,000 HCC-related deaths. In general, men are more likely to develop HCC than women. The incidence of HCC is highest in regions endemic for hepatitis B virus (HBV), such as People’s Republic of China and southeast Asia.1 HCC is highly malignant and deadly, mainly due to the fact that only a small fraction of HCC is operable at the time of diagnosis, when patients are already at an advanced stage of disease and consequently have a very poor prognosis.2 Therefore, early detection of HCC is essential for successful treatment and better survival.
Despite the great need for a sensitive and specific early diagnostic marker of HCC, the diagnosis still relies on the serum alpha-fetoprotein (AFP) level coupled with diagnostic ultrasound. AFP and ultrasound screening results in both underdiagnosis of a significant number of patients with HCC and a number of false positives in patients with more benign liver disease, such as hepatitis and cirrhosis.3–5 Therefore, identifying novel and noninvasive biomarkers to detect HCC with high sensitivity and specificity is a high priority to improve the prognosis and survival in patients with HCC.
MicroRNAs (miRNAs) are approximately 22-nucleotide, noncoding, endogenous RNA molecules with an important role in a number of biological processes, including embryonic development, cell differentiation, and tumorigenesis.6 Some miRNAs have been found to have oncogenic or tumor-suppressive properties.7 Moreover, miRNA expression profiles can be unique to malignant tissues, and also specific to certain types of cancer.3
An HCC-specific miRNA expression profile has been identified and found to distinguish liver tumors from normal liver tissue with high accuracy.4 A subsequent study found miRNA-221 to be elevated in HCC.8,9 Overexpression of miRNA-221 downregulated levels of cyclin-dependent kinase inhibitors, ie, CDKN1B/p27 and CDKN1C/p57,10,11 which are related to a poor prognosis in patients with HCC.12,13 In addition, expression of miRNA-122a, a liver-specific and the most abundant miRNA in the liver, is repressed in HCC.14,15 These decreased miRNA-122a levels are associated with a poor prognosis and metastasis in HCC,16,17 which also contributes to the malignant phenotype of HCC cells.18 However, these studies analyzed miRNA expression in patient tumor tissues, prompting a need for specific analysis of blood samples.
Given that circulating miRNAs can be used as blood-based markers for detection of cancer,19 we hypothesized that miRNA-122a and miRNA-221 could serve as noninvasive circulating biomarkers for diagnosis of HCC. We investigated miRNA-221 and miRNA-122a expression in serum samples from a group of patients with HCC and controls matched for age and sex. As a control for the diagnostic technique, we also measured AFP levels in the same cohort.
Materials and methods
Patient information
Eighty-five patients being treated for HCC at the Third Military Medical University Affiliated Daping Hospital from January 2011 to June 2012 were enrolled in the study. In all cases, a pathologist confirmed the clinical diagnosis of HCC. Eighty-five volunteers matched for age and sex were also recruited as controls. All patients and volunteers consented to the study and were investigated for family medical history. The study was reviewed and approved by the medical research ethics committee. Informed consent was obtained from each subject prior to data collection and drawing of blood. A questionnaire was used to collect the data from all study subjects, which included age, sex, family history, tobacco smoking, alcohol consumption, and HBV infection. Information on hepatitis C virus (HCV) infection was not collected.
Plasma preparation
First, 2 mL of peripheral venous blood was collected, supplemented with EDTA to avoid coagulation, and stored at room temperature for no longer than two hours before further processing. To isolate plasma, blood samples were centrifuged at 1200× g for 10 minutes at 4°C. The supernatants were then transferred to 1.5 mL Eppendorf tubes and centrifuged further at 12,000× g and 4°C to pellet the cellular components, which were then discarded. The purified plasma samples were transferred to fresh Eppendorf tubes and stored at −80°C for future miRNA extraction.
MiRNA extraction
Total RNA was extracted from plasma using BioZOL™ total RNA extraction reagent (Hangzhou Bioer Technology, Hangzhou, People’s Republic of China), according to the manufacturer’s instructions. The resulting RNA samples were dissolved in 40 μL of nuclease-free water and stored at −80°C for future use.
Quantification of plasma miRNA
Expression of serum miRNA-221 and miRNA-122a was measured using SYBR Green microRNA quantitative reverse transcription polymerase chain reaction reagents (CoWin Biotech Co, Ltd, Beijing, People’s Republic of China). Next, 2 μL of total RNA was reverse-transcribed at 42°C for 30 minutes using miRNA-specific reverse transcription primers. During real-time quantitative PCR reaction, 2 μL of reverse transcription products were used as the template. The primers used in the quantitative miRNA PCR were as follows: miRNA-221(5′-AGC TAC ATT GTC TGC TGG GTT TC-3′), miRNA-122a (5′-TGG AGT GTG ACA ATG GTG TTT GT-3′), and U6snRNA (5′-CAC CAC GTT TAT ACG CCG GTG-3′). Melting curve analysis was performed at the end of the PCR cycles in order to validate the specificity of the expected PCR product. The PCR reaction was performed in triplicate for the miRNA in each sample. The reaction conditions were as follows: 95°C for one minute, 50 cycles at 95°C for 15 seconds, and 60°C for 30 seconds. The Ct values were determined using SDS2.1 software. The quantitative value for a given sample of miRNA was calculated by subtracting the Ct value of U6 RNA, which served as the internal reference. The relative expression of specific miRNA was determined by the ΔCt value, which was defined as ΔCt = (Ct miRNA of sample × − Ct U6RNA of sample x).
Serum AFP detection
Serum AFP values were determined by AFP chemiluminescence assay using a Beckman i800 luminometer (Beckman Coulter, Chongqing, People’s Republic of China). The normal maximum AFP value is 9 ng/mL.
Statistical analysis
The statistical analysis was performed using Statistical Package for Social Sciences version 15.0 software (SPSS Inc, Chicago, IL). The relative expression of miRNA in our samples demonstrated a normal distribution, so statistically significant differences in miRNA levels between the cases and controls were determined by the independent samples test. Data on the relative expression of miRNA is shown as the mean ± standard deviation. Comparison of percentages was done using the Pearson Chi-square test or Fisher’s Exact test. Diagnostic values for the different classifiers (miRNAs or AFP) were determined by calculating the area under the receiver operating characteristic (ROC) curve. P < 0.05 was considered to be statistically significant.
Results
Patient characteristics
Eighty-five patients diagnosed with HCC as previously reported20 were included in this study, and 85 volunteers matched for age, sex, and HBV infection served as controls (Table 1). The AFP level was significantly higher in the HCC group compared with the controls (Table 1). Tumor-node-metastasis staging of the HCC patients20 is shown in Table 2.
Table 1.
Clinical and demographic characteristics of patients with hepatocellular carcinoma and healthy controls
| Variables | Cases (n = 85) | Controls (n = 85) | P-value | ||
|---|---|---|---|---|---|
|
|
|
||||
| n | % | n | % | ||
| Average age (years) | 53.6 ± 12.0 | 50.8 ± 13.0 | 0.154a | ||
| Sex | |||||
| Male | 70 | 82.4 | 69 | 81.2 | |
| Female | 15 | 17.6 | 16 | 18.8 | 0.843b |
| Hepatitis B virus | |||||
| Positive | 75 | 88.2 | 69 | 81.2 | |
| Negative | 10 | 11.8 | 16 | 18.8 | 0.201b |
| Alfa-fetoprotein | |||||
| Positive | 67 | 78.8 | 1 | 1.2 | |
| Negative | 18 | 21.2 | 84 | 98.8 | <0.001c |
Notes:
Independent samples test;
Pearson Chi-square test;
Fisher’s Exact test.
Table 2.
Clinical staging of patients with hepatocellular carcinoma
| TNM stage | Cases, n (%) |
|---|---|
| II | 11 (12.9) |
| IIIA | 16 (18.8) |
| IIIB | 19 (22.5) |
| IVA | 16 (18.8) |
| IVB | 16 (18.8) |
| Other* | 7 (8.2) |
Note:
Patients without enough medical investigation for staging.
Abbreviation: TNM, tumor-node-metastasis.
Serum miRNA-221 and miRNA-122a levels in cases and controls
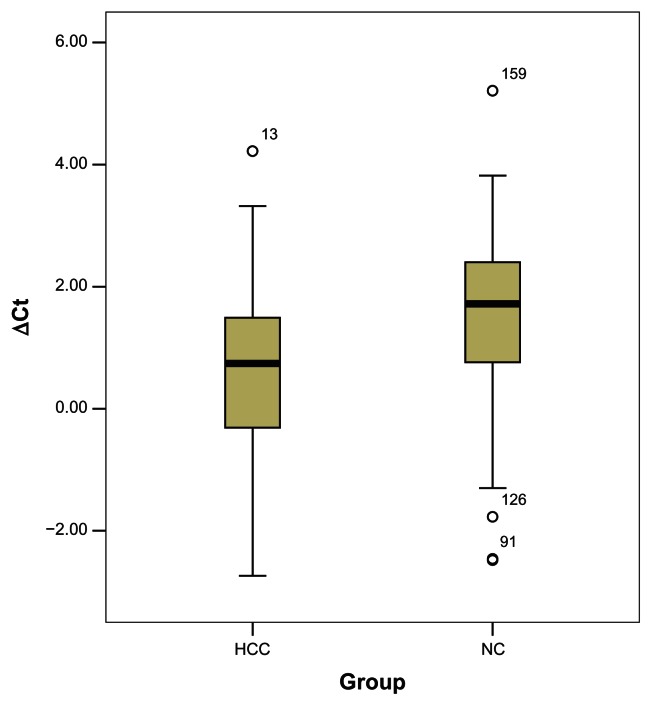
MiRNAs were isolated from the peripheral venous blood of patients and controls. miRNA-221 and miRNA-122a levels were quantified using the SYBR Green quantitative reverse transcription PCR assay with U6RNA as the internal control. Consistent with a previous report of miRNA-122 being suppressed in HCC tissue,14 we observed a significantly lower miRNA-122a level in serum samples from HCC patients in comparison with those from controls (Figure 1 and Table 3). We also observed higher miRNA-221 expression in the patient group, but the difference was not statistically significant (P = 0.225, data not shown).
Figure 1.
Relative expression of miRNA-122a in patients with hepatocellular carcinoma and controls.
Table 3.
Relative expression of miRNA-221 and miRNA-122a in patients with hepatocellular carcinoma and controls
| Variables | Cases (n = 85) | Controls (n = 85) | P value |
|---|---|---|---|
| ΔCt (221) | −2.10 ± 1.02 | −2.53 ± 1.43 | 0.225a |
| ΔCt (122a) | 0.74 ± 1.87 | 1.72 ± 1.77 | <0.001b |
Notes:
Independent samples test;
Mann–Whitney U test (median ± Q, skewed distribution).
Serum miRNA-122a level correlates with multiple risk factors for HCC
Given that serum miRNA-122a levels were significantly lower in HCC patients, we further analyzed the correlation between serum miRNA-122a level and classic risk factors for HCC. As shown in Table 4, miRNA-122a expression was significantly lower in men with HCC than in women with the disease. A trend of lower miRNA-122a expression was also found in patients who smoked cigarettes, consumed alcohol, had a family history of HCC, or were positive for HBV infection (Table 4). These results show that miRNA-122a expression is associated with known risk factors for HCC.
Table 4.
Correlation between serum miRNA-122a and known risk factors for HCC
| HCC risk factors | Group | n | miRNA-122aa | P valueb |
|---|---|---|---|---|
| Age, years | 40–60 | 51 | 0.59 ± 1.33 | 0.987 |
| <40 or >60 | 34 | 0.60 ± 1.35 | ||
| Sex | Male | 70 | 0.44 ± 1.34 | 0.018 |
| Female | 15 | 1.32 ± 0.98 | ||
| Smoking status | Ever/current | 35 | 0.30 ± 1.27 | 0.083 |
| Never | 50 | 0.80 ± 1.34 | ||
| Alcohol consumption | Ever/current | 28 | 0.45 ± 1.32 | 0.503 |
| Never | 57 | 0.66 ± 1.34 | ||
| Cirrhosis | Yes | 47 | 0.65 ± 1.25 | 0.676 |
| No | 38 | 0.53 ± 1.43 | ||
| Family history of HCC | Yes | 4 | 0.58 ± 1.28 | 0.069 |
| No | 81 | 0.65 ± 1.31 | ||
| HBV | Positive | 75 | 0.49 ± 1.25 | 0.054 |
| Negative | 10 | 1.35 ± 1.70 | ||
| AFP | Positive | 67 | 0.55 ± 1.28 | 0.572 |
| Negative | 18 | 0.75 ± 1.53 |
Notes:
Mean ± standard deviation of ΔCt miRNA-122a;
independent samples test.
Abbreviations: AFP, alfa-fetoprotein; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Diagnostic potential of circulating miRNA-122a and AFP in HCC
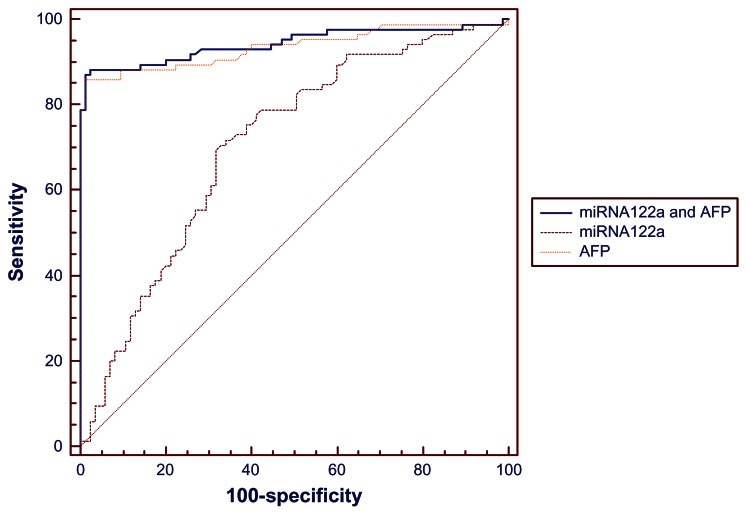
We used ROC curves to compare the ability of serum miRNA-122a to discriminate between patients with HCC and controls with that of circulating AFP. Consistent with being the classic HCC diagnostic marker, AFP showed a very high area under the concentration-time curve (AUC, 0.935), with a cutoff value was 4.69 ng/mL, resulting in a sensitivity of 85.9% and a specificity of 98.8% in separating cases and controls. The ROC curve for plasma miRNA-122a expression levels had an AUC of 0.707, with 70.6% sensitivity and 67.1% specificity, respectively, at a cutoff value of 1.205 (ΔCt). The ROC curve for the combined classifier (AFP and miRNA-122a) resulted in the highest AUC (0.943) with a sensitivity of 87.1% and a specificity of 98.8% (Figure 2 and Table 5), which was significantly better than that of miRNA-122a. However, the performance of the combined classifier was not significantly better than AFP alone.
Figure 2.
Receiver operating characteristic curves for serum AFP, miRNA-122a and the combination of AFP and miRNA-122a.
Abbreviation: AFP, alfa-fetoprotein.
Table 5.
Comparison of receiver operating characteristic curves for serum AFP, miRNA-122a, and the combination of AFP and miRNA-122a
| AUC | 95% CI | Sensitivity (%) | Specificity (%) | P value | |
|---|---|---|---|---|---|
| miRNA-122a and AFP | 0.943 | 0.897–0.973 | 87.1 | 98.8 | <0.0001a |
| miRNA-122a | 0.707 | 0.632–0.774 | 70.6 | 67.1 | <0.0001b |
| AFP | 0.935 | 0.887–0.967 | 85.9 | 98.8 | 0.4359c |
Notes:
miRNA-122a and AFP ~ miRNA-122a;
miRNA-122a ~ AFP;
miRNA-122a and AFP ~ AFP.
Abbreviations: AFP, alfa-fetoprotein; AUC, area under the concentration-time curve; CI, confidence interval.
Discussion
In recent years, miRNAs have emerged as a new class of diagnostic tools for identification of cancer. Because miRNA expression profiles are tissue-specific and differ between malignant and normal tissues, this class of small RNAs may be able to serve as a marker to discriminate tumor tissue from normal tissue or for prognostic purposes.21 Although several groups have investigated the role of miRNA as a tumor marker in HCC,22–26 most of the reports have used histological specimens, and so have limited clinical significance. Given that miRNAs are stable in the circulation, we propose that they may serve as ideal noninvasive biomarkers for diagnosis of HCC. Therefore, we analyzed plasma levels of two miRNAs in patients with HCC and controls, and evaluated their diagnostic performance in the context of AFP, the conventional diagnostic marker of HCC.
Although miRNA-221 levels have been reported to be elevated in HCC tumors,27 the difference in serum miRNA-221 levels between patients with HCC and controls was not statistically significant in our study. Given that miRNA-221 is expressed in organs as well as the liver,28 our results suggest that tumor tissue is not the only source of serum miRNA-221. Further studies are needed to investigate the relationship between tumor and serum miRNA-221 levels, and any differences in miRNA-221 levels between different organs to confirm the lack of a diagnostic value of serum miRNA-221 in HCC.
Expression of miRNA-122a is liver-specific and accounts for about 70% of total miRNA expression in the liver. Recent studies have demonstrated that miRNA-122a expression is decreased during liver tumorigenesis and miRNA-122a can act as a tumor suppressor. Consistent with these findings, we observed a significant decrease in serum miRNA-122a levels in patients with HCC compared with controls. In apparent contrast, Xu et al29 indicated that serum miRNA-122 is elevated in patients with HCC compared with healthy controls. However, the results obtained by Xu et al and our results are the same, with the only difference being the definition of ΔCt used. Further, serum miRNA-122a levels in our study were correlated with multiple well known risk factors for HCC, including sex, cigarette smoking, consumption of alcohol, HBV infection, and family history of HCC. All these data strongly suggest that serum miRNA-122a is a promising diagnostic marker for HCC.
Next, we explored the diagnostic potential of miRNA-122a by calculating AUC values on the ROC curve for both serum miRNA-122a and AFP. Interestingly, the AUC of AFP (0.935) in our study was much higher than the range previously reported (0.712–0.830).30–32 We reasoned that the high AUC observed in our study was probably due to the fact that most of our patients had late-stage HCC, and therefore had a significantly higher AFP level. The ROC curve for miRNA-122a gave a promising AUC value of 0.707, with a sensitivity of 70.6% and a specificity of 67.1%. Although lower than the AUC for AFP, the diagnostic performance of miRNA-122a in our study is comparable with that of AFP (AUC 0.712–0.830), the AFP immunocomplex (AUC 0.691), squamous cell carcinoma antigen (AUC 0.703), and squamous cell carcinoma immunocomplex (AUC 0.694) from previous studies in patients with HCC.33,34 Combining the miRNA-122a classifier with the AFP level, the AUC value was slightly increased compared with AFP alone, suggesting that miRNA-122a may be of some diagnostic value. However, in our study, the diagnostic power of AFP was significantly superior to that of miRNA-122a, and the combined classifier did not result in significantly better performance than AFP alone. These findings establish that serum miRNA-122a has some value as a diagnostic marker, both individually and in combination with AFP. However, AFP levels remain the gold standard for diagnosis of HCC. Further studies on circulating miR122a in patients with early-stage HCC will be essential to determine the diagnostic potential of miR122a in liver cancer.
Lanford et al35 found that miRNA-122a was involved in regulating viral replication in HCV-related liver cancer, suggesting that the miRNA-122a expression level may be associated with the rate of HCV infection. In our study, information on HCV infection was not collected. Unlike in western populations, HCV infection is not a major risk factor for HCC in the People’s Republic of China. Instead, HBV infection is a more significant risk factor for HCC in the Chinese population.36–39 Studies from several groups have demonstrated that endogenous miRNA-122a can inhibit expression of HBV, suggesting a role for miRNA-122a in HBV-related liver cancer.40,41 In our study, serum miRNA-122a levels in HBV-positive patients tended to be lower in comparison with those in HBV-negative patients (Table 4). This observation suggests that, in addition to being suppressed by miRNA-122a, HBV may inhibit miRNA-122a expression and presumably increase the risk of HCC in HBV-positive individuals. We know that people with HBV infection can develop chronic hepatitis, therefore some HBV-positive volunteers may could not be referred to as healthy control but because this study included only 85 controls, and we could not exclude HBV-positive controls or divide them into groups according to the presence or absence of HCC and chronic hepatitis.
In summary, our data suggest that the serum miRNA-122a level has some value as a diagnostic tool for HCC. It is possible that serum miRNA-221 levels do not correlate well with miRNA levels in tumor tissue, so miRNA-221 may not be able to serve as a noninvasive diagnostic marker for HCC. In the present study, we included only 85 patients and the same number of matched controls. Future investigations including larger patient populations and patients with early-stage HCC are needed to confirm the potential diagnostic value of miRNA-122a in HCC.
Footnotes
Disclosure
Medjaden Bioscience Limited assisted in the preparation of this manuscript. The authors report no other conflicts of interest in this work.
References
- 1.Curado MP, Edwards B, Shin HR, et al. Cancer Incidence in Five Continents. Lyon, France: International Agency for Research in Cancer Scientific Publications; 2007. [Accessed April 28, 2013]. Available from: http://www.iarc.fr/en/publications/pdfs-online/epi/sp160/CI5vol9-A.pdf. [Google Scholar]
- 2.Schwartz M, Roayaie S, Konstadoulakis M. Strategies for the management of hepatocellular carcinoma. Nat Clin Pract Oncol. 2007;4:424–432. doi: 10.1038/ncponc0844. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce MC. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 5.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Fu X, Wang Q, Chen J, et al. Clinical significance of miRNA-221 and its inverse correlation with p27(Kip1) in hepatocellular carcinoma. Mol Biol Rep. 2011;38:3029–3035. doi: 10.1007/s11033-010-9969-5. [DOI] [PubMed] [Google Scholar]
- 9.Gramantieri L, Fornari F, Ferracin M, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galardi S, Mercatelli N, Giorda E, et al. miRNA-221 and miRNA-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27 Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 11.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 12.Tannapfel A, Grund D, Katalinic A, et al. Decreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer. 2000;89:350–355. doi: 10.1002/1097-0215(20000720)89:4<350::aid-ijc6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Nakai S, Masaki T, Shiratori Y, et al. Expression of p57(KIP2) in hepatocellular carcinoma: relationship between tumor differentiation and patient survival. Int J Oncol. 2002;20:769–775. [PubMed] [Google Scholar]
- 14.Kutay H, Bai S, Datta J, et al. Downregulation of miRNA-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Chang J, Nicolas E, Marks D, et al. miRNA-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 16.Fornari F, Gramantieri L, Giovannini C, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- 17.Tsai WC, Hsu PW, Lai TC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2008;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 18.Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to Sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Cancer Society (ACS) Liver Cancer. 2012. [Accessed January 18, 2013]. http://www.cancer.org/acs/groups/cid/documents/webcontent/003114-pdf.pdf.
- 21.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 22.Chen YT, Kitabayashi N, Zhou XK, Fahey TJ, 3rd, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21:1139–1146. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 23.Taylor DD, Taylor CG. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Zhang C, Hu Z, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 26.Yanaihara N, Caplen N, Bowman E, et al. Unique micro-RNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 27.Rong M, Chen G, Dang Y. Increased MiR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang QE, Racicot KE, Kaucher AV, Oatley MJ, Oatley JM. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–290. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Wu C, Che X, et al. Circulating microRNAs, miRNA-21, miRNA-122, and miRNA-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 30.Porta C, De Amici M, Quaglini S, et al. Circulating interleukin-6 as a tumor marker for hepatocellular carcinoma. Ann Oncol. 2008;19:353–358. doi: 10.1093/annonc/mdm448. [DOI] [PubMed] [Google Scholar]
- 31.Mao Y, Yang H, Xu H, et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59:1687–1693. doi: 10.1136/gut.2010.214916. [DOI] [PubMed] [Google Scholar]
- 32.Shen Q, Fan J, Yang XR, et al. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 2012;13:817–826. doi: 10.1016/S1470-2045(12)70233-4. [DOI] [PubMed] [Google Scholar]
- 33.Giannelli G, Marinosci F, Trerotoli P, et al. SCCA antigen combined with alpha-fetoprotein as serologic markers of HCC. Int J Cancer. 2005;117:506–509. doi: 10.1002/ijc.21189. [DOI] [PubMed] [Google Scholar]
- 34.Giannelli G, Fransvea E, Trerotoli P, et al. Clinical validation of combined serological biomarkers for improved hepatocellular carcinoma diagnosis in 961 patients. Clin Chim Acta. 2007;383:147–152. doi: 10.1016/j.cca.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis c virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan DE, Reddy KR. Rising incidence of hepatocellular carcinoma: the role of hepatitis B and C; the impact on transplantation and outcomes. Clin Liver Dis. 2003;7:683–714. doi: 10.1016/s1089-3261(03)00060-6. [DOI] [PubMed] [Google Scholar]
- 37.El-Serag HB. Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 2002;35:S72–S78. doi: 10.1097/00004836-200211002-00002. [DOI] [PubMed] [Google Scholar]
- 38.El-Serag HB, Davila JA, Petersen NJ, McGlynn KA. The continuing increase in the incidence of hepatocellular carcinoma in the United States: an update. Ann Intern Med. 2003;139:128. doi: 10.7326/0003-4819-139-10-200311180-00009. [DOI] [PubMed] [Google Scholar]
- 39.Di Bisceglie AM, Lyra AC, Schwartz M, Reddy RK, Martin P, Gores G. Hepatitis C-related carcinoma in the United States: influence of ethnic status. Am J Gastroenterol. 2003;98:2060–2063. doi: 10.1111/j.1572-0241.2003.t01-1-07641.x. [DOI] [PubMed] [Google Scholar]
- 40.Qiu LP, Fan HX, Jin WS, et al. MiR-122-induced downregulation of HO-1 negatively affects miRNA-122-mediated suppression of HBV. Biochem Biophys Res Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 41.Fan G, Wang CM, Tian C, et al. MiR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep. 2011;26:1281–1286. doi: 10.3892/or.2011.1375. [DOI] [PubMed] [Google Scholar]