Abstract
Trichostrongylus colubriformis (Strongylida), a small intestinal nematode of small ruminants, is a major cause of production and economic losses in many countries. The aims of the present study were to define the transcriptome of the adult stage of T. colubriformis, using 454 sequencing technology and bioinformatic analyses, and to predict the main pathways that key groups of molecules are linked to in this nematode. A total of 21,259 contigs were assembled from the sequence data produced from a normalized cDNA library; 7,876 of these contigs had known orthologues in the free-living nematode Caenorhabditis elegans, and encoded, amongst others, proteins with ‘transthyretin-like’ (8.8%), ‘RNA recognition’ (8.4%) and ‘metridin-like ShK toxin’ (7.6%) motifs. Bioinformatic analyses inferred that relatively high proportions of the C. elegans homologues are involved in biological pathways linked to ‘peptidases’ (4%), ‘ribosome’ (3.6%) and ‘oxidative phosphorylation’ (3%). Highly represented were peptides predicted to be associated with the nervous system, digestion of host proteins or inhibition of host proteases. Probabilistic functional gene networking of the complement of C. elegans orthologues (n = 2,126) assigned significance to particular subsets of molecules, such as protein kinases and serine/threonine phosphatases. The present study represents the first, comprehensive insight into the transcriptome of adult T. colubriformis, which provides a foundation for fundamental studies of the molecular biology and biochemistry of this parasitic nematode as well as prospects for identifying targets for novel nematocides. Future investigations should focus on comparing the transcriptomes of different developmental stages, both genders and various tissues of this parasitic nematode for the prediction of essential genes/gene products that are specific to nematodes.
Keywords: Trichostrongylus colubriformis, Transcriptome, Next-generation sequencing, Bioinformatics, Peptidases, Ancylostoma-secreted proteins
1. Introduction
Parasitic nematodes of livestock animals are of major socio-economic importance worldwide due to the diseases and associated production losses that they cause. The nematode Trichostrongylus colubriformis (Strongylida, Trichostrongylidae), is amongst the most important parasites of small ruminants, and can be a major cause of economic losses (O’Connor et al., 2006). Its life cycle is direct, with morulated eggs being passed in the faeces of the host. Under suitable environmental conditions (i.e. 18 to 21°C, 100% humidity; Olsen, 1986), the first-stage larvae (L1s) hatch from eggs to then develop (via the second stage, L2) to infective, third-stage larvae (L3s). The cuticle of the L2 is retained as a sheath around the L3 and protects it from desiccation (Olsen, 1986). Infective L3s are ingested with herbage by the host, pass through the forestomachs and undergo an exsheathment process. This process is triggered by the pepsin/HCl in the abomasum, stimulating receptors in the L3 to produce exsheathment fluids (Olsen, 1986). The exsheathed L3 penetrate the mucosa of the small intestine and moult to the fourth-stage larvae (L4), which return to the intestinal lumen and develop to adult males and females within ~3 weeks following the ingestion of L3s (Olsen, 1986). Adult T. colubriformis live in mucus-covered tunnels in the mucosal surface of the small intestine, where they feed on chyme components (Holmes, 1985). Heavy infections are associated with severe enteritis, characterized by extensive villus atrophy, mucosal thickening and erosion and infiltration of lymphocytes and neutrophils into affected mucosal areas (Holmes, 1985). Clinical signs of trichostrongylosis include malabsorption, weight loss and diarrhoea (= scouring or “black scour”).
Traditionally, the control of T. colubriformis infection and trichostrongylosis has relied heavily on the administration of anthelmintics. The excessive and suppressive use of such drugs (Kaplan, 2002, 2004) has led to major problems with anthelmintic resistance (Waller, 1985; Sangster, 1996). Attempts to develop effective vaccines to circumvent resistance problems have largely been unsuccessful to date (Sangster, 1996; Maass et al., 2009). Therefore, there is a continuous need to identify molecular targets for the development of new and efficacious nematocides. A detailed understanding of the complement of molecules transcribed in the adult stage of this parasitic nematode could provide a basis for the identification or prevalidation of essential genes and gene products for the subsequent design of such nematocides.
Investigations of the transcriptome of parasitic nematodes using different approaches (see Ranganathan et al., 2009) is gradually leading to a better understanding of the biochemical and molecular processes involved in parasite development, reproduction and interactions with their host/s (Campbell et al., 2008; Huang et al., 2008; Jacob et al., 2008; Nisbet et al., 2008; Cantacessi et al., 2009a; Ranganathan et al., 2009; Cantacessi et al., 2010a). In particular, next-generation sequencing technologies, such as 454-Roche (www.454.com; Margulies et al., 2005), ABI-SOLiD (www.appliedbiosystems.com; Pandey et al., 2008), Illumina-Solexa (www.illumina.com; Bentley et al., 2008) and Helicos (www.helicosbio.com; Harris et al., 2008) are changing the way we discover and define parasite transcriptomes and genomes (see Droege and Hill, 2008; Jex et al., 2010). These advances in sequencing techniques are reflected in the development of enhanced computational methods for the pre-processing, assembly and annotation of sequence data (Nagaraj et al., 2007a,b, 2008). Furthermore, the availability of the entire genome sequences of other helminths, such as the free-living nematode Caenorhabditis elegans, for which detailed information of, for example, molecular and biochemical aspects of development, metabolism and reproduction is available (see www.wormbase.org) for comparative purposes, are allowing the elucidation of fundamental aspects of the biology of parasitic nematodes of public and veterinary health importance (see Nisbet et al., 2008; Ranganathan et al., 2009; Rabelo et al., 2009; Cantacessi et al., 2010a).
Despite the substantial economic impact of trichostrongylosis in livestock (see Sackett and Holmes, 2006), no genomic and transcriptomic information for T. colubriformis is available in public databases. Gaining an improved understanding of fundamental molecular pathways linked to parasite survival in the environment, development and reproduction in the vertebrate host and host-parasite interactions will assist in finding new ways of disrupting these pathways and thus facilitate the identification of new drug targets. In the present study, we (i) produced the first, large-scale transcriptomic dataset for adult T. colubriformis using a next-generation sequencing-based approach, (ii) subjected these data to detailed bioinformatic exploration, and (iii) predicted key pathways and groups of molecules involved in fundamental metabolic pathways of the biology in this nematode.
2. Materials and methods
2.1. Parasite material
Merino lambs (8–12 weeks of age), maintained under helminth-free conditions, were inoculated intra-ruminally with 10,000 infective third-stage larvae (L3) of T. colubriformis (McMaster strain; Animal Ethics Approval Number 707528, The University of Melbourne). The patency of the infection (~21–25 days) was established based on the detection of strongylid eggs in the faeces using the McMaster flotation method (MAFF, 1977). For the collection of adult worms, infected lambs were euthanized with an overdose of pentobarbitone sodium (Lethobarb, Virbac Pty. Ltd.), administered intravenously 30 days after inoculation. Adult worms were immediately collected from the first 4 m of the small intestine, washed extensively in phosphate-buffered saline (PBS; pH 7.4), and snap frozen in liquid nitrogen for subsequent storage at −70 °C.
2.2. Preparation of 3′-cDNA from Trichostrongylus colubriformis for 454 sequencing
Total RNA from adult female and male worms was prepared using TRIzol Reagent (GibcoBRL, Life Technologies or Invitrogen, Carlsbad, CA) following the manufacturers’ instructions and treated with Ambion Turbo DNase (Ambion/Applied Biosystems, Austin, TX). The integrity of the RNA was verified using the Bioanalyzer 2100 (Agilent Technologies, Cedar Creek, Texas), and the yield determined using the NanoDrop ND-1000 UV-VIS spectrophotometer v.3.2.1 (NanoDrop Technologies, Wilmington, DE). The cDNA library was constructed using the SMART™ kit (Clontech/Takara Bio, CA). An optimized PCR cycling protocol (over 20 cycles) was used to amplify full-length cDNAs employing primers complementary to the SMART IIA-Probe and custom oligo(dT) and the Advantage-HF 2 polymerase mix (Clontech/Takara). The cDNA was then normalized by denaturation-reassociation, treated with duplex-specific nuclease (Trimmer kit, Evrogen, CA) and amplified over 14 cycles. Subsequently, the 5′- and 3′- adaptors were removed by digestion with the exonuclease Mme1 and streptavidin-coated paramagnetic beads (Mitreva and Mardis, 2009). The normalized cDNA (500-700 bases) was then amplified using 9 cycles of Long and Accurate (LA)-PCR (Barnes, 1994) and then sequenced in a Genome Sequencer™ (GS) FLX instrument (Roche Diagnostics), employing a standard protocol (Margulies et al., 2005).
2.3. Bioinformatic analysis of sequence data
Sequences from the normalized cDNA library for T. colubriformis were aligned and clustered using the Contig Assembly Program v.3, CAP3 (Huang and Madan, 1999), employing a minimum sequence overlap length cut-off of 30 bases and an identity threshold of 95%, and assembled. Following the pre-processing of the expressed sequence tags (ESTs), T. colubriformis contigs in the dataset were subjected to BLASTx (NCBI; www.ncbi.nlm.nih.gov) and BLASTn (EMBL-EBI Parasite Genome Blast Server; www.ebi.ac.uk) to identify putative homologues in C. elegans, other nematodes and organisms other than nematodes (e-value cut-off: ≤ 1e-05). WormBase WS200 (www.wormbase.org) was interrogated extensively for relevant information on C. elegans orthologues/homologues, including RNA interference (RNAi) phenotypic, transcriptomic, proteomic and interactomic data. T. colubriformis contigs were conceptually translated into peptides using the program ESTScan (Nagaraj et al., 2007a). Peptides were classified using InterProScan (domain/motifs) and gene ontology (GO; Conesa et al., 2005), and mapped to respective pathways in C. elegans using the KEGG Orthology-Based Annotation System (KOBAS; Wu et al., 2006). The open reading frames (ORFs) inferred from ESTs with orthologues in C. elegans were also subjected to “secretome analysis” using the program SignalP v.2.0 (available at www.cbs.dtu.dk/services/SignalP/), employing both the neural network and hidden Markov models to predict signal peptides and/or anchors (Nielsen et al., 1997; Nielsen and Krogh, 1998; Bendtsen et al., 2004). Also, transmembrane domains were predicted using the program TMHMM (www.cbs.dtu.dk/services/TMHMM/; Sonnhammer et al., 1998; Krogh et al., 2001; Moller et al., 2001). Sequence comparisons of peptides predicted from the ESTs of T. colubriformis with those available for C. elegans (WormPep v. 202), other parasitic nematodes and organisms other than nematodes in current databases (i.e. www.wormbase.org and www.ncbi.nlm.nih.gov), was performed using SimiTri (Parkinson and Blaxter, 2003), which provides a two-dimensional display of similarity relationships.
The method developed by Zhong and Sternberg (2006) was used to predict the interaction networks among C. elegans orthologues of T. colubriformis contigs. Genomic data (regarding interactions, phenotypes, expression and GO) from C. elegans gene orthologues/homologues, also incorporating data from Drosophila melanogaster (vinegar fly), Saccharomyces cerevisiae (yeast), Mus musculus (mouse) and Homo sapiens (human), were integrated using a naïve Bayesian model to predict genetic interactions among C. elegans genes using the recommended, stringent cut-off value of 4.6 (Zhong and Sternberg, 2006; Campbell et al., 2008). The predicted networks resulting from the analyses were saved in a graphic display file (gdf) format and examined using the graph exploration system available at http://graphexploration.cond.org/.
3. Results
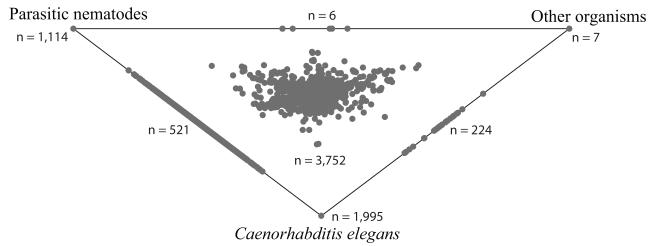
A total of 2,674,406 ESTs (average of 328.4 bp ± 276.9 bases in length) were generated (NCBI Sequence Read Archive [SRA] accession ID SRP002574). After filtering of the sequences <100 nucleotides in length, the CAP3 assembly yielded 21,259 contigs (average length: 495 bp ± 224.9; Supplementary Fig. 1; sequences available from http://www.nematode.net/ or http://research.vet.unimelb.edu.au/gasserlab/index.html). A total of 2,692 contigs (13%) matched known nucleotide sequences available in current databases, and 7,876 (37%) had known C. elegans homologues. The results of the conceptual translation of nucleotide into amino acid sequences, signal peptide and transmembrane domains predictions and InterProScan, GO and KOBAS (pathway mapping) analyses are listed in Table 1. The findings from homology searches of the proteins predicted for T. colubriformis with those available for C. elegans, other parasitic nematodes and organisms other than nematodes are displayed in Fig. 1. In total, 15,475 proteins were inferred from all 21,259 contigs, of which 6,492 and 5,393 matched known C. elegans and other parasitic nematode homologues, respectively (Fig. 1); 15,291 predicted peptides of T. colubriformis mapped to known proteins with 2,417 different domains (Table 1; Supplementary Table 1). ‘Transthyretin-like’ (IPR001534; 8.8%), ‘RNA recognition motif, RNP-1’ (IPR000504; 8.4%) and ‘metridin-like ShK toxin’ (IPR003582; 7.6%) were the domains most commonly detected (Table 2). The GO annotation revealed that 5,860 predicted peptides of T. colubriformis could be assigned to 320 ‘biological process’, 3,892 to 124 ‘cellular component’ and 10,941 to 446 ‘molecular function’ terms (Table 3; Supplementary Table 2). The most represented GO terms were ‘translation’ (GO:0006412; 11.5%) and ‘metabolic process’ (GO:0008152; 8.4%) for ‘biological process’, ‘intracellular’ (GO:0005622; 25.7%) and ‘ribosome’ (GO:0005840; 14.6%) for ‘cellular component’ and ‘ATP binding’ (GO:0005524; 6.5%) and ‘structural constituent of ribosome’ (GO:0003735; 5.6%) for ‘molecular function’ (Table 3; Supplementary Table 2). Pathway mapping using KOBAS predicted 5,150 peptides of T. colubriformis to be involved in 235 distinct pathways, of which most were represented by ‘peptidases’ (n = 211; 4%), ‘ribosome’ (n = 184; 3.6%), and ‘oxidative phosphorylation’ (n = 153; 3%) (Supplementary Table 3). The 211 predicted peptides of T. colubriformis which were assigned to biological pathways linked to ‘peptidases’ (Supplementary Table 3) had significant homology (at the amino acid level; e-value cut-off: < 1e-05) to a total of 38 unique C. elegans peptidases (listed in Table 4).
Table 1.
Number of ESTs determined from the cDNA library of the adult stage of Trichostrongylus colubriformis prior to and after assembly, and the results of bioinformatic analyses.
| No. of EST clusters | 21,259 |
| Average length (± range) | 495 bp ± 224.9 |
| Containing an Open Reading Frame (no. of full-length sequences) | 15,475 (12,472) |
| Signal peptides | 274 |
| Containing transmembrane domains | 588 |
| Returning InterProScan results | 15,475 (2,417 domains) |
| Gene Ontology | |
| Biological process | 5,860 (320 terms) |
| Cellular component | 3,892 (124 terms) |
| Molecular function | 10,941 (446 terms) |
| Prediction of biological pathways (KOBAS) | 235 |
| Homology searches | |
| With known orthologues | 2,692 (13%) |
| With known homologues | 7,876 (37%) |
Fig. 1.
Relationships of individual contigs of expressed sequence tags (EST) from the adult stage of Trichostrongylus colubriformis with protein homologues from Caenorhabditis elegans, other parasitic nematodes and organisms other than nematodes, displayed in a SimiTri plot (Parkinson and Blaxter, 2003).
Table 2.
The thirty most represented protein domains inferred from peptides, conceptually translated from individual contigs of expressed sequence tags (ESTs) for the adult stage of Trichostrongylus colubriformis.
| InterPro code | InterPro description | No. of ESTs (%)a |
|---|---|---|
| IPR001534 | Transthyretin-like | 212 (8.8) |
| IPR000504 | RNA recognition motif, RNP-1 | 202 (8.3) |
| IPR003582 | Metridin-like ShK toxin | 187 (7.6) |
| IPR001680 | WD40 repeat | 166 (6.9) |
| IPR016040 | NAD(P)-binding domain | 141 (5.8) |
| IPR001304 | C-type lectin | 136 (5.6) |
| IPR002068 | Heat shock protein Hsp20 | 132 (5.5) |
| IPR002223 | Proteinase inhibitor I2, Kunitz metazoa | 131 (5.4) |
| IPR002110 | Ankyrin | 123 (5.1) |
| IPR019781 | WD40 repeat, subgroup | 115 (4.7) |
| IPR007087 | Zinc finger, C2H2-type | 110 (4.6) |
| IPR019782 | WD40 repeat 2 | 104 (4.3) |
| IPR018249 | EF-HAND 2 | 95 (4) |
| IPR013032 | EGF-like region, conserved site | 92 (3.8) |
| IPR012677 | Nucleotide-binding, alpha-beta plait | 84 (3.5) |
| IPR011009 | Protein kinase-like | 84 (3.5) |
| IPR014044 | SCP-like extracellular | 79 (3.3) |
| IPR002130 | Peptidyl-prolyl cis-trans isomerase, cyclophilin-type | 77 (3.2) |
| IPR002423 | Chaperonin Cpn60/TCP-1 | 75 (3.1) |
| IPR001395 | Aldo/keto reductase | 69 (2.9) |
| IPR011046 | WD40 repeat-like | 68 (2.8) |
| IPR008978 | HSP20-like chaperone | 66 (2.7) |
| IPR001650 | DNA/RNA helicase, C-terminal | 65 (2.7) |
| IPR016187 | C-type lectin fold | 63 (2.6) |
| IPR001128 | Cytochrome P450 | 60 (2.5) |
| IPR009072 | Histone-fold | 60 (2.5) |
| IPR000535 | Major sperm protein | 60 (2.5) |
| IPR000242 | Protein-tyrosine phosphatase, receptor/non-receptor type | 59 (2.4) |
| IPR017986 | WD40 repeat, region | 59 (2.4) |
| IPR000557 | Calponin repeat | 58 (2.4) |
Percentage is calculated on the total number of InterPro domains that could be mapped in the present dataset.
Table 3.
Functions predicted for proteins encoded in the transcriptome of the adult stage of Trichostrongylus colubriformis based on GO. The parental (= level 2) GO categories were assigned according to InterPro domains with homology to functionally annotated molecules.
| GO category | GO code | GO description | No. of ESTs (%) |
|---|---|---|---|
| Biological process | GO:0008152 | Metabolic process | 2,034 (13.3) |
| GO:0009987 | Cellular process | 1,846 (12.1) | |
| GO:0051179 | Localization | 431 (2.9) | |
| GO:0065007 | Biological regulation | 282 (1.8) | |
| GO:0050789 | Regulation of biological process | 270 (1.8) | |
| GO:0016043 | Cellular component organization | 150 (1) | |
| GO:0044085 | Cellular component biogenesis | 109 (0.7) | |
| GO:0010926 | Anatomical structure formation | 87 (0.6) | |
| GO:0050896 | Response to stimulus | 45 (0.3) | |
| GO:0022610 | Biological adhesion | 13 (0.1) | |
| GO:0032501 | Multicellular organismal process | 7 (0.05) | |
| GO:0032502 | Developmental process | 5 (0.03) | |
| GO:0002376 | Immune system process | 1 (0.01) | |
| GO:0016265 | Death | 1 (0.01) | |
| Cellular component | GO:0005623 | Cell | 1,713 (11.2) |
| GO:0044464 | Cell part | 1,713 (11.2) | |
| GO:0043226 | Organelle | 761 (5) | |
| GO:0032991 | Macromolecular complex | 589 (3.9) | |
| GO:0005576 | Extracellular region | 58 (0.4) | |
| GO:0031975 | Envelope | 48 (0.3) | |
| GO:0031974 | Membrane-enclosed lumen | 40 (0.3) | |
| GO:0044456 | Synapse part | 10 (0.01) | |
| GO:0045202 | Synapse | 10 (0.01) | |
| GO:0044421 | Extracellular region part | 9 (0.01) | |
| Molecular function | GO:0005488 | Binding | 2,358 (15.4) |
| GO:0003824 | Catalytic activity | 2,032 (13.3) | |
| GO:0005198 | Structural molecule activity | 284 (1.9) | |
| GO:0005215 | Transporter activity | 202 (1.3) | |
| GO:0009055 | Electron carrier activity | 75 (0.5) | |
| GO:0030234 | Enzyme regulator activity | 67 (0.4) | |
| GO:0030528 | Transcription regulator activity | 67 (0.4) | |
| GO:0045182 | Translator regulator activity | 35 (0.2) | |
| GO:0060089 | Molecular transducer activity | 33 (0.2) | |
| GO:0016209 | Antioxidant activity | 15 (0.1) | |
| GO:0010860 | Proteasome regulator activity | 2 (0.01) | |
| GO:0016530 | Metallochaperone activity | 1 (0.01) |
Table 4.
Caenorhabditis elegans peptidase homologues of Trichostrongylus colubriformis predicted peptides.
|
C. elegans homologue (gene code) |
Description | No. of T. colubriformis homologues |
|---|---|---|
| NAS-31 (F58B4.1) | Meprin A metalloprotease | 43 |
| NEP-1 (ZK20.6) | Thermolysin-like zinc metallopeptidases | 28 |
| PAS-5 (F25H2.9) | Subunit of the core 20S proteasome subcomplex | 20 |
| CPR-6 (C25B8.3) | Cysteine proteinase Cathepsin L | 18 |
| CLP-1 (C06G4.2) | Ca2+-dependent cysteine protease (calpain) | 17 |
| PAS-3 (Y110A7A.14) | Subunit of the core 20S proteasome subcomplex | 15 |
| CPZ-1 (F32B5.8) | Cysteine proteinase Cathepsin L | 15 |
| NAS-5 (T23H4.3) | Meprin A metalloprotease | 6 |
| TRY-1 (ZK546.15) | Trypsin | 5a |
| F44E7.4 | Zn2+-dependent endopeptidases | 5 |
| PBS-3 (Y38A8.2) | Subunit of the core 20S proteasome subcomplex | 4 |
| PBS-2 (C47B2.4) | Subunit of the core 20S proteasome subcomplex | 3 |
| PBS-7 (F39H11.5) | Subunit of the core 20S proteasome subcomplex | 3 |
| UBH-1 (F46E10.8) | Ubiquitin C-terminal hydrolase | 3 |
| IMP-1 (C36B1.12) | Uncharacterized conserved protein, contains PA domain |
2 |
| PES-9 (R11H6.1) | Metalloexopeptidases | 1 |
| PAM-1 (F49E8.3) | Puromycin-sensitive aminopeptidase | 1 |
| PAS-7 (ZK945.2) | Subunit of the core 20S proteasome subcomplex | 1 |
| PBS-4 (T20F5.2) | Subunit of the core 20S proteasome subcomplex | 1 |
| APP-1 (W03G9.4) | Xaa-Pro aminopeptidase | 1 |
| SPG-7 (Y47G6A.10) | Metalloprotease | 1 |
| CLP-2 (T04A8.16) | Ca2+-dependent cysteine protease | 1 |
| BLI-4 (K04F10.4) | Subtilisin-like proprotein convertase | 1 |
| CPR-1 (C52E4.1) | Cysteine proteinase Cathepsin L | 1 |
| TRY-4 (F31D4.6) | Trypsin | 1 |
| EGL-21 (F01D4.4) | Zinc carboxypeptidase | 1 |
| ULP-3 (Y48A5A.2) | Sentrin-specific cysteine protease | 1 |
| ADM-4 (ZK154.7) | Tumor necrosis factor-alpha-converting enzyme | 1 |
| MATH-33 (H19N07.2) | Ubiquitin carboxyl-terminal hydrolase | 1 |
| DPF-1 (T23F1.7) | Dipeptidyl aminopeptidase | 1 |
| DPF-3 (K02F2.1) | Dipeptidyl aminopeptidase | 1 |
| MAP-2 (Y116A8A.9) | Metallopeptidase | 1 |
| CLPP-1 (ZK970.2) | ATP-dependent Clp protease | 1 |
| CLP-1 (C06G4.2) | Ca2+-dependent cysteine protease | 1 |
| DPF-5 (R11E3.8) | Dipeptidyl aminopeptidase | 1 |
| CPL-1 (T03E6.7) | Cysteine proteinase Cathepsin L | 1 |
| NAS-9 (C37H5.9) | Meprin A metalloprotease | 1 |
| SEL-12 (F35H12.3) | Presenilin | 1a |
| IMP-2 (T05E11.5) | Uncharacterized conserved protein, contains PA domain |
1a |
Protein/s with predicted signal peptides
Probabilistic functional gene networking predicted 2,126 orthologues in C. elegans to interact directly with 2,847 other genes (range: 1-271; cf. Supplementary Fig. 2). In particular, a subset of 42 of these orthologues were each predicted to interact directly with ≥ 100 other genes (Table 5). The majority of these orthologues had embryonic (n = 29; 69%), larval (n = 18; 43%) and/or adult (n = 8; 19%) lethal RNAi phenotypes in C. elegans. Genes encoding serine/threonine protein kinases had the highest representation (n = 6; 14.3%), followed by GTPases (n = 5; 11.9%), serine/threonine protein phosphatases (n =3; 7.1%), hedgehog proteins (n = 3; 7.1%), transcription and translation factors (n = 2; 6.5%) and other proteins (n = 1; ~2.4% for each) (Table 5).
Table 5.
Description and gene ontology (GO) classifications (according to the categories ‘biological process’, ‘cellular component’ and ‘molecular function’) of the Caenorhabditis elegans orthologues of Trichostrongylus colubriformis molecules predicted to interact with ≥100 other genes.
| Gene name (code) |
Description | No. predicted interacting genes |
Gene Ontology | RNAi phenotypesa | ||
|---|---|---|---|---|---|---|
| Biological process | Cellular component |
Molecular function | ||||
| glp-1 (F02A9.6) | Member of the LIN- 12/Notch family of receptors |
271 | Emb, Ste, Lvl | |||
| lin-12 (R107.8) | Member of the LIN- 12/Notch family of receptors |
260 | Cell differentiation | Integral to membrane |
Calcium-ion binding | Slo, Ste, Unc |
| cdc-42 (R07G3.1) | RHO GTPase | 255 | Small GTPase mediated signal transduction |
Intracellular | GTP-binding | Emb, Lvl, Ste |
| let-60 (ZK792.6) | Member of the GTP- binding RAS protooncogene family |
236 | Small GTPase mediated signal transduction |
Intracellular | GTP-binding | Let, Emb, Lvl, Lva, Ste |
| let-23 (ZK1067.1) | Transmembrane tyrosine kinase |
203 | Protein amino acid phosphorylation |
Membrane | ATP-binding | Unclassified |
| ced-10 (C09G12.8) | GTPase | 194 | Small GTPase mediated signal transduction |
Intracellular | GTP-binding | Let, Emb, Lvl, Lva |
| crb-1 (F11C7.4) | Homolog of Drosophila CRUMBS |
190 | Calcium-ion binding | Slo | ||
| ima-3 (F32E10.4) | Importin alpha nuclear transport factor |
189 | Intracellular protein transport |
Nucleus | Protein transporting activity |
Emb, Lva, Ste |
| gei-4 (W07B3.2) | Glutamine/asparagine- rich protein |
159 | Let, Emb, Lvl, Lva | |||
| dbl-1 (T25F10.2) | Member of the transforming growth factor beta (TGFbeta) superfamily |
148 | Growth factor activity | Slo | ||
| hmp-2 (K05C4.6) | Beta-catenin | 143 | Binding | Emb, Slo | ||
| act-4 (M03F4.2) | Actin | 134 | Emb, Lvl, Lva, Ste | |||
| gsp-2 (F56C9.1) | Serine/threonine protein phosphatase |
134 | Hydrolase activity | Let, Emb, Sck | ||
| mig-2 (C35C5.4) | Member of the Rho family of GTP-binding proteins |
132 | Small GTPase mediated signal transduction |
Intracellular | GTP-binding | Unclassified, embryonic defects |
| ras-1 (C44C11.1) | Ras-related GTPase | 128 | Small GTPase mediated signal transduction |
Intracellular | GTP-binding | |
| wrt-6 (ZK377.1) | Hedgehog-like protein | 125 | Cell communication | Peptidase activity | Unc, small | |
| kin-19 (C03C10.1) | Casein kinase (serine/threonine/tyrosin e protein kinase) |
123 | Protein amino acid phosphorylation |
ATP binding | Emb, Ste, Slo | |
| cdk-1 (T05G5.3) | Protein kinase | 122 | Protein amino acid phosphorylation |
ATP binding | Emb, Ste | |
| fib-1 (T01C3.7) | Fibrillarin | 122 | RNA processing | Nucleus | Emb, Lva, Ste | |
| cye-1 (C37A2.4) | G1/S-specific cyclin E | 121 | Emb, Lvl, Lva, Ste, Slo |
|||
| psa-4 (F01G4.1) | DNA/RNA helicase | 121 | Regulation of transcription |
Nucleus | ATP binding | Emb, Sck, Ste |
| rho-1 (Y51H4A.3) | Ras-related small GTPase |
120 | Protein transport | GTP binding | Emb, Lvl, Ste, Unc | |
| grd-11 (K02E2.2) | Hedgehog-like protein | 119 | Proteolysis | Peptidase activity | ||
| grd-2 (F46B3.5) | Hedgehog-like protein | 119 | Proteolysis | Peptidase activity | Small, Unc | |
| let-92 (F38H4.9) | Serine/threonine protein phosphatase |
118 | Hydrolase activity | Emb, Lvl, Ste, Sck | ||
| kin-3 (B0205.7) | Casein kinase II | 116 | Protein amino acid phosphorylation |
ATP binding | Emb, Ste, Slo | |
| goa-1 (C26C6.2) | G-protein alpha subunit | 116 | G-protein coupled receptor protein signaling pathway |
GTP binding | Emb, Ste | |
| act-2 (T04C12.5) | Actin | 113 | Protein binding | Emb, Lvl, Lva, Ste | ||
|
psa-1 (Y113G7B.23) |
Chromatin remodeling factor |
112 | DNA binding | Let, Emb, Lvl, Lva, Ste |
||
| F33H2.5 | DNA polymerase | 112 | Nucleotide and nucleic acid metabolic process |
Nucleic acid binding | Emb, Ste, Unc | |
|
hmg-1.2 (F47D12.4) |
HMG box-containing protein |
110 | Nucleus | DNA binding | Emb, Lvl, Lva, Ste | |
| par-5 (M117.2) | Multifunctional chaperone |
110 | Protein domain specific binding |
Emb, Lvl, Slo | ||
| rac-2 (K03D3.10) | Ras-related small GTPase |
109 | Small GTPase mediated signal transduction |
Intracellular | GTP-binding | Unclassified |
| nmy-2 (F20G4.3) | Myosin class II heavy chain |
106 | Myosin complex |
ATP binding | Emb, Lvl, Lva, Ste, Slo |
|
| pal-1 (C38D4.6) | Transcription factor | 104 | Nucleus | Transcription factor activity |
Emb, Slo | |
| ife-3 (B0348.6) | Translation initiation factor |
104 | Emb | |||
| ftt-2 (F52D10.3) | Multifunctional chaperone |
103 | Protein domain specific binding |
Let, Emb, Lvl, Lva, Ste, Slo |
||
| air-2 (B0207.4) | Serine/threonine protein kinase |
102 | Protein amino acid phosphorylation |
ATP binding | Emb, Ste | |
| his-47 (B0035.7) | Histone 2A | 101 | Nucleosome assembly | Nucleosome | DNA binding | Let, Emb, Ste |
| plk-1 (C14B9.4) | Serine/threonine protein kinase |
101 | Protein amino acid phosphorylation |
ATP binding | Let, Emb, Lvl, Ste | |
| his-46 (B0035.9) | Histone H4 | 100 | Nucleosome assembly | Nucleosome | DNA binding | Let, Emb, Lvl, Ste |
| air-1 (K07C11.2) | Serine/threonine protein kinase |
100 | Protein amino acid phosphorylation |
ATP binding | Emb, Ste | |
| gsp-1 (F29F11.6) | Serine/threonine protein phosphatase |
100 | Hydrolase activity | Emb, Lvl, Ste, Sck | ||
Abbreviations of RNAi phenotypes (alphabetical): adult lethal (Let), embryonic lethal (Emb), larval arrest (Lva), larval lethal (Lvl), sick (Sck), slow growth (Slo), sterile (Ste), uncoordinated (Unc).
4. Discussion
The present study has provided the first, detailed analysis of the transcriptome of adult T. colubriformis and identified some groups of molecules predicted to play pivotal roles in essential biological processes in this parasite. The percentage (~40%) of T. colubriformis sequences with orthologues/homologues in public databases was similar to that reported in similar transcriptomic studies of other animal parasitic helminths (Cantacessi et al., 2010a,b; Young et al., 2010), and is likely to reflect the paucity of sequence data available for this group. In addition, of the ~22,000 contigs assembled here, ~25% did not have predicted ORFs. The likely explanation for this result is technical and would appear to relate to a 3′-bias in sequences derived from the normalized cDNA library for T. colubriformis. Future investigations should compare the data from non-normalized libraries with those from the present study. Among the T. colubriformis proteins encoded in the transcriptome, those with ‘transthyretin-like’, ‘RNA recognition motif, RNP-1′ and ‘metridin-like ShK toxin’ motifs were the most commonly identified. The ‘transthyretin-like’ proteins (TTLs; Jacob et al., 2007) represent one of the largest protein families encoded by genes specific to nematodes (Parkinson et al., 2004). The function of the TTLs differs from that of the ‘transthyretin’ and the ‘transthyretin-related’ proteins, which are known to be carriers of lipophilic substances or hormones (Jacob et al., 2007). Members of the TTL family have been detected in nematodes, including Xiphinema index, Meloidogyne incognita and Radophilis similis of plants (McCarter et al., 2004; Parkinson et al., 2004; Furlanetto et al., 2005), and Brugia malayi of humans (Hewitson et al., 2008), and Ostertagia ostertagi (related to T. colubriformis) of ruminants (Vercauteren et al., 2003; Saverwyns et a., 2008). In nematodes, a role of TTL proteins in the nervous system has been hypothesized, supported also by the observation that ttl genes are expressed in the ventral nerve of R. similis (i.e. Rs-ttl-2), in the tail and head neurons as well as the hypodermis of C. elegans (gene code R13A5.6; www.wormbase.org), and based on the sequence similarity between TTL proteins and the C. elegans ‘neuropeptide-like proteins’ (NLP) (Jacob et al., 2007).
Interestingly, 12 proteins predicted in the transcriptome of T. colubriformis showed high sequence similarity to NLPs of C. elegans, including NLP-12 (gene code M01D7.5) (see www.wormbase.org). Based on the observation that nlp-12 mRNA has been localized to a single neurone in the posterior end of T. colubriformis (McVeigh et al., 2006), it was speculated that nlp-12 is expressed in a cell of the pre-anal ganglion, which includes both motor- and inter-neurones (McVeigh et al., 2006). Transcription of nlp-12 was also detected in L3s, and adult males and females of T. colubriformis, suggesting that the expression of NLP-12 might not be developmentally regulated and supporting the hypothesis for a key role of this protein in the nervous system of nematodes (McVeigh et al., 2006). Neuropeptides have been the focus of a number of studies of parasitic nematodes (see, for instance, Johnston et al., 2009; Mühlfeld et al., 2009), particularly because the neuromuscular system represents the target of several anthelmintic compounds, such as piperazine, levamisole and macrocyclic lactones (Holden-Dye and Walker, 2007). In the potato cyst nematode, Globodera pallida, the silencing by RNAi of five characterized genes encoding FMRFamide-like peptides (FLPs, a family of neuropeptides similar to the NLPs; cf. Johnston et al., 2009) resulted in impaired locomotory behaviour of the infective juvenile stage, which led to the hypothesis that RNAi-mediated flp gene silencing might represent a novel approach for the control of plant parasites (Kimber et al., 2007). Some success with RNAi-based silencing of selected genes in T. colubriformis L3s (Issa et al., 2005) and some nlp genes (e.g. nlp-10, nlp-11, nlp-12) expressed in neural tissues of C. elegans, which resulted in non-wildtype phenotypes such as defects in embryonic development and aldicarb resistance (www.wormbase.org), suggests that there is scope for investigating the functions of nlp genes in the neuromuscular system in this trichostrongylid.
The ‘metridin-like ShK toxin’ domain was the second most represented protein motif amongst the peptides inferred from the transcriptome of T. colubriformis (see Table 2). This domain, which is named after ‘metridin’, a toxin from the brown sea anemone, Metridium senile, and ‘ShK’, a structurally defined polypeptide from the sea anemone, Stoichactis helianthus (Kalman et al., 1998), is found in one or more copies as a C-terminal domain in the metallo-peptidases of C. elegans (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR003582#PUB00023590; Suzuki et al., 2004). Indeed, the vast majority of peptides of T. colubriformis were predicted to be involved in protein catabolism (see Table 4). Previously, serine- and metallo-proteases have been identified in the excretory/secretory products (ES) from L4s and adults of T. vitrinus (MacLennan et al., 1997, 2005), and shown to be active at various pHs (MacLennan et al., 1997). It was suggested that these proteases might facilitate the survival of the parasite in the host by mediating, for example, tissue penetration, feeding and/or immuno-evasion by (i) digesting antibodies (Hotez and Prichard, 1995); (ii) cleaving cell-surface receptors for cytokines (Björnberg et al., 1995) and/or (iii) causing the direct lysis of immune cells (Robinson et al., 1990). Proteases expressed on the epithelial surface of the gut of nematodes have been the focus of a number of studies, aimed at exploring their potential as vaccine candidates, particularly in blood-feeding nematodes (Knox et al., 2003; Williamson et al., 2003; Loukas et al., 2005a; Bethony et al., 2006; Pearson et al., 2009; Ranjit et al., 2009). In one of these studies (Loukas et al., 2005a), antibodies raised against the gut proteases of hookworms were demonstrated to bind to the nematode intestine during the blood-meal and shown to neutralize the proteolytic activity of these enzymes in vitro. Although attempts have been made to neutralize gut proteases of strongylid nematodes that are not obligate blood-feeders, the results have been not conclusive (Geldhof et al., 2002; De Maere et al., 2005); this outcome suggests that these nematodes do not ingest sufficient (IgG) antibodies for the vaccine to be effective (cf. Knox et al., 2003). It has been proposed that secreted proteases might be attractive targets for the development of vaccines against strongylid nematodes that are not obligate blood-feeders (Loukas et al., 2005a). Interestingly, the predicted peptidases (n = 211; see Table 4 and Supplementary Table 3) identified in the transcriptome of T. colubriformis did not have a signal peptide indicative of excretion/secretion, with the exception of the C. elegans homologues TRY-1, SEL-12 and IMP-2 (see Table 4). A possible explanation for the absence of such a signal is that these proteases could be excreted/secreted using a “non-classical” pathway that does not involve signal peptide cleavage (Nickel, 2003). Alternatively, these proteases could be released bound to secreted proteinase inhibitors (MacLennan et al., 2005). Indeed, the ‘proteinase inhibitor I2, Kunitz metazoa’ was amongst the ten most represented protein motifs identified for T. colubriformis (Table 2). Some protease inhibitors have been studied in T. colubriformis. For instance, an aspartyl protease inhibitor (Tco-api-1) has been identified as a major allergen by immunochemical analysis of somatic antigens of T. colubriformis L3 using IgE purified from the serum of sheep grazed on worm-contaminated pastures (Shaw et al., 2003), and is proposed to play a key role in the establishment of parasites in the vertebrate host by inhibiting pepsin activity during the transit of L3s through the gastric environment. In adult worms, proteinase inhibitors may also be implicated in the evasion of the host immune response through the cleavage of leucocyte elastases, mast cell proteases and cathepsins released from stimulated polymorphonuclear neutrophils (Björnberg et al., 1995). Given that a significant mucosal (IgA and IgE) immune response can be induced in sheep vaccinated with T. colubriformis native or recombinant antigens delivered across the epithelium of the jejunal or rectal lymphoid tissue (McClure, 2009), further investigations of the potential of secreted protease inhibitors as vaccine targets are warranted.
Other secreted proteins, such as those containing a ‘sperm-coating protein (SCP)-like extracellular domain’ (InterPro: IPR014044), also called SCP/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS; Pfam accession number no. PF00188), were amongst the top 20 most represented molecules in the transcriptome of T. colubriformis (see Table 2 and Supplementary Table 1). This is the first record of SCP/TAPS proteins in this parasitic nematode. SCP/TAPS belong to a large group of proteins that include the Ancylostoma-secreted proteins (ASPs; Cantacessi et al., 2009b). In parasitic nematodes, ASPs were first characterized for hookworms (Hawdon et al., 1996, 1999) and subsequently from related strongylid nematodes (Visser et al., 2008). As ASPs are abundant in the excretory/secretory (ES) products of the infective L3, they are thought to play an important role in the transition from the free-living to the parasitic stage of a nematode during its invasion of the host (Hawdon et al., 1996, 1999; Moser et al., 2005; Datu et al., 2008). In adult hookworms, ASPs have been proposed to play an immunomodulatory function during the invasion of the host, the migration through tissues, attachment to the intestinal wall and blood-feeding (Loukas et al., 2005b). In trichostrongylid nematodes that do not feed on blood, homologues of the hookworm ASPs have been identified in the Ostertagia ostertagi (brown stomach worm). In this species, two molecules (designated Oo-ASP-1 and Oo-ASP-2), were characterized as major antigens in a protective ES-thiol fraction of partially purified ES products. These molecules were shown to be highly expressed in L4s and adult males of O. ostertagi, a finding that was consistent with their transcriptional profile (Geldhof et al., 2003). Similarly, in T. vitrinus and Oesophagostomum dentatum (the nodule worm of pigs), ESTs with high sequence similarity to different types of asps were demonstrated to be male-enriched (Nisbet and Gasser, 2004; Cottee et al., 2006). The function of the ASPs in trichostrongylid parasites is currently unknown (Cantacessi et al., 2009b). A clear understanding of the expression patterns of ASPs in larval stages and both genders of these nematodes would provide clues as to the roles of these molecules in the life cycle and interactions with the host.
A probabilistic functional gene network of protein-encoding C. elegans orthologues inferred from the transcriptome of T. colubriformis was constructed to link genes that are known to function together in C. elegans. Amongst the orthologues predicted to each interact with >100 other genes, serine/threonine protein kinase and phosphatase genes were the most abundantly represented group overall (n = 9; 21.4%) (Table 5). In nematodes these molecules are known or inferred to be involved in sperm production by the adult males, as suggested from previous studies of C. elegans (Reinke et al., 2000), T. vitrinus (Nisbet and Gasser, 2004), Haemonchus contortus (the ‘barber pole’ worm of small ruminants) (Campbell et al., 2008, 2010) and O. dentatum (Boag et al., 2003; Cottee et al., 2006). In the latter nematode, a catalytic subunit of a serine/threonine protein phosphatase (PP1) was characterized (Od-mpp1), and gene silencing by RNAi of the corresponding C. elegans homologue resulted in a significant reduction (30-40%) in the numbers of F2 progeny produced (Boag et al., 2003). In an independent study (Hanazawa et al., 2001), gene silencing of these genes by double-stranded RNAi in C. elegans hermaphrodites resulted in impaired sperm function. Indeed, protein kinases and phosphatases are known to be highly expressed in the sperm-producing germline tissue of C. elegans (Hanazawa et al., 2001), thus suggesting key roles in regulating sperm maturation, following the expulsion of the organelles involved in protein synthesis from maturing spermatids (Hanazawa et al., 2001). In T. vitrinus, a serine/threonine protein phosphatase (designated Tv-stp-1), was shown to be transcribed specifically in adult males, but not in adult females or any of the larval stages (Hu et al., 2007), whereas the transcript corresponding to the H. contortus homologue of Tv-stp-1 (i.e. Hc-stp-1) was detected in L4s and adult males of this species (Campbell et al., 2010). In addition, sequence conservation among Od-mpp1, Tv-stp-1, Hc-stp-1 and genes encoding serine/threonine protein phosphatases in C. elegans suggest a similar biological function (Hu et al., 2007; Campbell et al., 2010). Similarly, the T. colubriformis gene orthologues of serine/threonine phosphatases identified in the present study are likely to play key roles in the reproductive processes of this species, but still need detailed investigation.
The transcriptomic dataset described here constitutes a basis for future investigations of essential pathways of development and reproduction in T. colubriformis, a statement supported by functional gene networking inferring that 95% of C. elegans orthologues that interacted with >100 other genes were linked to lethality, growth defects or sterility based on gene silencing (see Table 5). Clearly, next-generation sequencing technologies, particularly 454-Roche (www.454.com), Illumina-Solexa (www.illumina.com) and Helicos (www.helicosbio.com), might assist future genomic and transcriptomic studies, aimed, for instance, at exploring the sequence variability of mRNA transcripts encoding surface antigens in different populations of infective L3s of T. colubriformis (Maass et al., 2009), thus assisting in the elucidation of an aspect of immune evasion in this nematode. Complemented by proteomic exploration, the characterisation of the transcriptomes of all developmental stages and both sexes of T. colubriformis from non-normalized cDNA libraries will allow a global study of differential gene expression. In addition, the development of methods for RNAi in the L3 of T. colubriformis (Issa et al., 2005) offers opportunities for investigating the function(s) of molecules (e.g. serine/threonine phosphatases) predicted to play crucial roles in parasitic nematodes. Future investigations should also focus on inferring the functions of orthologous molecules from trichostrongylid nematodes using genetic complementation in C. elegans (cf. Hu et al., 2010). Improved knowledge of fundamental molecular pathways in nematodes should provide a sound basis for the discovery and prevalidation of targets for drug design.
Supplementary Material
Supplementary Fig. 1. Length distribution of contigs following the assembly of the Trichostrongylus colubriformis nucleotide sequence data.
Supplementary Fig. 2. Graphic representation of the genetic interaction networks predicted for Caenorhabditis elegans orthologues (blue dots) of Trichostrongylus colubriformis genes (red dots).
Supplementary Table 1. List of InterPro domains in the predicted peptide sequences encoded in the adult stage of Trichostrongylus colubriformis.
Supplementary Table 2. List of gene ontology (GO) terms (according to the categories ‘biological process’, ‘cellular component’ and ‘molecular function’) linked to proteins inferred to be encoded by the adult stage of Trichostrongylus colubriformis.
Supplementary Table 3. Predicted biological pathways linked to molecules inferred to be encoded in the adult stage of Trichostrongylus colubriformis.
Acknowledgements
This research was supported by grants from the National Human Genome Research Institute (NHGRI) and National Institutes of Health (MM) and the Australian Research Council, the Australian Academy of Science and the Australian-American Fulbright Commission (RBG). CC is the grateful recipient of International Postgraduate Research Scholarship (IPRS) from the Australian Government and a fee-remission scholarship through the University of Melbourne as well as the Clunies Ross (2008) and Sue Newton (2009) awards from the School of Veterinary Science of the same university. The authors thank Gareth Hutchinson for providing T. colubriformis, Weiwei Zhong for advice on genetic interactions and the staff of WormBase for assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc. Natl. Acad. Sci. U S A. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethony JM, Loukas A, Hotez PJ, Knox DP. Vaccines against blood-feeding nematodes of humans and livestock. Parasitology. 2006;133:S63–S79. doi: 10.1017/S0031182006001818. [DOI] [PubMed] [Google Scholar]
- Björnberg F, Lantz M, Gullberg U. Metalloproteases and serineproteases are involved in the cleavage of the two tumour necrosis factor (TNF) receptors to soluble forms in the myeloid cell lines U-937 and THP-1. Scand. J. Immunol. 1995;42:418–424. doi: 10.1111/j.1365-3083.1995.tb03675.x. [DOI] [PubMed] [Google Scholar]
- Boag PR, Ren P, Newton SE, Gasser RB. Molecular characterisation of a male-specific serine/threonine phosphatase from Oesophagostomum dentatum (Nematoda: Strongylida), and functional analysis of homologues in Caenorhabditis elegans. Int. J. Parasitol. 2003;33:313–325. doi: 10.1016/s0020-7519(02)00263-1. [DOI] [PubMed] [Google Scholar]
- Campbell BE, Nagaraj SH, Hu M, Zhong W, Sternberg PW, Ong EK, Loukas A, Ranganathan S, Beveridge I, McInnes RL, Hutchinson GW, Gasser RB. Gender-enriched transcripts in Haemonchus contortus-- predicted functions and genetic interactions based on comparative analyses with Caenorhabditis elegans. Int. J. Parasitol. 2008;38:65–83. doi: 10.1016/j.ijpara.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Campbell BE, Rabelo EM, Hoffmann A, Hu M, Gasser RB. Characterization of a Caenorhabditis elegans Glc seven-like phosphatase (gsp) orthologue from Haemonchus contortus (Nematoda) Mol. Cell. Probes. 2010;24:178–189. doi: 10.1016/j.mcp.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Zou FC, Hall RS, Zhong W, Jex AR, Campbell BE, Ranganathan S, Sternberg PW, Zhu XQ, Gasser RB. Bioinformatic analysis of abundant, gender-enriched transcripts of adult Ascaris suum (Nematoda) using a semi-automated workflow platform. Mol. Cell. Probes. 2009a;23:205–217. doi: 10.1016/j.mcp.2009.03.003. 2009a. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Campbell BE, Visser A, Geldhof P, Nolan MJ, Nisbet AJ, Matthews JB, Loukas A, Hofmann A, Otranto D, Sternberg PW, Gasser RB. A portrait of the “SCP/TAPS” proteins of eukaryotes--developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 2009b;27:376–388. doi: 10.1016/j.biotechadv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Campbell BE, Young ND, Jex AR, Hall RS, Presidente PJA, Zawadzki JL, Zhong W, Aleman-Meza B, Loukas A, Sternberg PW, Gasser RB. Differences in transcription between free-living and CO2-activated third-stage larvae of Haemonchus contortus. BMC Genomics. 2010a;11:266. doi: 10.1186/1471-2164-11-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C, Mitreva M, Jex AR, Young ND, Campbell BE, Hall RS, Doyle MA, Ralph SA, Rabelo EM, Ranganathan S, Sternberg PW, Loukas A, Gasser RB. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl. Trop. Dis. 2010b;4:e684. doi: 10.1371/journal.pntd.0000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cottee PA, Nisbet AJ, Abs El-Osta YG, Webster TL, Gasser RB. Construction of gender-enriched cDNA archives for adult Oesophagostomum dentatum by suppressive-subtractive hybridization and a microarray analysis of expressed sequence tags. Parasitology. 2006;132:691–708. doi: 10.1017/S0031182005009728. [DOI] [PubMed] [Google Scholar]
- Datu BJ, Gasser RB, Nagaraj SH, Ong EK, O’Donoghue P, McInnes R, Ranganathan S, Loukas A. Transcriptional changes in the hookworm, Ancylostoma caninum, during the transition from a free-living to a parasitic larva. PLoS Negl. Trop. Dis. 2008;2:e130. doi: 10.1371/journal.pntd.0000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maere V, Vercauteren I, Gevaert K, Vercruysse J, Claerebout E. An aspartyl protease inhibitor of Ostertagia ostertagi: molecular cloning, analysis of stage and tissue specific expression and vaccine trial. Mol. Biochem. Parasitol. 2005;141:81–88. doi: 10.1016/j.molbiopara.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Droege M, Hill B. The Genome Sequencer FLX System--longer reads, more applications, straight forward bioinformatics and more complete data sets. J. Biotechnol. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Furlanetto C, Cardle L, Brown DJF, Jones JT. Analysis of expressed sequence tags from the ectoparasitic nematode Xiphinema index. Nematology. 2005;7:95–104. [Google Scholar]
- Geldhof P, Claerebout E, Knox D, Vercauteren I, Looszova A, Vercruysse J. Vaccination of calves against Ostertagia ostertagi with cysteine proteinase enriched protein fractions. Parasite Immunol. 2002;24:263–270. doi: 10.1046/j.1365-3024.2002.00461.x. [DOI] [PubMed] [Google Scholar]
- Geldhof P, Vercauteren I, Gevaert K, Staes A, Knox DP, Vandekerckhove J, Vercruysse J, Claerebout E. Activation-associated secreted proteins are the most abundant antigens in a host protective fraction from Ostertagia ostertagi. Mol. Biochem. Parasitol. 2003;128:111–114. doi: 10.1016/s0166-6851(03)00044-6. [DOI] [PubMed] [Google Scholar]
- Hanazawa M, Mochii M, Ueno N, Kohara Y, Iino Y. Use of cDNA subtraction and RNA interference screens in combination reveals genes required for germ-line development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 2001;98:8686–8691. doi: 10.1073/pnas.141004698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. Single molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- Hawdon JM, Jones BF, Hoffman DR, Hotez PJ. Cloning and characterization of Ancylostoma-secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. J. Biol. Chem. 1996;271:6672–6678. doi: 10.1074/jbc.271.12.6672. [DOI] [PubMed] [Google Scholar]
- Hawdon JM, Narasimhan S, Hotez PJ. Ancylostoma secreted protein 2: cloning and characterization of a second member of a family of nematode secreted proteins from Ancylostoma caninum. Mol. Biochem. Parasitol. 1999;99:149–165. doi: 10.1016/s0166-6851(99)00011-0. [DOI] [PubMed] [Google Scholar]
- Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, Ashton PD, Wilson A, Maizels RM. The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol. Biochem. Parasitol. 2008;160:8–21. doi: 10.1016/j.molbiopara.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Holden-Dye L, Walker RJ. Anthelmintic drugs. WormBook. 2007;2:1–13. doi: 10.1895/wormbook.1.143.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes PH. Pathogenesis of trichostrongylosis. Vet. Parasitol. 1985;18:89–101. doi: 10.1016/0304-4017(85)90059-7. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Prichard DI. Hookworm infection. Sci. Am. 1995;6:42–48. doi: 10.1038/scientificamerican0695-68. [DOI] [PubMed] [Google Scholar]
- Hu M, Abs EL-Osta YG, Campbell BE, Boag PR, Nisbet AJ, Beveridge I, Gasser RB. Trichostrongylus vitrinus (Nematoda: Strongylida): molecular characterization and transcriptional analysis of Tv-stp-1, a serine/threonine phosphatase gene. Exp. Parasitol. 2007;117:22–34. doi: 10.1016/j.exppara.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Hu M, Lok JB, Ranjit N, Massey HC, Jr, Sternberg PW, Gasser RB. Structural and functional characterisation of the fork head transcription factor-encoding gene, Hc-daf-16, from the parasitic nematode Haemonchus contortus (Strongylida) Int. J. Parasitol. 2010;40:405–415. doi: 10.1016/j.ijpara.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CQ, Gasser RB, Cantacessi C, Nisbet AJ, Zhong W, Sternberg PW, Loukas A, Mulvenna J, Lin RQ, Chen N, Zhu XQ. Genomic-bioinformatic analysis of transcripts enriched in the third-stage larva of the parasitic nematode Ascaris suum. PLoS Negl. Trop. Dis. 2008;2:e246. doi: 10.1371/journal.pntd.0000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa Z, Grant WN, Stasiuk S, Shoemaker CB. Development of methods for RNA interference in the sheep gastrointestinal parasite, Trichostrongylus colubriformis. Int. J. Parasitol. 2005;35:935–940. doi: 10.1016/j.ijpara.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Jacob J, Vanholme B, Haegeman A, Gheysen G. Four transthyretin-like genes of the migratory plant-parasitic nematode Radopholus similis: members of an extensive nematode-specific family. Gene. 2007;402:9–19. doi: 10.1016/j.gene.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Jacob J, Mitreva M, Vanholme B, Gheysen G. Exploring the transcriptome of the burrowing nematode Radopholus similis. Mol. Genet. Genomics. 2008;280:1–17. doi: 10.1007/s00438-008-0340-7. [DOI] [PubMed] [Google Scholar]
- Jex AR, Littlewood DT, Gasser RB. Toward next-generation sequencing of mitochondrial genomes--focus on parasitic worms of animals and biotechnological implications. Biotechnol. Adv. 2010;28:151–159. doi: 10.1016/j.biotechadv.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Johnston MJ, McVeigh P, McMaster S, Fleming CC, Maule AG. FMRFamide-like peptides in root knot nematodes and their potential role in nematode physiology. J. Helminthol. 2009;21:1–13. doi: 10.1017/S0022149X09990630. [DOI] [PubMed] [Google Scholar]
- Kalman K, Pennington MW, Lanigan MD, Nguyen A, Rauer H, Mahnir V, Paschetto K, Kem WR, Grissmer S, Gutman GA, Christian EP, Cahalan MD, Norton RS, Chandy KG. ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide. J. Biol. Chem. 1998;273:32697–32707. doi: 10.1074/jbc.273.49.32697. [DOI] [PubMed] [Google Scholar]
- Kaplan RM. Anthelmintic resistance in nematodes of horses. Vet. Res. 2002;33:491–507. doi: 10.1051/vetres:2002035. [DOI] [PubMed] [Google Scholar]
- Kaplan RM. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kimber MJ, McKinney S, McMaster S, Day TA, Fleming CC, Maule AG. flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. FASEB J. 2007;21:1233–1243. doi: 10.1096/fj.06-7343com. [DOI] [PubMed] [Google Scholar]
- Knox DP, Redmond DL, Newlands GF, Skuce PJ, Pettit D, Smith WD. The nature and prospects for gut membrane proteins as vaccine candidates for Haemonchus contortus and other ruminant trichostrongyloids. Int. J. Parasitol. 2003;33:1129–1137. doi: 10.1016/s0020-7519(03)00167-x. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Loukas A, Bethony JM, Mendez S, Fujiwara RT, Goud GN, Ranjit N, Zhan B, Jones K, Bottazzi ME, Hotez PJ. Vaccination with recombinant aspartic hemoglobinase reduces parasite load and blood loss after hookworm infection in dogs. PLoS Med. 2005a;2:10. doi: 10.1371/journal.pmed.0020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukas A, Constant SL, Bethony JM. Immunobiology of hookworm infection. FEMS Immunol. Med. Microbiol. 2005b;43:115–124. doi: 10.1016/j.femsim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Maass DR, Harrison GB, Grant WN, Hein WR, Shoemaker CB. Intraspecific epitopic variation in a carbohydrate antigen exposed on the surface of Trichostrongylus colubriformis infective L3 larvae. PLoS Pathog. 2009;5:e1000597. doi: 10.1371/journal.ppat.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAFF . Technical Bulletin 18. Her Majesty’s Stationary Office; London, UK: 1977. Manual of Veterinary Parasitological Laboratory Techniques. [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan K, Gallagher MP, Knox DP. Stage-specific serine and metallo-proteinase release by adult and larval Trichostrongylus vitrinus. Int. J. Parasitol. 1997;27:1031–1036. doi: 10.1016/s0020-7519(97)00074-x. [DOI] [PubMed] [Google Scholar]
- MacLennan K, McLean K, Knox DP. Serpin expression in the parasitic stages of Trichostrongylus vitrinus, an ovine intestinal nematode. Parasitology. 2005;130:349–357. doi: 10.1017/s003118200400616x. [DOI] [PubMed] [Google Scholar]
- McCarter JP, Mitreva MD, Martin J, Dante M, Wylie T, Rao U, Pape D, Bowers Y, Theising B, Murphy CV, Kloek AP, Chiapelli BJ, Clifton SW, Bird DM, Waterston RH. Analysis and functional classification of transcripts from the nematode Meloidogyne incognita. Genome Biol. 2003;4:R26. doi: 10.1186/gb-2003-4-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure SJ. Mucosal delivery of native and recombinant protein vaccines against Trichostrongylus colubriformis. Int. J. Parasitol. 2009;39:599–606. doi: 10.1016/j.ijpara.2008.09.010. [DOI] [PubMed] [Google Scholar]
- McVeigh P, Leech S, Marks NJ, Geary TG, Maule AG. Gene expression and pharmacology of nematode NLP-12 neuropeptides. Int. J. Parasitol. 2006;36:633–640. doi: 10.1016/j.ijpara.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Mardis ER. Large-scale sequencing and analytical processing of ESTs. Methods Mol. Biol. 2009;533:153–187. doi: 10.1007/978-1-60327-136-3_8. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Moser JM, Freitas T, Arasu P, Gibson G. Gene expression profiles associated with the transition to parasitism in Ancylostoma caninum larvae. Mol. Biochem. Parasitol. 2005;143:39–48. doi: 10.1016/j.molbiopara.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Mühlfeld S, Schmitt-Wrede HP, Harder A, Wunderlich F. FMRFamide-like neuropeptides as putative ligands of the latrophilin-like HC110-R from Haemonchus contortus. Mol. Biochem. Parasitol. 2009;164:162–164. doi: 10.1016/j.molbiopara.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Nagaraj SH, Deshpande N, Gasser RB, Ranganathan S. ESTExplorer: an expressed sequence tag (EST) assembly and annotation platform. Nucleic Acids Res. 2007a;35:W143–W147. doi: 10.1093/nar/gkm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj SH, Gasser RB, Ranganathan S. A hitchhiker’s guide to expressed sequence tag (EST) analysis. Brief. Bioinform. 2007b;8:6–21. doi: 10.1093/bib/bbl015. [DOI] [PubMed] [Google Scholar]
- Nagaraj SH, Gasser RB, Nisbet AJ, Ranganathan S. In silico analysis of expressed sequence tags from Trichostrongylus vitrinus (Nematoda): comparison of the automated ESTExplorer workflow platform with conventional database searches. BMC Bioinf. 2008;9:S10. doi: 10.1186/1471-2105-9-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot. Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6); Menlo Park, California: AAAI Press; 1998. pp. 122–30. [PubMed] [Google Scholar]
- Nickel W. The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur. J. Biochem. 2003;270:2109–2119. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Gasser RB. Profiling of gender-specific gene expression for Trichostrongylus vitrinus (Nematoda: Strongylida) by microarray analysis of expressed sequence tag libraries constructed by suppressive-subtractive hybridisation. Int. J. Parasitol. 2004;34:633–643. doi: 10.1016/j.ijpara.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Cottee PA, Gasser RB. Genomics of reproduction in nematodes: prospects for parasite intervention? Trends Parasitol. 2008;24:89–95. doi: 10.1016/j.pt.2007.12.001. [DOI] [PubMed] [Google Scholar]
- O’Connor LJ, Walkden-Brown SW, Kahn LP. Ecology of the free-living stages of major trichostrongylid parasites of sheep. Vet. Parasitol. 2006;142:1–15. doi: 10.1016/j.vetpar.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Olsen OW. The Quarterly Review of Biology. University of Chicago Press; 1986. Animal Parasites. Their Life Cycles and Ecology. [Google Scholar]
- Pandey V, Nutter RC, Prediger E. Applied Biosystems SOLiD™ System: Ligation-Based Sequencing. In: Jantz M, editor. Next Generation Genome Sequencing: Towards Personalized Medicine. Wiley; Milton, Australia: 2008. pp. 29–41. [Google Scholar]
- Parkinson J, Blaxter ML. SimiTri--visualizing similarity relationships for groups of sequences. Bioinformatics. 2003;19:390–395. doi: 10.1093/bioinformatics/btf870. [DOI] [PubMed] [Google Scholar]
- Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, Schmid R, Hall N, Barrell B, Waterston RH, McCarter JP, Blaxter ML. A transcriptomic analysis of the phylum Nematoda. Nat. Genet. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- Pearson MS, Bethony JM, Pickering DA, de Oliveira LM, Jariwala A, Santiago H, Miles AP, Zhan B, Jiang D, Ranjit N, Mulvenna J, Tribolet L, Plieskatt J, Smith T, Bottazzi ME, Jones K, Keegan B, Hotez PJ, Loukas A. An enzymatically inactivated hemoglobinase from Necator americanus induces neutralizing antibodies against multiple hookworm species and protects dogs against heterologous hookworm infection. FASEB J. 2009;23:3007–3019. doi: 10.1096/fj.09-131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabelo EM, Hall RS, Loukas A, Cooper L, Hu M, Ranganathan S, Gasser RB. Improved insights into the transcriptomes of the human hookworm Necator americanus--fundamental and biotechnological implications. Biotechnol. Adv. 2009;27:122–132. doi: 10.1016/j.biotechadv.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Ranganathan S, Menon R, Gasser RB. Advanced in silico analysis of expressed sequence tag (EST) data for parasitic nematodes of major socio-economic importance--fundamental insights toward biotechnological outcomes. Biotechnol. Adv. 2009;27:439–448. doi: 10.1016/j.biotechadv.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Ranjit N, Zhan B, Hamilton B, Stenzel D, Lowther J, Pearson M, Gorman J, Hotez P, Loukas A. Proteolytic degradation of hemoglobin in the intestine of the human hookworm Necator americanus. J. Infect. Dis. 2009;199:904–912. doi: 10.1086/597048. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in. C. elegans. Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Robinson BW, Venaille TJ, Mendis AH, McAleer R. Allergens as proteases: an Aspergillus fumigatus proteinase directly induces human epithelial cell detachment. J. Allergy Clin. Immunol. 1990;86:726–731. doi: 10.1016/s0091-6749(05)80176-9. [DOI] [PubMed] [Google Scholar]
- Sackett D, Holmes P. Assessing the economic cost of endemic disease on the profitability Australian beef cattle and sheep producers. Meat and Livestock Australia (MLA) Limited; Sydney, Australia: 2006. [Google Scholar]
- Sangster N. Pharmacology of anthelmintic resistance. Parasitology. 1996;113:S201–S216. doi: 10.1017/s0031182000077982. [DOI] [PubMed] [Google Scholar]
- Saverwyns H, Visser A, Van Durme J, Power D, Morgado I, Kennedy MW, Knox DP, Schymkowitz J, Rousseau F, Gevaert K, Vercruysse J, Claerebout E, Geldhof P. Analysis of the transthyretin-like (TTL) gene family in Ostertagia ostertagi - Comparison with other strongylid nematodes and Caenorhabditis elegans. Int. J. Parasitol. 2008;38:1545–1546. doi: 10.1016/j.ijpara.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, McNeill MM, Maass DR, Hein WR, Barber TK, Wheeler M, Morris CA, Shoemaker CB. Identification and characterisation of an aspartyl protease inhibitor homologue as a major allergen of Trichostrongylus colubriformis. Int. J. Parasitol. 2003;33:1233–1243. doi: 10.1016/s0020-7519(03)00157-7. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology; Menlo Park, CA. AAAI; 1998. pp. 175–182. [PubMed] [Google Scholar]
- Suzuki M, Sagoh N, Iwasaki H, Inoue H, Takahashi K. Metalloproteases with EGF, CUB, and thrombospondin-1 domains function in molting of Caenorhabditis elegans. Biol. Chem. 2004;385:565–568. doi: 10.1515/BC.2004.069. [DOI] [PubMed] [Google Scholar]
- Vercauteren I, Geldhof P, Peelaers I, Claerebout E, Berx G, Vercruysse J. Identification of excretory-secretory products of larval and adult Ostertagia ostertagi by immunoscreening of cDNA libraries. Mol. Biochem. Parasitol. 2003;126:201–208. doi: 10.1016/s0166-6851(02)00274-8. [DOI] [PubMed] [Google Scholar]
- Visser A, Van Zeveren AM, Meyvis Y, Peelaers I, Van den Broeck W, Gevaert K, Vercruysse J, Claerebout E, Geldhof P. Gender-enriched transcription of activation associated secreted proteins in Ostertagia ostertagi. Int. J. Parasitol. 2008;38:455–465. doi: 10.1016/j.ijpara.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Waller PJ. Resistance to anthelmintics and the implications for animal production. In: Anderson N, Waller PJ, editors. Resistance in Nematodes to Anthelmintic Drugs. CSIRO/ Australian Wool Corporation; Melbourne: 1985. pp. 1–11. [Google Scholar]
- Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 2003;19:417–423. doi: 10.1016/s1471-4922(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Hall RS, Jex AR, Cantacessi C, Gasser RB. Elucidating the transcriptome of Fasciola hepatica - a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol. Adv. 2010;28:222–231. doi: 10.1016/j.biotechadv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Length distribution of contigs following the assembly of the Trichostrongylus colubriformis nucleotide sequence data.
Supplementary Fig. 2. Graphic representation of the genetic interaction networks predicted for Caenorhabditis elegans orthologues (blue dots) of Trichostrongylus colubriformis genes (red dots).
Supplementary Table 1. List of InterPro domains in the predicted peptide sequences encoded in the adult stage of Trichostrongylus colubriformis.
Supplementary Table 2. List of gene ontology (GO) terms (according to the categories ‘biological process’, ‘cellular component’ and ‘molecular function’) linked to proteins inferred to be encoded by the adult stage of Trichostrongylus colubriformis.
Supplementary Table 3. Predicted biological pathways linked to molecules inferred to be encoded in the adult stage of Trichostrongylus colubriformis.