Abstract
Background
Vibriosis caused by V. anguillarum is a commonly encountered disease in Atlantic cod farms and several studies indicate that the initiation of infection occurs after the attachment of the pathogen to the mucosal surfaces (gut, skin and gills) of fish. Therefore it is necessary to investigate the role of different mucosal components in fish upon V. anguillarum infection. The present study has two parts; in the first part we analyzed the differential expression of skin mucus proteins from Atlantic cod naturally infected with V. anguillarum using two dimensional gel electrophoresis coupled with mass spectrometry. In the second part, a separate bath challenge experiment with V. anguillarum was conducted to assess the mRNA levels of the genes in skin tissue, corresponding to the selected proteins identified in the first part.
Results
Comparative proteome analysis of skin mucus of cod upon natural infection with V. anguillarum revealed key immune relevant proteins like calpain small subunit 1, glutathione-S-transferase omega 1, proteasome 26S subunit, 14-kDa apolipoprotein, beta 2-tubulin, cold inducible RNA binding protein, malate dehydrogenase 2 (mitochondrial) and type II keratin that exhibited significant differential expression. Additionally a number of protein spots which showed large variability amongst individual fish were also identified. Some of the proteins identified were mapped to the immunologically relevant JNK (c-Jun N-terminal kinases) signalling pathway that is connected to cellular events associated with pathogenesis. A bath challenge experiment with V. anguillarum showed differential expression of beta 2-tubulin, calpain small subunit 1, cold inducible RNA binding protein, flotillin1, and glutathione S-transferase omega 1 transcripts in the skin tissue of cod during early stages of infection.
Conclusions
Differentially expressed proteins identified in the cod skin mucus point towards their possible involvement in V. anguillarum pathogenesis. The role of some of these proteins in vibriosis in cod described in this paper can be considered unconventional with respect to their established functions in higher vertebrates. Based on the differential expression of these proteins they are possibly important components of fish defence against bacteria and innate immunity at large. The feasibility of utilizing these proteins/genes as markers of bacterial infection or stress in cod needs to be explored further.
Keywords: Comparative proteome, Skin mucosa, Atlantic cod, Vibrio anguillarum, Bath challenge
Background
Fish like Atlantic cod, of the gadoid lineage, are thought to rely extensively on innate immune components [1,2] to ward off infection. Skin mucus is considered to be one of the foremost barriers against pathogen invasion and several mucosal components like lectins, proteases, mucins etc. have been shown to be relevant in the survival of fish [3].
Vibrio anguillarum strains O2α and O2β are commonly associated with vibriosis in cod [4,5] and these infections can cause bleeding skin lesions/ulcers and septicemia, resulting in mass mortalities of farmed cod [6]. The ubiquitous distribution of V. anguillarum in various marine habitats, their opportunistic nature, and the sporadic recurrences of vibriosis caused by deviating serotypes make the complete eradication of this disease in fish farms infeasible [7,8].
Vibrio infection in mammals is initiated at gut mucosal surfaces [9]. This information spurred investigations on the role of mucosal surfaces in fish, especially in the gut during the progress of vibriosis. Early studies on carp, anally intubated with V. anguillarum bacterins, have shown antigen uptake in the second gut segment and subsequent detection of specific antibodies in the gut mucus, skin mucus and serum, indicating a common mucosal response to the antigen [10]. Other studies have shown that establishment of bacteria in the gut is facilitated by chemotaxis, induced by mucosal components such as amino acids, carbohydrate moieties on proteins like high molecular weight mucins, and bile components [11]. Skin is also considered as a potential infection route for V. anguillarum, besides gills and gut [7]. Bath challenge experiments have shown that the primary site of attachment and colonization of V. anguillarum is the skin and intestine of zebrafish (Danio rerio) [12], and skin and fins of juvenile rainbow trout [13]. V. anguillarum virulence is associated with extracellular components like proteases which can cause localized inflammation at the skin surface [6] and it has been shown that epithelial cells in rainbow trout skin can phagocytose V. anguillarum[14]. Further, mucosal antibody production has been quantified upon V. anguillarum infection [15]. These findings indicate that the pathogen invasion can trigger immune responses in skin and its associated mucosal surface.
Modern techniques like proteomics and transcriptomics can be made use of to study the proteins and genes in the skin mucosa of Atlantic cod afflicted with vibriosis. Such approaches would help us understand host inflammatory responses during disease progression and may pave way for the discovery of vibriosis-related biomarkers. Therefore in the present study we used comparative proteomics to identify differentially expressed proteins from the skin mucus of cod during a natural outbreak of vibriosis. Further, following a V. anguillarum challenge, we examined the transcriptional profiles of the genes, corresponding to the selected proteins, in the skin of cod.
Methods
V. anguillarum infected Atlantic cod juveniles
Atlantic cod juveniles hatched (eggs obtained from Cod Farmers ASA, Norway) at Mørkvedbukta Research Station, University of Nordland, Bodø, Norway in early spring 2011 had a natural outbreak of vibriosis during late summer 2011. These were non-vaccinated fish of size 35-50 g, maintained at 7°C, and fed commercial juvenile feed (Amber Neptun) from Skretting (Stavanger, Norway). Fish from 3 tanks that were diagnosed with vibriosis by Norwegian Veterinary Institute, North-Norway (Harstad; primary agency for aquaculture disease diagnosis in northern Norway) were used for the first study. The fish in these tanks had experienced stress due to handling of fish while grading and a rise in water temperature (7°C to 10°C). The cumulative mortalities recorded over a period of 15 days in the three tanks were 28, 41 and 52%. The fish samples were collected at the end of the recorded mortality period of 15 days starting from first mortality. These fish appeared to be moribund and had bloodshot fins with faint hemorrhages around the fin base. Six fish that were randomly sampled from these tanks to obtain skin mucus were infected with V. anguillarum, as confirmed by identifying the bacteria grown on TSA-blood agar with salt [16]. Mucus from six fish of the same stock maintained in separate tanks in a different fish hall, which were confirmed to be not infected, was also sampled. There were no mortalities observed in these control fish group over the same period.
Mucus sample
Six fish each from infected and non-infected groups were anesthetized in MS-222 and killed by a blow to the head for sampling. Mucus collection was done from normal skin surface avoiding collection of blood along with mucus. Mucus was scraped off with sterile micro slides, transferred into an eppendorf tube and were immediately frozen at 80°C. All the protocols employed in this study were in accordance with the guidelines of Norwegian Animal Research Authority (FDU, http://www.fdu.no/fdu/).
Sample preparation and two dimensional gel electrophoresis
Skin mucus samples were directly solubilized in sample buffer (Bio-Rad, CA, USA) containing 7 M urea, 2 M thiourea, 1% (w/v) ASB-14, 40 mM Tris, 0.001% Bromophenol Blue modified with 0.5% Bio-Lyte 3/10 ampholyte (Bio-Rad) and 50 mM DTT, vortexed for 15 min at room temperature (RT) and centrifuged at 20000 × g for 30 min. The supernatant was transferred to a fresh tube and kept on ice. An aliquot of the supernatant solution was dialysed using nanosep omega 3 kDa cut off columns (Pierce, IL, USA) and the protein quantity in the original solution was measured using Qubit protein assay kit (Invitrogen, Paisley, UK). Hundred μg protein sample was rehydrated in 17 cm 3–10 IPG strips (Bio-Rad) and isoelectric focusing (IEF) was carried out using protean IEF cell (Bio-Rad). After IEF, the strips were immersed in equilibration buffer (6 M urea, 0.375 M Tris–HCl, pH 8.8, 2% SDS, 20% glycerol) containing 0.2% DTT and again in the same buffer containing 0.25% iodoacetamide. The equilibrated strips were loaded on to 12% SDS-PAGE gels and run on PROTEAN IIxi system (Bio-Rad). The gels were stained overnight with SYPRO Ruby (Invitrogen) according to supplier’s protocol and imaging and documentation was carried out using ChemiDoc™ XRS imaging system (Bio-Rad). Gel images were analysed using PD Quest Basic software (Bio-Rad). The gel image analysis workflow included spot detection, spot normalization using local regression mode available in the software and finally spot matching and differential expression analysis. Datasets were generated for spots showing significant differential expression using two-tailed Student’s t-test for means with 95% confidence level and p <0.05. A fold change threshold of 1.5 was also set to generate a separate dataset. The density values for the spots were then exported to Microsoft Excel (Microsoft, WA, USA) to recalculate the means and find the exact p values as well as creating bar charts.
LC-MSMS analysis
Eight protein spots showing significant differences and 19 spots that were showing –differential expression trend were excised from preparative SYPRO Ruby stained gels loaded with 500 μg protein. The protein spots were in-gel reduced, alkylated and trypsinised as described elsewhere [17] and subjected to LC-MSMS analysis (ESI Quad TOF; Micromass/Waters, MA, USA). Peak lists (pkl) generated by the Protein Lynx Global server software (version 2.1, Micromass/Waters, MA, USA) were adjusted with an internal trypsin standard and further employed for protein database searching.
Bioinformatic analysis
The peak list files obtained from LC-MSMS analyses were submitted to MASCOT [http://www.matrixscience.com] search engine and searched against NCBInr with the following parameters: maximum one missed cleavage by trypsin, peptide mass tolerance 100 ppm, MS/MS ion tolerance set to 0.1 Da, fixed carbamidomethyl modification and variable oxidation modification. Protein hits not satisfying a minimum score of 40 and with a low sequence coverage were further searched against vertebrate EST database, taxonomy Actinopterygii. An interaction network of selected proteins was generated based on their mammalian orthologues (generated through BLASTp searches in NCBI database) using Ingenuity pathway analysis [http://www.ingenuity.com].
Study on experimentally infected fish
The bacterial challenge experiment was conducted at the Fish Health Laboratory of the Institute of Marine Research (IMR), Bergen, Norway in cooperation with University of Nordland (UiN), Bodø, Norway. The study was approved by Norwegian Animal Research Authority (FDU; FDU case number 09/112472, approved 22.09.2009) and protocols employed were in accordance with their guidelines. The fish used were juvenile, non vaccinated Atlantic cod procured at 30 g size (0-yr class) from a commercial hatchery. Prior to the challenge study, groups of 24 fish each of average weight 90 g were introduced into two 500 L experimental tanks. The fish were fed on a laboratory prepared fishmeal-based feed with the nutrient profiles similar to that of commercial formulations. For performing the bacterial challenge, the water flow into the tanks was stopped for a period of one hour. The fish were then either bath challenged with Vibrio anguillarum (strain H610 from the collection of the Fish Health Group at IMR) or mock challenged with culture media (tryptic soy broth with 1.5% NaCl). The bacterial amount employed for the challenge was 1.6x107cfu ml-1. Skin and gill samples were collected at 0 h (4 fish, initial sample) and from both treatment groups (5 fish from each group) at 4 h and 48 h post challenge and were immediately snap frozen in liquid nitrogen.
RNA extraction and cDNA synthesis
RNA was extracted from cod skin and gills using Trizol method as described elsewhere [18]. Total RNA was quality checked on 1% agarose gels, quantified using Qubit RNA assay (Invitrogen) and treated with gDNA wipe out buffer. cDNA was synthesized from 1 μg RNA using QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany).
Primer design and qPCR
A tool box of genes identified from the proteome experiment were employed for quantitative real-time PCR. Primers were designed using Primer3 software available online [http://frodo.wi.mit.edu/], after retrieving their full length or partial sequences from Atlantic cod genome database [http://www.ensembl.org] or NCBI dbEST and they were then evaluated using Netprimer (PREMIER Biosoft, USA). The oligonucleotide primers and their attributes are shown in Table 1. Novel full/partial EST sequences were cloned by TA Topo cloning kit (Invitrogen) and sequenced as previously reported [18].
Table 1.
Oligonucleotide primers employed in this study
|
Gene name |
|
Sequence (5’-3’) |
Primer |
Purpose |
Acc. |
|---|---|---|---|---|---|
| efficiency (%) | number | ||||
| Profilin2 (Pfn2) |
F1 |
CCAGCCACTCAATATGTCGT |
|
cloning |
[GenBank:KC460541] |
| R2 |
CAAGAACCTGGGAAACACAC |
||||
| |
F |
ATCGCAGCACTTTATTCACC |
95.18 |
qPCR |
|
| R |
GCGTCCTCCTTGTAGAGGTT |
||||
| Peroxiredoxin 6 (Prdx6) |
F |
TATCGGTGAGACAGGACCAT |
|
cloning |
[GenBank:KC460539] |
| R |
TGGCGGTTTATTCTAAGTGC |
||||
| |
F |
GATGAGATCGACAAGGATGG |
96.99 |
qPCR |
|
| R |
TCTATGACCCTCAGCAGCTC |
||||
| Flotillin1 (Flot1) |
F |
TCGCTGAAATAGGTCTCTGG |
|
cloning |
[GenBank:KC460542] |
| R |
ACTCAGCCTTCTTGGTGTTG |
||||
| |
F |
ACCAGGACTACCTCCACTCC |
91.88 |
qPCR |
|
| R |
TACTGGGCAGACACCTTCTC |
||||
| Calpain small subunit 1 (Capns1) |
F |
TTCTCTTCTCACCGCAGAAC |
|
cloning |
[GenBank:KC460540] |
| R |
GACTTTTCCAGCTCCTCCTC |
||||
| |
F |
AACATGGCAACATGGACTTT |
94.05 |
qPCR |
|
| R |
TTGACATCAAGGGAGATGGT |
||||
| Beta 2 Tubulin (Tubb2) |
F |
CAGCTACTTCGTGGAATGGA |
94.47 |
qPCR |
[GenBank:AAD56401] |
| R |
CTGTTGCCGATGAAGGTTAC |
||||
| Cold inducible RNA binding protein (Cirbp) |
F |
CTCTTCGGAACTCTCAACCA |
|
cloning |
[GenBank:KC460543] |
| R |
AATACAGCCAGCCAAGTGAC |
||||
| |
F |
GTATGGAAACATCGCCAAAG |
94.17 |
qPCR |
|
| R |
CTCGTCGGCATTATCAAACT |
||||
| Glutathione S-transferase omega1(Gsto1) |
F |
ATTGCGTCTTGGTCATTCAT |
|
cloning |
[GenBank:KC460544] |
| R |
ACAGGCCATAGTCGTAGTCG |
||||
| |
F |
CTGAAACACTTCCTGGATGG |
95.60 |
qPCR |
|
| R |
GCGTGCATGGTTTCTTTAAC |
||||
| Ubiquitin (Ubi) |
F |
GGCCGCAAAGATGCAGAT |
96.10 |
qPCR |
[GenBank:EX735613] |
| R |
CTGGGCTCGACCTCAAGAGT |
||||
| Acidic ribosomal protein (Arp) |
F |
TGATCCTCCACGACGATGAG |
95.37 |
qPCR |
[GenBank:EX741373] |
| R |
CAGGGCCTTGGCGAAGA |
||||
| Interleukin 1β (il1β) |
F |
GGAGAACACGGACGACCTGA |
|
qPCR |
[GenBank:AJ535730] |
| R | CGCACCATGTCACTGTCCTT |
1Forward primer.
2 Reverse primer.
qPCR was performed with StepOne thermocycler (Applied Biosystems (AB), CA, USA). A 10 μL PCR mix per reaction was prepared with 1 μL of primers (5 pM each), 5 μL of Fast SYBR Green Master Mix (AB) and 4 μL of 25x diluted cDNA template. The reaction mix was dispensed into 96 well plates and covered with adhesive optical film (AB) and amplification was performed according to the following conditions: holding stage for 20 sec at 95°C for activation of fast Taq DNA polymerase followed by 40 cycles of denaturation at 95°C for 3 sec and annealing/extension for 30 sec at 60°C. Fluorescence data was acquired during the annealing/extension step. The samples were run in duplicates and pooled DNase treated RNA and no template negative controls were also included in the scheme. A 3-fold dilution series from pooled cDNA were included in each plate for each gene and efficiency (E) of PCR reaction for each set of primers was calculated based on standard curve method according to the formula E = 10(-1/slope)-1*100 . The Ct values obtained for each gene was transformed into relative quantities by setting the highest value to 1 according to the requirement of geNORM software [http://medgen.ugent.be/~jvdesomp/genorm/] to obtain the normalization values for each sample calculated based on the geometric mean of reference genes ubiquitin (Ubi) and acidic ribosomal protein (Arp). The normalized values of individual samples were used for statistical analysis.
Statistical analysis
The statistical analyses of qPCR data were carried out using GraphPad Prism software (GraphPad Software, Inc. CA, USA). The sample sets were tested for normality using Kolmogorov-Smirnov test and data was log transformed where necessary. Transcript profiles for each gene from bacterial challenge group were compared with the respective values from mock challenge control groups, at each time point, using unpaired student’s t-test. Zero hour samples that are included in the bar charts as a basal reference point were not included for statistical comparisons.
Results & discussion
Proteomic studies on skin mucus from naturally infected fish
Skin mucus is a critical component of piscine immune system. Various antibacterial and inflammatory factors like immunoglobulin, lectins, acute phase proteins, and proteases have been identified in fish mucus [19-21]. We have earlier reported a proteome reference map of Atlantic cod skin mucus, identifying among others innate immune components such as lysozyme, apolipoproteins, galectin-1 and serpins [22]. A separate study showing the presence of antimicrobial components in different tissues including skin mucosa of cod is also available [23]. These studies indicate that the cod skin mucus harbours a significant number of immune relevant proteins. In the present study the skin mucus proteome of healthy cod was compared with that of V. anguillarum infected cod. The skin mucus 2D gels showed good resolution and reproducibility (Figure 1). Through gel image analysis, only 8 protein spots were found to be differentially expressed (Figure 2). An additional 19 spots were identified based on high changes observed in individual infected fish (based on variation in spot intensities). There is however, no statistical support for their identification as indicated in Table 2. Nevertheless, this information could be used for further studies, especially targeted transcriptomics or proteomics under natural and bath infection scenarios in fish. The spot details along with accession numbers and other parameters are shown in Table 2.
Figure 1.

Representative 2D gels of Atlantic cod skin mucus A) Healthy and B) V. anguillarum infected. Annotated spots were identified using LC-MSMS. The mucus proteins were isoelectrically focused on 17cm IPG strips (pI 3–10) in first dimension, run on a 12.5% SDS-PAGE in second dimension and stained with SYPRO Ruby fluorescent dye. Note that the spot numbers correspond to the protein identities outlined in Table 2. N = 6.
Figure 2.

Normalized spot intensities of protein showing statistically significant differences (p < 0.05) in healthy vs V. anguillarum infected cod skin mucus. Data based on comparative 2D gel analysis employing PD Quest Basic software. N = 6.
Table 2.
Proteins identified in Atlantic cod skin mucus
|
N |
Protein name (Species) |
Acc. no. |
Theoretical pI/MW |
IS |
SC (%) |
Cod EST |
FCx` |
p |
|---|---|---|---|---|---|---|---|---|
| Acc. no | ||||||||
| 1 |
Profilin-2 (Danio rerio) |
NP_958493 |
6.09/14.6 |
474 |
35 |
GO379352 |
<1.5 |
>0.05 |
| 2 |
Profilin-2 ( Salmo salar ) |
ACI66182 |
6.72/14.5 |
393 |
33 |
FG312066 |
1.6 |
>0.05 |
| 3 |
Nucleoside diphosphate kinase (G. morhua) |
AET05826 |
7.84/15.5 |
623 |
77 |
NA |
<1.5 |
>0.05 |
| 4 |
Inositol monophosphatase 1(Anguilla anguilla) |
CBI68709 |
5.35/30.6 |
699 |
35 |
GW854257 |
<1.5 |
>0.05 |
| 5 |
Guanine nucleotide binding protein polypeptide 2 like-1 (S. salar) |
ACH70647 |
7.64/28.6 |
675 |
44 |
GW858107 |
<1.5 |
>0.05 |
| 6 |
Voltage dependent anion selective channel protein 1 (S. salar) |
ACI33832 |
8.69/30.9 |
664 |
27 |
GW858279 |
<1.5 |
>0.05 |
| 7 |
Tropomyosin-alpha-4 chain (S. salar) |
ACN10541 |
4.57/27.9 |
760 |
39 |
FF409290 |
<1.5 |
>0.05 |
| 8 |
Calmodulin (S. salar) |
ACI68592 |
4.09/16.7 |
103 |
19 |
NA |
<1.5 |
>0.05 |
| 9 |
Calpain small subunit-1 ( S. salar ) |
ACI167026 |
5.34/24.6 |
428 |
44 |
EX730492 |
<1.5 |
0.0233 |
| 10 |
Calpain small subunit-1 (S. salar) |
ACI167026 |
5.34/24.6 |
731 |
42 |
GW858828 |
<1.5 |
>0.05 |
| 11 |
Peroxiredoxin-6 (Scophthalmus maximus) |
ADJ57694 |
5.46/24.3 |
306 |
44 |
ES239904 |
<1.5 |
>0.05 |
| 12 |
PREDICTED: Glutathione S-transferase omega-1 like ( Oreochromis niloticus ) |
XP_003438319 |
6.01/27.4 |
201 |
22 |
FCO73610 |
<1.5 |
0.0409 |
| 13 |
Malate dehydrogenase 2-NAD (mitochondrial) ( S. salar ) |
NP_001133198 |
8.15/35.8 |
327 |
19 |
NA |
1.52 |
0.0221 |
| 14 |
Proteasome (macropain) 26S subunit ( S. salar) |
ACH85257 |
6.81/44.4 |
246 |
19 |
NA |
<1.5 |
0.0130 |
| 15 |
PREDICTED: alpha-enolase-like isoform 1 (S. salar) |
XP_003444858 |
5.99/47.5 |
791 |
46 |
NA |
<1.5 |
>0.05 |
| 16 |
Flotillin-1 (Larimichthys crocea) |
AEP68103 |
|
295 |
17 |
NA |
<1.5 |
>0.05 |
| 18 |
Type II keratin E3 ( Oncorhynchus mykiss ) |
NP_001123458 |
5.32/55.3 |
380 |
20 |
GW854186 |
<1.5 |
0.0413 |
| 19 |
14kDa apolipoprotein ( Epinephelus coioides ) |
ACM41842 |
6.26/15.4 |
249 |
35 |
EX189720 |
3.6 |
0.0238 |
| 20 |
PREDICTED: similar to ribosomal protein P2 (Taeniopygia guttata) |
XP_002199373 |
4.21/11.7 |
327 |
57 |
EX174193 |
<1.5 |
>0.05 |
| 21 |
PREDICTED: endoplasmin-like (O. niloticus) |
XP_003443932 |
4.75/91.6 |
339 |
24 |
GW849407 |
<1.5 |
>0.05 |
| 22 |
Serine/threonine-protein phosphatase2A 65 kDa regulatory subunit A beta isoform ( S. salar ) |
ACN58704 |
4.94/65.9 |
355 |
29 |
NA |
2.3 |
>0.05 |
| 23 |
Beta-2-tubulin ( G. morhua ) |
AAD56401 |
4.71/50.1 |
670 |
36 |
NA |
2.5 |
0.0419 |
| 24 |
Preproapolipoprotein A-1 ( G. morhua ) |
AAU87042 |
5.28/29.8 |
253 |
36 |
NA |
2.0 |
>0.05 |
| 25 |
Cold inducible RNA-binding protein ( S. salar ) |
ADM16300 |
5.46/16.5 |
391 |
33 |
GO383013 |
2.9 |
0.0029 |
| 26 |
FK-506 binding protein 1A (G. morhua) |
AEK21706 |
6.71/11.8 |
154 |
32 |
NA |
<1.5 |
>0.05 |
| 29 |
Proteasome activator complex subunit 1 (S. salar) |
ACM09791 |
5.54/28.62 |
473 |
18 |
GW854458 |
<1.5 |
>0.05 |
| 30 | PREDICTED: Calreticulin-like isform 2 (O. niloticus) | XP_003448811 | 4.43/47.146 | 325 | 41 | ES771164 | <1.5 | >0.05 |
N-spot number, Acc. No.-Accession number, pI-isoelectric point, MW, Molecular weight; IS, Ion score; SC, Sequence coverage; FC, Fold change; NA, Not applicable.
Protein spots were identified by LC-MSMS. Proteins either with p < 0.05 or fold change >1.5 or both are in bold letters.
The rather low number of significantly different protein spots identified in this study could be linked to the possible variation in the progression of V. anguillarum infection in individual fish. This could be a disadvantage while analyzing samples from naturally infected fish. Besides, comparative 2D gel analysis has its limitations with regard to high abundant proteins being overrepresented in the gels, which can mask the detection of low abundant inflammatory molecules like interleukins. Also V. anguillarum has been shown to have high levels (up to 70 fold) of protease activity in the presence of gastrointestinal mucus [24]. This coupled with the fact that protease inhibitors were not used in the present study might have resulted in degradation of host proteins thereby affecting their detection. Nevertheless, the present approach helps in understanding the involvement of protein components during the natural progression of vibriosis in cod.
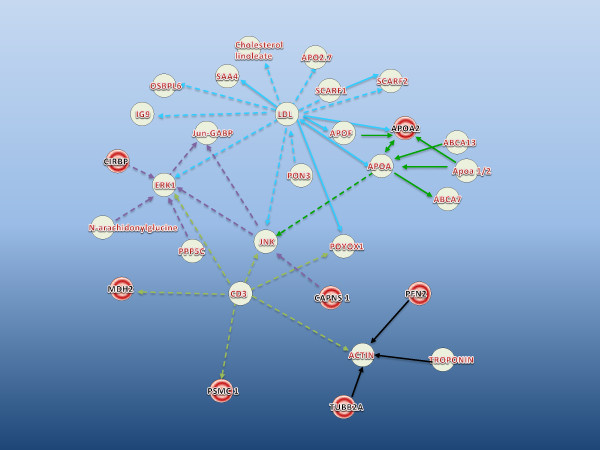
Proteomic studies to elucidate host responses in fish infected with V. anguillarum are limited. To gain a better understanding of the possible role of identified proteins in disease progression and inflammation, a protein-protein interaction network generated by Ingenuity Pathway Analysis (IPA) is shown in Figure 3. The network highlights the interaction of lipoproteins (LDL), mitogen activated protein kinases (ERK1, c-Jun N-terminal kinases (JNK)) and CD3 with proteins identified in the study. The afore mentioned protein families especially the JNKs are activated by stress and inflammatory signals [25] and might have been triggered by a combination of temperature/handling stress the fish had been subjected to, followed by the V. anguillarum infection.
Figure 3.
Ingenuity pathway analysis of some key proteins identified in the study. Each gene involved in the pathway is denoted by their ENTREZ gene symbol or in some cases full gene name. The proteins indicated in red circles showed statistically significant (p < 0.05) differential expression and/ or fold change >1.5 in comparative 2D gel analysis. Grey circles indicate other proteins involved in the pathway. Different coloured arrows are shown to highlight the central molecules involved in the pathway namely; CD3, JNK, ERK1, LDL and APOA. Solid arrows indicate direct protein interactions and dashed arrows indicate indirect protein interactions. Pathway analysis was done based on the mammalian orthologs of the proteins identified in cod mucus.
Calpain small subunit 1 (CAPNS1) was found to be downregulated in the present study (spot number 9, Figure 2). CAPNS1 is a regulatory component of the calcium dependent cysteine proteases and is ubiquitous in mammalian cells [26]. Calpain mediated proteolysis is limited and calpain usually modifies/alters the substrate proteins instead of bringing about complete degradation [27] indicating its ability to specifically trigger downstream physiological response. There is also evidence that inflammatory cytokines like TNF-α and interleukins can downregulate calpain in cells and tissues [28]. Upregulation of IL-1β and IL-8 has been observed in the gut of Atlantic cod after intraperitoneal injection with live V. anguillarum[29]. Similarly, upregulation of these genes in cod gut were reported after immersion challenge with V. anguillarum[30]. Therefore it is likely that inflammatory cytokines induced during V. anguillarum infection in cod might have resulted in low levels of CAPNS1 observed in the present study (Figure 2). As V. anguillarum infections are characterized by hemorrhagic lesions and wounds in the skin, the downregulation of calpain is logical as the presence of active proteases might lead to exacerbation of disease and irreversible tissue damage [31]. Besides, epidermal lesions that may cause calcium imbalance and disturb membrane integrity can affect the activation of calpain. Human calpain II has been found to be upregulated in psoriasis skin lesions, which are characterized by calcium deficiency [31]. Calpain has been reported in different tissues of fish [32]. Mucosal specific calpains present in higher vertebrates [26] are thought to play important roles in mucosal defense.
Cold inducible RNA binding proteins (CIRBP) are stress proteins with RNA binding domain that can regulate cellular activity under stressful conditions such as hypoxia [33] or during brief periods of osmotic stress [34] in fish. In the present study, Cirbp was significantlyup regulated in infected fish (spot number 25, Figure 2. CIRBP influence the MAPK pathway leading to downstream inflammatory responses as shown in Figure 3.
Malate dehydrogenase 2 (mitochondrial) (MDH2) is a mitochondrial isozyme of malate dehydrogenase that is primarily involved in citric acid cycle [35]. Conditions that alter metabolism can result in elevation of mitochondrial enzymes like malate dehydrogenase [36]. An upregulation of MDH2 was noted in the infected cod (spot number 13, Figure 2), possibly triggered by V. anguillarum lipopolysacharide (LPS) as shown in a comparative proteome study of Moraxella sp. infected kidney in sea bream (Sparus aurata) [37].
Keratin is a cytoskeletal protein whose primary function is to protect cells from mechanical and non mechanical injuries [38]. Recent reports have also shown that keratin from fish mucus possesses antibacterial activity owing to its pore-forming properties [39]. Keratin turnover is dependent on ubiquitin-proteasome pathway and its expression levels can be altered upon cell injury [40]. There was a downregulation of Keratin II in cod mucus following vibrio infection (spot number 18, Figure 2). Beta-2-tubulins (TUBB2) form integral components of cytoskeleton and are building blocks of microtubules [41]. Tubb2 downregulation has been reported in Chinese white shrimp (Fenneropenaeus chinensis) challenged with V. anguillarum[42]. Being an important component of cytoskeletal architecture, different expression levels of tubulin can be presumed to influence the formation of new phagosomes and thus affect phagocytic activity. Whether these changes could be pathogen mediated needs to be investigated. In the present study infected fish showed an increase in TUBB2 in the skin mucus (spot number 23, Figure 2), possibly due to increased expression of Tubb2 in the mucosal cells associated with phagocytic processes.
14-kDa apolipoprotein (apo-14kDa) is a fish specific high density lipoprotein (HDL) orthologous to mammalian apolipoproteinA-II (apoA-II) [43], the expression of which was significantly increased in infected fish (spot number 19, Figure 2). In carp (Cyprinus carpio) it has been shown to have antibacterial effects in vitro[44]. Further, apo-14kDa was upregulated in the liver of V. anguillarum infected European seabass (Dicentrarchus labrax) [45].
Glutathione-S-transferase omega-1 (GSTO1) is an enzyme involved in biotransformation of compounds including toxic substances and oxidative stress products, transport of ligands, and regulation of signaling pathways [46]. In cod, the Gsto1 transcripts were found in different tissues including skin [47]. In the present study, the protein amount was reduced in infected fish (spot number 12, Figure 2). In Manila clam (Venerupis philippinarum) infected with V. anguillarum[48] and Chinese mitten crab (Eriocheir sinensis) injected with Aeromonas hydrophila, an upregulation of novel gst transcripts was noted as early as 6 h post infection [49]. The upregulation of gst reported in the aforementioned studies could be due to reactive oxygen metabolites induced by the V. anguillarum infection as shown in a study where the measurement on enzyme activity complemented the gene expression data [50].
Proteasome 26 subunit is involved in ATP dependent degradation of ubiquitinated proteins, which are instrumental in oxidative stress response and partakes in generating peptide fragments of invading pathogen for antigen presentation [51]. Proteasome expression changes have been reported in response to V. anguillarum infection [42] and in Atlantic cod challenged with formalin killed A. salmonicida[52]. In the current study, Proteasome 26S levels were significantly higher in the infected fish (spot number 14, Figure 2).
Other important proteins that exhibited a differential expression trend in this study, including profilin-2, flotilin-1, peroxiredoxin-6, apolipoprotein AI are also important from an immune perspective. The genes corresponding to these proteins could also be considered as potential biomarkers in studies involving V. anguillarum infection.
In addition to bacterial infection, Atlantic cod may be susceptible to concurrent or secondary infections with parasites like Trichodina spp [53]. Attempts to identify ectoparasites in skin or gills was not done, hence teoretically such infections may be present and also trigger differential expression of certain proteins in the skin of cod.
Gene expression studies on skin from experimentally challenged fish
In order to understand if the changes in the mucus proteins, arising from natural infection, correspond to the alterations of the respective genes in skin, a controlled bath challenge experiment was performed. The observations were made only during early stages of infection. The V. anguillarum H610 challenge protocol at Institute of Marine Research (IMR), Bergen is robust and well established as reported in different studies [7,54].
Since the proteins were identified through ESTs, the genes had to be cloned. The qPCR expression patterns of selected genes are shown in Figure 4. Significant differential expression of mRNA was detected as early as 4 h post infection. Capns1 and Gsto1 were upregulated at 4 h post challenge whereas Flot1 was significantly downregulated compared to the mock challenged controls at 4 h and 48 h post infection (Figure 4). Expression of Cirbp and Tubb2 were significantly downregulated 48 h post infection (Figure 4). There was no significant difference in the expression patterns of Pfn2 and Prdx6 (Figure 4). To confirm infection after experimental bacterial challenge, interleukin-1β (il1β) transcripts were quantified in the skin and gill tissue of the infected fish. The il1β mRNA levels were found to be significantly upregulated in gills at 48 h post bath challenge compared to the control [see Additional file 1]. il1β transcripts of skin showed no changes at the two time points (data not shown).
Figure 4.
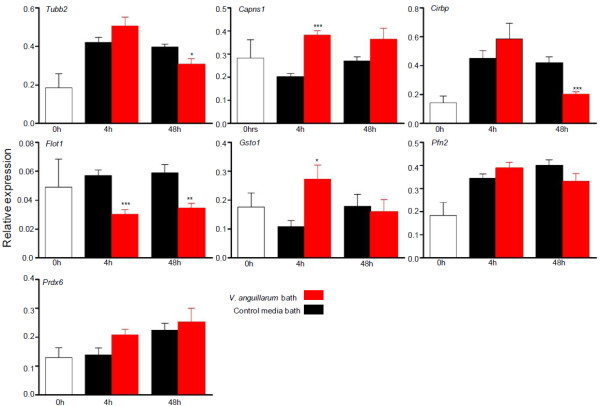
Relative expression levels of genes (based on the proteome study) in Atlantic cod skin upon V. anguillarum bath challenge. Quantification by real-time PCR. Values are indicated as means ± SEM (N = 5). Transcript levels normalized to reference genes: Ubiquitin (Ubi) and acidic ribosomal protein (Arp). Asterisk (*p < 0.05 or **p < 0.001 or ***p < 0.0001) above the error bars indicate statistically significant difference (Student’s t-test) between infected and control fish, at each time point. Initial samples are depicted as 0h samples. Standard gene abbreviations are used.
The gene expression data and proteomic data need not always show a one to one correspondence [55]. Nevertheless, as observed in the qPCR study these gene transcripts show a differential expression during the early stages of V. anguillarum infection and it can be surmised that they may play important role in the progression of vibriosis in cod. Fish skin has been seldom targeted for gene expression studies, in challenge experiments and infection scenarios, as most of them focused on systemic immune-relevant organs like spleen and head kidney. The expression of key immune genes in skin has been explored primarily in the case of ectoparasite infections [56]. A recent report on the expression of key immune relevant genes in naive Atlantic cod skin [57] taken together with the present study demonstrates the role of immune relevant components in the skin mucus/skin of cod both at protein and transcript level during bacterial infection.
Thus, through the proteomic approach we were able to identify differentially regulated proteins in skin mucus that might be relevant targets for gene expression studies as evident from this study. Going further, the ultimate objective of this study is to utilize the mucosal proteins as biomarkers for early assessment of fish health status, aimed at controlling bacterial diseases including vibriosis.
Conclusions
We identified a group of proteins, of which the involvement of some can be considered novel in V. anguillarum infection of cod. More importantly, these proteins are shown to be closely associated with key pathways that mount an immune response in the host. The early induction of mRNA transcripts of some of the selected genes also points towards their involvement along with other acute phase proteins as primary orchestrators in the fish mucosal immunity against bacterial infection.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Conceived and designed the experiments: BR, VK and MFB. Performed the experiments: BR and LJ. Analyzed the data: BR and MFB. Wrote the paper: BR, VK and MFB. All authors read and approved the final manuscript.
Supplementary Material
Relative expression of interleukin-1β (il1β) in gill tissue of Atlantic cod bath challenged with Vibrio anguillarum. Quantification of il1β mRNA level by real-time PCR at 4 h and 48 h post bath challenge. Values are indicated as means ± SEM (N = 5). Transcript levels normalized to reference genes: Ubiquitin (Ubi) and acidic ribosomal protein (Arp). Asterisk (*p < 0.05) above the error bars indicate statistically significant difference (Student’s t-test) between infected and control fish, at each time point.
Contributor Information
Binoy Rajan, Email: Binoy.Rajan@uin.no.
Jep Lokesh, Email: Lokesh.Jeppinamogeru@uin.no.
Viswanath Kiron, Email: Kiron.Viswanath@uin.no.
Monica F Brinchmann, Email: Monica.Brinchmann@uin.no.
Acknowledgements
The present study was supported by the projects ‘Preventive Health Care of Farmed Fish’ (176528) and ‘Mucosal Immune System of Atlantic Cod- Creating a knowledge Base’ (184703) from the Research Council of Norway, granted to VK. Dr. Øivind Bergh is thanked for his contributions in conducting the challenge experiments at Bergen. The authors thank Hilde Ribe and Kathrine Klippenberg, University of Nordland, Hari Rudra, Ann Cathrine Einen and Rolf Hetelid Olsen, Institute of Marine Research for their technical support in conducting the bacterial challenge experiment.
References
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilstrom L, Warr GW, Stromberg S. Why is the antibody response of Atlantic cod so poor? The search for a genetic explanation. Fish Sci. 2005;71:961–971. doi: 10.1111/j.1444-2906.2005.01052.x. [DOI] [Google Scholar]
- Esteban MA. An overview of the immunological defenses in fish skin. ISRN Immunol. 2012. [DOI]
- Larsen JL, Pedersen K, Dalsgaard I. Vibrio anguillarum serovars associated with vibriosis in fish. J Fish Dis. 1994;17:259–267. doi: 10.1111/j.1365-2761.1994.tb00221.x. [DOI] [Google Scholar]
- Pedersen K, Grisez L, Houdt R, Tiainen T, Ollevier F, Larsen JL. Extended serotyping scheme for Vibrio anguillarum with the definition and characterization of seven provisional O-serogroups. Curr Microbiol. 1999;38:183–189. doi: 10.1007/PL00006784. [DOI] [PubMed] [Google Scholar]
- Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, Rediers H. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J Fish Dis. 2011;34:643–661. doi: 10.1111/j.1365-2761.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- Engelsen AR, Sandlund N, Fiksdal IU, Bergh O. Immunohistochemistry of Atlantic cod larvae Gadus morhua experimentally challenged with Vibrio anguillarum. Dis Aquat Organ. 2008;80:13–20. doi: 10.3354/dao01926. [DOI] [PubMed] [Google Scholar]
- Mikkelsen H, Lund V, Larsen R, Seppola M. Vibriosis vaccines based on various sero-subgroups of Vibrio anguillarum O2 induce specific protection in Atlantic cod (Gadus morhua L.) juveniles. Fish Shellfish Immunol. 2011;30:330–339. doi: 10.1016/j.fsi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet. 2012;379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombout JWHM, Blok LJ, Lamers CHJ, Egberts E. Immunization of carp (Cyprinus carpio) with a Vibrio anguillarum bacterin: Indications for a common mucosal immune system. Dev Comp Immunol. 1986;10:341–351. doi: 10.1016/0145-305X(86)90024-8. [DOI] [PubMed] [Google Scholar]
- O’Toole R, Lundberg S, Fredriksson S-Å, Jansson A, Nilsson B, Wolf-Watz H. The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J Bacteriol. 1999;181:4308–4317. doi: 10.1128/jb.181.14.4308-4317.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole R, von Hofsten J, Rosqvist R, Olsson P-E, Wolf-Watz H. Visualisation of Zebrafish infection by GFP-labelled Vibrio anguillarum. Microb Pathog. 2004;37:41–46. doi: 10.1016/j.micpath.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Spanggaard B, Huber I, Nielsen J, Nielsen T, Gram L. Proliferation and location of Vibrio anguillarum during infection of rainbow trout, Oncorhynchus mykiss (Walbaum) J Fish Dis. 2000;23:423–427. doi: 10.1046/j.1365-2761.2000.00257.x. [DOI] [Google Scholar]
- Lindell K, Fahlgren A, Hjerde E, Willassen N-P, Fällman M, Milton DL. Lipopolysaccharide O-antigen prevents phagocytosis of Vibrio anguillarum by rainbow trout (Oncorhynchus mykiss) skin epithelial cells. PLoS One. 2012;7:e37678. doi: 10.1371/journal.pone.0037678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervarcke S, Ollevier F, Kinget R, Michoel A. Mucosal response in African catfish after administration of Vibrio anguillarum O2 antigens via different routes. Fish Shellfish Immunol. 2005;18:125–133. doi: 10.1016/j.fsi.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Rock JL, Nelson DR. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect Immun. 2006;74:2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øverbye A, Brinchmann MF, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3:300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- Fernandes JMO, Ruangsri J, Kiron V. Atlantic cod piscidin and its diversification through positive selection. PLoS One. 2010;5:e9501. doi: 10.1371/journal.pone.0009501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, MacKinnon SL, Ross NW. A comparative study on innate immune parameters in the epidermal mucus of various fish species. Comp Biochem Physiol B. 2007;148:256–263. doi: 10.1016/j.cbpb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Shephard KL. Functions for fish mucus. Rev Fish Biol Fish. 1994;4:401–429. doi: 10.1007/BF00042888. [DOI] [Google Scholar]
- Suzuki Y, Tasumi S, Tsutsui S, Okamoto M, Suetake H. Molecular diversity of skin mucus lectins in fish. Comp Biochem Physiol B. 2003;136:723–730. doi: 10.1016/S1096-4959(03)00178-7. [DOI] [PubMed] [Google Scholar]
- Rajan B, Fernandes JMO, Caipang CMA, Kiron V, Rombout JHWM, Brinchmann MF. Proteome reference map of the skin mucus of Atlantic cod (Gadus morhua) revealing immune competent molecules. Fish Shellfish Immunol. 2011;31:224–231. doi: 10.1016/j.fsi.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Ruangsri J, Fernandes JMO, Brinchmann M, Kiron V. Antimicrobial activity in the tissues of Atlantic cod (Gadus morhua L.) Fish Shellfish Immunol. 2010;28:879–886. doi: 10.1016/j.fsi.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Denkin SM, Nelson DR. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl Environ Microbiol. 1999;65:3555–3560. doi: 10.1128/aem.65.8.3555-3560.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahopoulos S, Zoumpourlis VC. JNK: A key modulator of intracellular signaling. Biochemistry (Mosc) 2004;69:844–854. doi: 10.1023/b:biry.0000040215.02460.45. [DOI] [PubMed] [Google Scholar]
- Sorimachi H, Hata S, Ono Y. Calpain chronicle-an enzyme family under multidisciplinary characterization. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87:287–327. doi: 10.2183/pjab.87.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saido TC, Sorimachi H, Suzuki K: Calpain: C. new perspectives in molecular diversity and physiological-pathological involvement. FASEB J. 1994;8:814–822. [PubMed] [Google Scholar]
- Ishikawa H, Nakagawa Y, Shimizu K, Nishihara H, Matsusue Y, Nakamura T. Inflammatory cytokines induced down-regulation of m-calpain mRNA expression in fibroblastic synoviocytes from patients with osteoarthritis and rheumatoid arthritis. Biochem Biophys Res Commun. 1999;266:341–346. doi: 10.1006/bbrc.1999.1819. [DOI] [PubMed] [Google Scholar]
- Inami M. The immune system of Atlantic cod: with emphasis on intestinal immunology. University of Tromsø, Faculty of Biosciences, Fisheries and Economics: PhD thesis; 2011. [Google Scholar]
- Lokesh J, Fernandes JMO, Korsnes K, Bergh Ø, Brinchmann MF, Kiron V. Transcriptional regulation of cytokines in the intestine of Atlantic cod fed yeast derived mannan oligosaccharide or β-glucan and challenged with Vibrio anguillarum. Fish Shellfish Immunol. 2012;33:626–631. doi: 10.1016/j.fsi.2012.06.017. [DOI] [PubMed] [Google Scholar]
- Matsushita Y, Shimada Y, Kawara S, Takehara K, Sato S. Autoantibodies directed against the protease inhibitor calpastatin in psoriasis. Clin Exp Immunol. 2005;139:355–362. doi: 10.1111/j.1365-2249.2005.02701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M, Nath J, Killefer J. Cloning of the calpain regulatory subunit cDNA from fish reveals a divergent domain-V. Anim Biotechnol. 2004;15:145–157. doi: 10.1081/ABIO-200036706. [DOI] [PubMed] [Google Scholar]
- Wellmann S, Bührer C, Moderegger E, Zelmer A, Kirschner R, Koehne P, Fujita J, Seeger K. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- Pan F, Zarate J, Choudhury A, Rupprecht R, Bradley TM. Osmotic stress of salmon stimulates upregulation of a cold inducible RNA binding protein (CIRP) similar to that of mammals and amphibians. Biochimie. 2004;86:451–461. doi: 10.1016/j.biochi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Minárik P, Tomásková N, Kollárová M, Antalík M. Malate dehydrogenases–structure and function. Gen Physiol Biophys. 2002;21:257–265. [PubMed] [Google Scholar]
- Cripps RA, Reish DJ. The effect of environmental stress on the activity of malate dehydrogenase and lactate dehydrogenase in Neanthes arenaceodentata (Annelida: Polychaeta) Comp Biochem Physiol B. 1973;46:123–133. doi: 10.1016/0300-9629(73)90565-3. [DOI] [PubMed] [Google Scholar]
- Addis M, Cappuccinelli R, Tedde V, Pagnozzi D, Viale I, Meloni M, Salati F, Roggio T, Uzzau S. Influence of Moraxella sp. colonization on the kidney proteome of farmed gilthead sea breams (Sparus aurata L.) Proteome Sci. 2010;8:50. doi: 10.1186/1477-5956-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/S0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- Molle V, Campagna S, Bessin Y, Ebran N, Saint N, Molle G. First evidence of the pore-forming properties of a keratin from skin mucus of rainbow trout (Oncorhynchus mykiss, formerly Salmo gairdneri) Biochem J. 2008;411:33–40. doi: 10.1042/BJ20070801. [DOI] [PubMed] [Google Scholar]
- Rogel MR, Jaitovich A, Ridge KM. The role of the ubiquitin proteasome pathway in keratin intermediate filament protein degradation. Proc Am Thorac Soc. 2010;7:71–76. doi: 10.1513/pats.200908-089JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li F, Jiang H, Yu Y, Liu C, Li S, Wang B, Xiang J. Proteomic analysis of differentially expressed proteins in lymphoid organ of Fenneropenaeus chinensis response to Vibrio anguillarum stimulation. Fish Shellfish Immunol. 2010;29:186–194. doi: 10.1016/j.fsi.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Choudhury M, Yamada S, Komatsu M, Kishimura H, Ando S. Homologue of mammalian apolipoprotein A-II in non-mammalian vertebrates. Acta Biochim Biophys Sin. 2009;41:370–378. doi: 10.1093/abbs/gmp015. [DOI] [PubMed] [Google Scholar]
- Concha MI, Smith VJ, Castro K, Bastías A, Romero A, Amthauer RJ. Apolipoproteins A-I and A-II are potentially important effectors of innate immunity in the teleost fish Cyprinus carpio. Eur J Biochem. 2004;271:2984–2990. doi: 10.1111/j.1432-1033.2004.04228.x. [DOI] [PubMed] [Google Scholar]
- Sarropoulou E, Sepulcre P, Poisa-Beiro L, Mulero V, Meseguer J, Figueras A, Novoa B, Terzoglou V, Reinhardt R, Magoulas A. Profiling of infection specific mRNA transcripts of the European seabass Dicentrarchus labrax. BMC Genomics. 2009;10:157. doi: 10.1186/1471-2164-10-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister C, Lüersen K, Heinick A, Hussein A, Domagalski M, Walter RD, Liebau E. Oxidative stress in Caenorhabditis elegans: protective effects of the Omega class glutathione transferase (GSTO-1) FASEB J. 2008;22:343–354. doi: 10.1096/fj.06-7426com. [DOI] [PubMed] [Google Scholar]
- Olsvik PA, Nordtug T, Altin D, Lie KK, Overrein I, Hansen BH. Transcriptional effects on glutathione S-transferases in first feeding Atlantic cod (Gadus morhua) larvae exposed to crude oil. Chemosphere. 2010;79:905–913. doi: 10.1016/j.chemosphere.2010.03.026. [DOI] [PubMed] [Google Scholar]
- Li C, Su X, Li Y, Li T, Sun C, Zhou T, Liu H. Two classes of glutathione S-transferase genes with different response profiles to bacterial challenge in Venerupis philippinarum. Fish Shellfish Immunol. 2012;32:219–222. doi: 10.1016/j.fsi.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Zhao D, Chen L, Qin C, Zhang H, Wu P, Zhang F. A delta-class glutathione transferase from the Chinese mitten crab Eriocheir sinensis: cDNA cloning, characterization and mRNA expression. Fish Shellfish Immunol. 2010;29:698–703. doi: 10.1016/j.fsi.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Canesi L, Barmo C, Fabbri R, Ciacci C, Vergani L, Roch P, Gallo G. Effects of vibrio challenge on digestive gland biomarkers and antioxidant gene expression in Mytilus galloprovincialis. Comp Biochem Physiol C. 2010;152:399–406. doi: 10.1016/j.cbpc.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Kloetzel P-M, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Feng CY, Johnson SC, Hori TS, Rise M, Hall JR, Gamperl AK, Hubert S, Kimball J, Bowman S, Rise ML. Identification and analysis of differentially expressed genes in immune tissues of Atlantic cod stimulated with formalin-killed, atypical Aeromonas salmonicida. Physiol Genomics. 2009;37:149–163. doi: 10.1152/physiolgenomics.90373.2008. [DOI] [PubMed] [Google Scholar]
- Bricknell IR, Bron JE, Bowden TJ. Diseases of gadoid fish in cultivation: a review. ICES J Mar Sci. 2006;63:253–266. doi: 10.1016/j.icesjms.2005.10.009. [DOI] [Google Scholar]
- Samuelsen OB, Bergh Ø. Efficacy of orally administered florfenicol and oxolinic acid for the treatment of vibriosis in cod (Gadus morhua) Aquaculture. 2004;235:27–35. doi: 10.1016/S0044-8486(03)00446-0. [DOI] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigh J, Lindenstrøm T, Buchmann K. Expression of pro-inflammatory cytokines in rainbow trout (Oncorhynchus mykiss) during an infection with Ichthyophthirius multifiliis. Fish Shellfish Immunol. 2004;17:75–86. doi: 10.1016/j.fsi.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Caipang CMA, Lazado CC, Brinchmann MF, Rombout JHWM, Kiron V. Differential expression of immune and stress genes in the skin of Atlantic cod (Gadus morhua) Comp Biochem Physiol D. 2011;6:158–162. doi: 10.1016/j.cbd.2011.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative expression of interleukin-1β (il1β) in gill tissue of Atlantic cod bath challenged with Vibrio anguillarum. Quantification of il1β mRNA level by real-time PCR at 4 h and 48 h post bath challenge. Values are indicated as means ± SEM (N = 5). Transcript levels normalized to reference genes: Ubiquitin (Ubi) and acidic ribosomal protein (Arp). Asterisk (*p < 0.05) above the error bars indicate statistically significant difference (Student’s t-test) between infected and control fish, at each time point.