Abstract
Background
Vitamin D insufficiency has been associated with the occurrence of various types of cancer, but causal relationships remain elusive. We therefore aimed to determine the relationship between genetic determinants of vitamin D serum levels and the risk of developing hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC).
Methodology/Principal Findings
Associations between CYP2R1, GC, and DHCR7 genotypes that are determinants of reduced 25-hydroxyvitamin D (25[OH]D3) serum levels and the risk of HCV-related HCC development were investigated for 1279 chronic hepatitis C patients with HCC and 4325 without HCC, respectively. The well-known associations between CYP2R1 (rs1993116, rs10741657), GC (rs2282679), and DHCR7 (rs7944926, rs12785878) genotypes and 25(OH)D3 serum levels were also apparent in patients with chronic hepatitis C. The same genotypes of these single nucleotide polymorphisms (SNPs) that are associated with reduced 25(OH)D3 serum levels were found to be associated with HCV-related HCC (P = 0.07 [OR = 1.13, 95% CI = 0.99–1.28] for CYP2R1, P = 0.007 [OR = 1.56, 95% CI = 1.12–2.15] for GC, P = 0.003 [OR = 1.42, 95% CI = 1.13–1.78] for DHCR7; ORs for risk genotypes). In contrast, no association between these genetic variations and liver fibrosis progression rate (P>0.2 for each SNP) or outcome of standard therapy with pegylated interferon-α and ribavirin (P>0.2 for each SNP) was observed, suggesting a specific influence of the genetic determinants of 25(OH)D3 serum levels on hepatocarcinogenesis.
Conclusions/Significance
Our data suggest a relatively weak but functionally relevant role for vitamin D in the prevention of HCV-related hepatocarcinogenesis.
Introduction
Chronic hepatitis C is associated with important morbidity, resulting in liver cirrhosis and its complications in a significant proportion of infected individuals [1]. Due to the rising age of the hepatitis C virus (HCV)-infected population in the Western world, a dramatic increase of cases with advanced liver cirrhosis and hepatocellular carcinoma (HCC) has been predicted for the next decade [1]. Due to the insufficient treatment options for advanced HCC as well as limited number of liver allograft donors for patients with curable disease, improved strategies to screen for early HCC or to prevent the development of HCC are warranted [2]. Recently, two genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in the region of DEPDC5 and MICA as susceptibility loci for HCC development in patients with chronic hepatitis C [3], [4]. Although these genetic variations were strongly associated with HCV-induced HCC, with potentially important implications for the identification of patients at high risk of developing HCC, it might be challenging to translate these findings into novel therapeutic strategies for HCC.
Vitamin D insufficiency is common in different populations worldwide and has been associated with the presence of various types of cancer, including HCC [5]–[7]. These observations attracted considerable interest because vitamin D can be easily supplemented, with only infrequent side-effects and little costs. Yet, it has remained mostly unclear whether there is a causal association between vitamin D insufficiency and cancer development, or whether reduced vitamin D serum levels are simply surrogates for other circumstances in cancer patients (e.g. malnutrition, limited exposure to sunlight) [5]–[7]. Recently, two large GWAS have identified SNPs at three loci (GC, encoding the vitamin D binding protein; DHCR7, encoding 7-dehydrocholesterol reductase; and CYP2R1, encoding a liver 25-hydroxylase) as genetic determinants of reduced 25(OH)D3 serum levels [8], [9]. We hypothesized, that associations between these genetic variations and the occurrence of malignant or other diseases may provide a stronger argument for a causal relationship between vitamin D and the development of such diseases than the investigation of (punctual) vitamin D serum levels. In view of the high prevalence of severe vitamin D deficiency in patients with chronic hepatitis C [10], we therefore sought to investigate the association between genetic variations in CYP2R1, GC, and DHCR7 and HCV-induced HCC.
Methods
Objectives
Genetic variations in three independent genes, CYP2R1, GC, and DHCR7, are associated with life-long reduced 25(OH)D3 serum levels [9]. Vitamin D insufficiency has been associated with the occurrence of various types of cancer, but causal relationships remain elusive. Therefore, we hypothesized that genetic determinants of 25(OH)D3 serum levels may be associated with HCV-related HCC if there is a causal relationship between vitamin D metabolism and HCC development in chronic hepatitis C patients. Hence, we assessed associations between the presence of HCC in HCV infected patients and genetic variants in CYP2R1, GC, and DHCR7 in the primary analysis of the present study.
Participants
For the primary analysis of the present study, the association between HCV-induced HCC and genetic variants in CYP2R1, GC, and DHCR7, four independent cohorts of HCV-infected individuals were investigated, two cohorts including Caucasians and two cohorts including Japanese individuals. All cohorts include consecutive patients from various outpatient clinics; hence the prevalence of HCC in our cohorts is not representative of the prevalence of HCC in HCV-infected patients in general.
The first Caucasian cohort was selected from patients enrolled in the Swiss Hepatitis C Cohort Study (SCCS). The SCCS is a multicenter study pursued at 8 major Swiss hospitals and their local affiliated centers, including a total of 3,648 patients with chronic or resolved HCV infection [11]. For the present analysis, SCCS patients were included in our study if they had chronic hepatitis C, had provided written informed consent for genetic testing, had genomic DNA available for testing, if they were Caucasian, and if the duration of infection with HCV was known. Patients with hepatitis B virus infection were excluded from the discovery cohort, as well as from the replication cohorts. A second, independent cohort of Caucasian patients with chronic HCV infection with or without HCC was identified (designated as Berlin/Bonn cohort in the following); these patients were recruited at the University Hospital Departments of Gastroenterology and Hepatology in Bonn, Berlin and Leipzig in Germany, as described in Nischalke et al [12]. In addition, two independent cohorts of Japanese patients with chronic hepatitis C with or without HCC were included (a detailed description of these cohorts is provided in Miki et al. [4]; the cohorts are designated as Japanese GWAS and Japanese Replication cohort, as in Miki et al.). Importantly, the duration of infection with HCV was known in patients of the SCCS but not in the three additional cohorts.
In addition to the primary analysis of this study, a number of sub-analyses were performed in order to validate our research strategy. These sub-analyses investigated possible associations between genetic variations in CYP2R1, GC and DHCR7 and i) liver fibrosis progression rate (FPR), ii) response to treatment with pegylated interferon-α (PEG-IFN-α) and ribavirin, and iii) 25(OH)D3 serum levels. These sub-analyses were performed in SCCS patients only because of the thorough documentation of these end-points together with the known duration of infection in the SCCS. FPR was defined as a dichotomized phenotype (< vs. ≥ sex-adjusted median FPR), which was calculated on the basis of the ratio of the METAVIR fibrosis score to the estimated duration of infection in years until liver biopsy (METAVIR units per year), as described previously [13]. Hence, all SCCS patients with at least one available liver biopsy with fibrosis staging prior to antiviral treatment and with known date of infection were included in the analyses of FPR. The treatment response analyses was restricted to SCCS patients who were treated under clinical practice conditions with either PEG-IFN-α2a or PEG-IFN-α2b in combination with weight-based ribavirin, with standard treatment durations (48 weeks for HCV genotype 1 and 4, 24 weeks for HCV genotype 2 and 3), and if they had received ≥80% of the recommended dose of both agents during the first 12 weeks of therapy. SVR was defined as HCV RNA below the limit of detection in a sensitive assay ≥24 weeks after treatment completion, and all patients who failed to achieve SVR were classified as nonresponders. Serum concentrations of 25(OH)D3 were determined in all SCCS patients with chronic hepatitis C in whom a plasma sample at baseline of antiviral therapy or at the time of a liver biopsy was available. Demographic and clinical characteristics were extracted from clinical databases. High alcohol intake was defined as consumption >40 g per day over a period of ≥5 years. Liver biopsies were evaluated by experienced local pathologists. Liver fibrosis was classified according to the METAVIR score. Necroinflammatory activity was stratified into two groups, absent to mild activity vs. moderate to high activity. Steatosis was classified as absent or present. HCC was diagnosed either by biopsy or, in selected cases, by typical presentation in two independent imaging modalities. The study was approved by local ethical committees.
Description of Investigations Undertaken
In a recent GWAS, Wang et al. have identified three loci (CYP2R1, GC, and DHCR7) to be associated with reduced 25(OH)D3 serum levels [9]. Importantly, a second independent GWAS by Ahn et al. yielded similar results and confirmed the association between the three loci and 25(OH)D3 serum levels [8]. From these studies, we selected the most significant SNPs for each locus (rs1993116 and rs10741657 in CYP2R1; rs2282679 in GC; rs7944926 and rs12785878 in DHCR7). rs10741657 and rs1993116 in CYP2R1 as well as rs7944926 and rs12785878 in DHCR7 are in high LD (Table S1). Hence, these SNPs can be substituted by each other for an investigation of the association between the indicated loci and HCV-related HCC, and either rs7944926 or rs12785878 in DHCR7 and either rs10741657 or rs1993116 in CYP2R1 were genotyped in each of the four cohorts. Specific SNPs were selected according to availability in our databases, as genotyping of these SNPs was either performed in the context of previous GWA studies of our cohorts [4], [14], or by using a fluorescent-based competitive allele-specific PCR genotyping system (KBioscience, UK) using the primers listed in Table S2. SNPs in CYP27B1, which we have previously shown to be associated with SVR [10], [15], were not included in the present study as they are believed to be predominantly important during the rapid regulation of calcitriol tissue levels in inflammatory responses, and because they have no impact on 25(OH)D3 serum levels.
Measurement of 25(OH)D3 was performed as described previously [10].
Ethics
The study was approved by local ethical committees of each affiliation (Switzerland: Universitätsspital Basel, Basel; Inselspital, Bern; University Hospital Geneva, Geneva; CHUV, Lausanne; Ospedale Moncucco, Lugano; Hôpital Neuchâtelois, Neuchatel; Kantonsspital St. Gallen, St. Gallen; Universitätsspital Zürich, Zürich; Germany: University of Berlin, Berlin; University of Bonn, Bonn; Japan: University of Hiroshima, Hiroshima), and written informed consent was received from all participants.
Statistical Analyses
Testing for Hardy-Weinberg equilibrium and linkage disequilibrium (LD) was performed with the genhw and pwld packages in Stata (version 9.1, StataCorp, College Station, TX). Associations of SNPs with risk of HCV-induced HCC (dichotomic variable HCC vs. no HCC) were assessed with χ2 contingency tables, and – in the SCCS in patients with known duration of infection – in uni- and multivariate Cox regression models. Associations between SNPs and treatment outcome (SVR vs. no SVR) and with FPR (below vs. above sex-adjusted median FPR) were assessed in logistic regression models and Cox regression models, respectively. In regression models, SNPs were analyzed using an additive model (none, one or two copies of the minor allele were coded 0, 1 and 2, respectively, assuming greater effect with increased copy number of the minor allele), unless otherwise specified.
Results
Patient Characteristics
Out of a total of 3,648 patients enrolled in the SCCS, 1661 patients with known duration of infection were included in the primary analyses of the present study based on the selection criteria defined above. Out of these patients, 50 individuals developed HCC. The three additional cohorts included a total number of 1229 patients with chronic hepatitis C and HCC, as well as 2714 patients with chronic hepatitis C without HCC at the time of the analysis. Thus, the primary analysis of the present study included 1279 chronic hepatitis C patients with HCC and 4325 without HCC. Baseline characteristics of these patients are summarized in Table 1.
Table 1. Baseline characteristics of included patients.
| SCCS | Japanese GWAS | Japanese Repl. | Bonn/Berlin | |||||
| HCC | Control | HCC | Control | HCC | Control | HCC | Control | |
| N | 50 | 1611 | 310 | 1252 | 803 | 1253 | 116 | 209 |
| Age (SD)1 | 28* (15) | 21 (10) | – | – | – | – | 61*(10.6) | 48(12.4) |
| Male sex, n (%)1 | 31 (62) | 991(62) | 230 (74)* | 725 (58) | 515 (64)* | 570 (45) | 73 (58) | 114 (55) |
| Alcohol (≥40 g/d ≥5 years), n (%)2 | 3 (8) | 220 (17) | – | – | – | – | – | – |
| Diabetes, n (%) | 9 (18)* | 91 (6) | – | – | 254 (32)* | 219 (18) | – | – |
| HCV Genotype, n (%)3 | ||||||||
| 1, 4 | 27 (60) | 1014 (64) | 307 (99) | 1241 (99) | 540 (72)* | 837 (67) | – | – |
| 2, 3 | 17 (40) | 570 (36) | 2 (1) | 10 (1) | 213 (28)* | 416 (33) | – | – |
These comparisons between HCC cases and controls are statistically significant (P<0.05). Repl, replication. The cohort labeling “Japanese GWAS” and “Japanese Replication” is based on the initial description of the cohorts in Miki et al., for the present study both cohorts served as replication cohorts.
Age and sex data was missing in 5 patients in the Bonn/Berlin cohort. For the SCCS, age of infection is shown. Age of infection was unknown for the Bonn/Berlin cohort, for this cohort age at diagnosis is shown.
Alcohol consumption data was missing in 12 HCC and 290 non-HCC patients from the SCCS.
HCV genotype was missing in 5 HCC and 23 non-HCC patients from the SCCS, in 1 HCC and 2 non-HCC patients from the Japanese GWAS, and 50 HCC patients from the Japanese replication cohorts.
In addition, 963 and 750 SCCS patients were eligible to assess the impact of genetic variations in CYP2R1, GC, and DHCR7 on FPR and on the outcome of standard treatment with PEG-IFN-α and ribavirin, respectively. Serum levels of 25(OH)D3 were only available in a minority for 496 SCCS patients.
Association between Genetic Determinants of 25(OH)D3 Serum Levels and HCV-induced HCC
Median 25(OH)D3 serum levels in patients with chronic hepatitis C with or without HCC in the SCCS were 12.7 and 14.3 ng/mL, (range 4.9–40.9 and 3.9–76.9) respectively (P = 0.19, values available in 496 patients). However, 25(OH)D3 serum levels fluctuate strongly during seasons and as a consequence of numerous circumstances such as exposure to sunlight, nutrition, accompanying diseases, vitamin D supplementation and others. We therefore believe that genetic variants in CYP2R1, GC, and DHCR7, with their proven impact on 25(OH)D3 serum levels, should be used as surrogates for long-term 25(OH)D3 serum levels. Hence, we genotyped the most relevant tagging SNPs for these loci (rs1993116/rs10741657 for CYP2R1, rs2282679 for GC, rs7944926/rs12785878 for DHCR7; data on LD and Hardy Weinberg equilibrium for these SNPs are shown in Table S1), and assessed their association with HCV-related HCC.
Table 2 summarizes results of the primary analysis for associations between HCV-related HCC and genetic variations in CYP2R1, GC, and DHCR7 in the four independent patient cohorts. In the combined analyses of these cohorts, the strongest association with HCV-induced HCC was found for GC (P = 0.007, OR = 1.56, 95% CI = 1.12–2.15) and DHCR7 (P = 0.003, OR = 1.42, 95% CI = 1.13–1.78), whereas CYP2R1 was almost significantly associated with HCV-induced HCC (P = 0.07, OR = 1.13, 95% CI = 0.99–1.28). Remarkably, in each independent cohort, frequencies and ORs for the risk alleles at each locus showed an association in the similar direction, even though statistical significance was not reached for all subgroups.
Table 2. Summary of associations between SNPs in CYP2R1, GC, and DHCR7, and HCV-related hepatocellular carcinoma development.
| CYP2R1 | ||||||||||||
| Cases | Controls | Risk allele frequencies | ||||||||||
| SNP | Study | Allele 1/2 | 11 | 12 | 22 | 11 | 12 | 22 | Case | Control | P | OR (95% CI) |
| rs1993116 | SCCS | A/G | 6 | 16 | 28 | 199 | 774 | 634 | 0.72 | 0.64 | 0.02 | 1.95 (1.18–3.41) |
| rs1993116 | JapaneseGWAS | A/G | 41 | 136 | 133 | 163 | 621 | 468 | 0.65 | 0.62 | 0.07 | 1.26 (0.98–1.61) |
| rs107416571 | Japanese Replication | A/G | 106 | 377 | 320 | 174 | 597 | 482 | 0.63 | 0.62 | 0.5 | 1.06 (0.88–1.27) |
| rs1993116 | Bonn-Berlin | A/G | 17 | 48 | 47 | 25 | 98 | 81 | 0.63 | 0.64 | 0.7 | 1.10 (0.67–1.76) |
| Combined | A/G | 170 | 577 | 528 | 561 | 2090 | 1665 | 0.64 | 0.63 | 0.07 | 1.13 (0.99–1.28) | |
Allele 2 indicates the risk allele, according to Wang et al. [9]. P-values and ORs were calculated for risk genotypes using favorable genotypes as a reference, i.e. for CYP2R1 by comparing GG vs. GA/AA genotypes, for GC by comparing TT/TG vs. GG genotypes, and for DHCR7 by comparing TT vs. TC/CC genotypes.
Only patients from the SCCS and Japanese GWAS were included in the combined analysis for this locus, because of the different allele frequencies for rs12785878 in Japanese patients compared to Caucasian patients. Data remain significant after inclusion of the Bonn-Berlin cohort (P = 0.018, OR = 1.27 [95% CI = 1.04–1.56]). *Genotyping of this SNP failed in the Japanese Replication cohort due to limited amounts of DNA. Please note that the total number of patients with available genotypes is not equal between different loci due to limited amount of DNA or genotyping failure in some cases.
rs1993116 and rs10741657 are in complete LD in the Caucasian and Japanese population (R2 = 0.95 and = 1.00, respectively), the major alleles of both SNPs have a similar impact on 25(OH)D3 serum levels, indicating that both SNPs can be used equivalently [9].
rs7944926 and rs12785878 are in complete LD in the Caucasian and Japanese population (R2 = 1.00 and = 1.00, respectively), the major alleles of both SNPs have a similar impact on 25(OH)D3 serum levels, indicating that both SNPs can be used equivalently [9].
Of note, genotyping of rs1287578 in DHCR7 revealed huge differences of T allele frequencies between Caucasian and Japanese cohorts. These differences are in line with allele frequencies reported in HapMap. Since previous GWAS on vitamin D serum levels did not include relevant numbers of Asian individuals, a functional interpretation of rs1287578 genotyping data in Japanese appears to be difficult, as the risk allele for this SNP in Asians has not yet been clearly identified. Therefore, data for rs1287578 genotype in the Japanese replication cohort were not included in the combined analysis shown in Table 2.
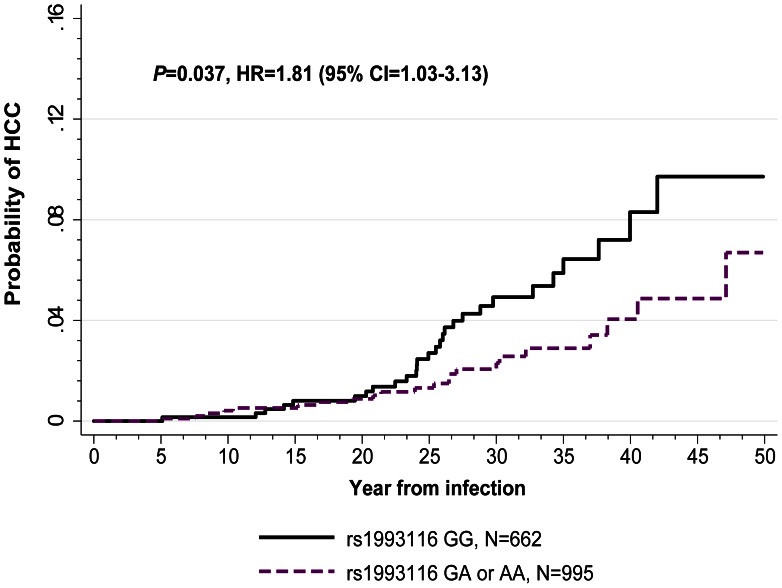
The known duration of infection allowed an additional cox regression analyses of the risk of HCC in patients from the SCCS. Figure 1 shows the cumulative incidence of HCC in patients with the CYP2R1 risk genotype rs1993116 GG compared to those with the favorable genotypes GA and AA (P = 0.037, hazard ratio (HR) = 1.81 (95% confidence interval (CI) = 1.03–3.13).
Figure 1. Risk of hepatocellular carcinoma (HCC) development in SCCS patients with chronic hepatitis C and known duration of infection, according to CYP2R1 rs1993116 genotypes.
The probability to develop HCC from the time of hepatitis C virus infection by CYP2R1 rs1993116 genotypes (GG vs. GA/AA) was assessed by using cumulative incidence curves, with censoring of data at the date of last follow-up or death. Statistics are shown for univariate Cox regression analysis. CI, confidence interval; HR, hazard ratio.
To exclude a possible selection bias within the SCCS, a subanalysis was performed in which the inclusion criterion “known duration of infection”, which was specific for the SCCS, was omitted. As shown in Table S3, results of this case-control study are largely comparable to the primary analysis of our study.
Association between Genetic Determinants of 25(OH)D3 Serum Levels and Liver Fibrosis Progression Rate (FPR) and Treatment Outcome
Thus far, we cannot completely exclude that the above described associations between genetic determinants of reduced 25(OH)D3 serum levels and HCV-induced HCC are primarily mediated by an effect of the indicated SNPs on FPR or treatment outcome. We therefore performed sub-analyses to test whether FPR or treatment outcome are associated with variations in CYP2R1, GC and DCHR7. In 963 SCCS patients in whom FPR could be calculated, none of the SNPs was significantly associated with slow vs. fast FPR (P = 0.2 for rs1993116 in CYP2R1; P = 0.5 for rs2282679 in GC, and P = 0.3 for rs7944926 in DHCR7; Table 3). In addition, in 750 SCCS patients who had received standard therapy with PEG-IFN-α and ribavirin, no significant associations were found between SNPs in CYP2R1, GC and DHCR7 and treatment outcome (SVR vs. no SVR; P = 0.9, 0.4, 0.2, respectively; Table 4), suggesting that the observed associations between these loci and HCC are specific for (HCV-induced) hepatocarcinogenesis. Finally, we calculated 25(OH)D3 serum levels according to CYP2R1, GC, and DHCR7 genotypes. 25(OH)D3 serum levels were 14.9, 13.4 and 12.4 ng/mL in patients with CYP2R1 rs1993116 genotype AA, AG, and GG (P = 0.41), respectively; 14.4, 14.2, 11.7 in patients with GC rs2282679 genotype TT, TG, GG (P = 0.037); and 14.3, 14.4, and 13.1 in patients with DHCR7 rs7944926 genotype TT, TC, and CC (P = 0.44).
Table 3. Association of SNPs in CYP2R1, GC, and DHCR7 with liver fibrosis progression rate (FPR) in patients with chronic hepatitis C.
| Patients, n (%) | ||||||
| Gene | SNP | Gt | Slow FPR* | Fast FPR* | P | OR (95% CI) |
| CYP2R1 | rs1993116 | GG | 183 (41) | 192 (37) | ||
| GA | 213 (48) | 263 (51) | 0.2 | 1.20 (0.93–1.56) | ||
| AA | 47 (11) | 65 (13) | ||||
| GC | rs2282679 | TT | 236 (53) | 266 (51) | ||
| TG | 175 (40) | 217 (42) | 0.5 | 1.08 (0.84–1.39) | ||
| GG | 31 (7) | 34 (7) | ||||
| DHCR7 | rs7944926 | TT | 160 (54) | 221 (58) | ||
| TC | 115 (39) | 139 (36) | 0.3 | 0.85 (0.63–1.16) | ||
| CC | 22 (7) | 22 (6) | ||||
FPR, liver fibrosis progression rate. Slow vs. fast FPR was defined as FPR<vs. ≥ sex-adjusted median FPR in METAVIR-units per year. Statistics are shown for logistic regression analyses based on the additive model of inheritance. The usage of other models of inheritance (recessive, dominant) did not result in significant associations as well (not shown).
Table 4. Association of SNPs in CYP2R1, GC, and DHCR7 with response to treatment of pegylated interferon-α and ribavirin in patients with chronic hepatitis C.
| Patients, n (%) | ||||||
| Gene | SNP | Gt | without SVR | with SVR | P | OR (95% CI) |
| CYP2R1 | rs1993116 | GG | 109 (38) | 171 (37) | ||
| GA | 138 (48) | 238 (52) | 0.9 | 1.03 (0.76–1.39) | ||
| AA | 42 (15) | 52 (11) | ||||
| GC | rs2282679 | TT | 166 (57) | 248 (54) | ||
| TG | 105 (36) | 186 (40) | 0.42 | 1.14 (0.85–1.54) | ||
| GG | 19 (7) | 26 (6) | ||||
| DHCR7 | rs7944926 | TT | 127 (53) | 230 (58) | ||
| TC | 95 (40) | 143 (36) | 0.2 | 0.81 (0.59–1.12) | ||
| CC | 17 (7) | 22 (6) | ||||
Statistics are shown for logistic regression analyses based on the additive model of inheritance. The usage of other models of inheritance (recessive, dominant) did not result in significant associations as well (not shown).
Discussion
The present study shows that genetic variations in CYP2R1, GC, and DHCR7 are associated with progression to HCC in patients with chronic hepatitis C. The SNPs investigated in CYP2R1, GC, and DHCR7 have been identified by two independent GWAS as relevant genetic determinants of reduced 25(OH)D3 serum levels [8], [9]. Although genetic association studies cannot prove causal relationships between genetic variations and specific phenotypes, the association here identified between three genetically independent, but functionally related susceptibility loci of vitamin D deficiency and HCV-induced HCC suggest that an impaired vitamin D metabolism (functionally) contributes to hepatocarcinogenesis in HCV-infected patients. Importantly, the lack of association of these SNPs in CYP2R1, GC, and DHCR7 with FPR and response to treatment with PEG-IFN-α and ribavirin suggests a specific effect of these genetic variations on HCV-induced hepatocarcinogenesis.
Vitamin D insufficiency has been previously linked to the development of HCC [16]. However, causal relationships remained mostly unclear because these studies were small or concentrated on the assessment of 25(OH)D3 serum levels at the date of HCC, which may result in false-positive associations due the influence of impaired liver function on circulating 25(OH)D3 [5], [16]–[18]. In addition, randomized controlled clinical trials evaluating the effect of vitamin D or its analogues on HCC development are lacking. Thus, the results of the present, large-scale genetic association study add a significant argument to evaluate the possible chemo-preventive effect of vitamin D supplementation in HCV-infected patients. However, although our data suggest a causal role of vitamin D metabolism in HCV-induced HCC, there is currently no published evidence that vitamin D supplementation would translate into a benefit in HCV-infected patients with advanced liver disease. In this regard, caution is advisable since some placebo-controlled studies evaluating the supplementation of other vitamins have reported a reduced overall survival rate in some verum groups [19]. Yet, in view of our present data, and in view of the high prevalence of severe vitamin D deficiency in patients with chronic hepatitis C [10], [20], [21], randomized controlled clinical trials in HCV-infected patients with advanced liver disease appear to be justified.
Limitations
Our study has potential limitations. First, 25(OH)D3 serum levels were only available in a relatively small subgroup of patients. Nevertheless, we observe the same trends for 25(OH)D3 serum levels according to CYP2R1, GC, and DHCR7 genotypes as described previously, and HCV-infected patients with HCC had slightly lower 25(OH)D3 serum levels compared to those without HCC. More important, we believe that analyses of associations between punctual 25(OH)D3 serum levels and endpoints such as HCC can be misleading, since 25(OH)D3 serum levels strongly fluctuate during seasons, with age, and as a consequence of numerous other conditions (liver fibrosis, diabetes, obesity, supplementation, etc.) [6], [7]. A second limitation of our study is the relatively weak level of statistical significance in the analyses of the individual cohorts, with the consequence that associations for the three loci with HCC became only fully significant in the combined analyses of all included patients. Most likely, this can be explained by the relatively low frequency of the risk genotypes (especially of GC and DHCR7), as well as by the numerous variables with influence 25(OH)D3 serum levels in a given individual [9]. In this regard, it is important to note that ORs for the risk genotype of all genes (CYP2R1, GC, and DHCR7) were similarly directed in the discovery cohort as well as in all replication cohorts, confirming a true association between these loci and HCV-induced HCC. In addition, the strengths of the associations between these loci and HCV-induced HCC were largely comparable to the strength of the associations between these loci and 25(OH)D3 serum levels (strong effect of GC and DHCR7, moderate effect of CYP2R1, according to Wang et al.) [9]. Nevertheless, the observed subtle differences between the different cohorts included in our study may not only be explained by the relatively small sample size of individual cohorts, but also by specific cohort features such as different recruitment strategies (e.g. cohort study versus case-control studies). In this regard, our study also cannot fully rule out whether all investigated genes (CYP2R1, GC, DHCR7) have the same impact on progression to HCV-related HCC at different stages of liver disease or in populations of different ancestries. Furthermore, our study cannot clearly characterize whether the observed findings apply for patients who did or did not respond to antiviral therapy. Though we show in the SCCS (in a minority of the whole study population), that SNPs in CYP2R1, GC, and DHCR7 were not associated with treatment outcome, it remains unclear whether these genetic variations are associated with HCC in both individuals with or without treatment-induced eradication of HCV.
The heterogeneity of the four independent cohorts included in our analyses might be perceived as another limitation. This applies for important cohort features such as race, recording of treatment modalities, HCC frequency, or different allele frequencies for some SNPs (especially rs1278578).
Conclusions
In conclusion, we provide evidence for a functionally relevant contribution of reduced 25(OH)D3 serum levels to HCV-induced HCC. Controlled clinical trials to evaluate the impact of vitamin D supplementation on HCC risk and overall survival in patients with chronic hepatitis C appear to be justified.
Supporting Information
Linkage disequilibrium of SNPs in CYP2R1 , GC , and DHCR7 investigated in the present study.
(DOC)
Primers for SNP genotyping assays.
(DOC)
Summary of associations between SNPs in CYP2R1 , GC , and DHCR7 , and HCV-related hepatocellular carcinoma development, considering the SCCS as case-control study.
(DOC)
Acknowledgments
The authors express their gratitude to Doris Kärger for expert technical assistance.
The members of the Swiss Hepatitis C Cohort Study Group are Francesco Negro (Geneva, Chairman), Antoine Hadengue (Geneva, Chairman of Scientific Committee), Laurent Kaiser, Laura Rubbia-Brandt (Geneva); Darius Moradpour, Cristina Cellerai (Lausanne); Martin Rickenbach (Lausanne Data Center); Andreas Cerny, Gladys Martinetti (Lugano); Jean-François Dufour, Meri Gorgievski, Virginie Masserey Spicher (Berne); Markus Heim, Hans Hirsch (Basel); Beat Müllhaupt, Beat Helbling, Stephan Regenass (Zurich); Raffaele Malinverni (Neuchâtel); David Semela, Guenter Dollenmaier (St Gallen); Gieri Cathomas (Liestal).
Funding Statement
This work was supported by the Swiss National Science Foundation (3100A0-122447 to DM, 32003B-127613 to PYB as well as 3347C0-108782/1 and 33CSC0-108782/2 to the SCCS), the Leenaards Foundation (to PYB), the European Community's FP7 (260844, to PYB), and the Santos-Suarez Foundation (to PYB). CML is supported by the Deutsche Forschungsgemeinschaft (LA 2806/2-1), by the Johann Wolfgang Goethe University (Förderung Nachwuchsforscher 2012), and by the Deutsche Leberstifung (S163/10087/2012). TB was supported by the German Competence Network for Viral Hepatitis (Hep-Net), funded by the German Ministry of Education and Research (BMBF, Grant No. 01 KI 0437, Project No. 10.1.3 and Core Project No. 10.1 Genetic host factors in viral hepatitis and Genetic Epidemiology Group in viral hepatitis), and by the BMBF Project: Host and viral determinants for susceptibility and resistance to hepatitis C virus infection (Grant No. 01KI0787). HDN and US were funded by the Deutsche Krebshilfe (107865). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nature Outlook (2011) Hepatitis C. Nature. 474: S1–S21. [DOI] [PubMed] [Google Scholar]
- 2. Forner A, Llovet JM, Bruix J (2012) Hepatocellular carcinoma. Lancet 371: 1245–55. [DOI] [PubMed] [Google Scholar]
- 3. Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, et al. (2011) Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet 43: 455–458. [DOI] [PubMed] [Google Scholar]
- 4. Miki D, Ochi H, Hayes CN, Abe H, Yoshima T, et al. (2011) Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet 43: 797–800. [DOI] [PubMed] [Google Scholar]
- 5. Campbell FC, Xu H, El-Tanani M, Crowe P, Bingham V (2010) The yin and yang of vitamin D receptor (VDR) signaling in neoplastic progression: operational networks and tissue-specific growth control. Biochem Pharmacol 79: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holick MF (2007) Vitamin D deficiency. N Engl J Med 357: 266–281. [DOI] [PubMed] [Google Scholar]
- 7. Rosen CJ (2011) Clinical practice. Vitamin D insufficiency. N Engl J Med 364: 248–254. [DOI] [PubMed] [Google Scholar]
- 8. Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, et al. (2010) Genome-wide association study of circulating vitamin D levels. Hum Mol Genet 19: 2739–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, et al. (2010) Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 376: 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lange CM, Bojunga J, Ramos-Lopez E, von Wagner M, Hassler A, et al. (2011) Vitamin D deficiency and a CYP27B1–1260 promoter polymorphism are associated with chronic hepatitis C and poor response to interferon-alfa based therapy. J Hepatol 54: 887–893. [DOI] [PubMed] [Google Scholar]
- 11. Prasad L, Spicher VM, Zwahlen M, Rickenbach M, Helbling B, et al. (2007) Cohort Profile: the Swiss Hepatitis C Cohort Study (SCCS). Int J Epidemiol 36: 731–737. [DOI] [PubMed] [Google Scholar]
- 12. Nischalke HD, Coenen M, Berger C, Aldenhoff K, Muller T, et al. (2012) The toll-like receptor 2 (TLR2) −196 to −174 del/ins polymorphism affects viral loads and susceptibility to hepatocellular carcinoma in chronic hepatitis C. Int J Cancer. 130: 1470–1475. [DOI] [PubMed] [Google Scholar]
- 13. Bochud PY, Cai T, Overbeck K, Bochud M, Dufour JF, et al. (2009) Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J Hepatol. 51: 655–666. [DOI] [PubMed] [Google Scholar]
- 14. Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, et al. (2010) Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138: 1338–1345. [DOI] [PubMed] [Google Scholar]
- 15. Lange CM, Bibert S, Kutalik Z, Burgisser P, Cerny A, et al. (2012) A genetic validation study reveals a role of vitamin D metabolism in the response to interferon-alfa-based therapy of chronic hepatitis C. PLoS One. 7: e40159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chiang KC, Yeh CN, Chen MF, Chen TC (2011) Hepatocellular carcinoma and vitamin D: a review. J Gastroenterol Hepatol 26: 1597–1603. [DOI] [PubMed] [Google Scholar]
- 17. Dalhoff K, Dancey J, Astrup L, Skovsgaard T, Hamberg KJ, et al. (2003) A phase II study of the vitamin D analogue Seocalcitol in patients with inoperable hepatocellular carcinoma. Br J Cancer 89: 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Falleti E, Bitetto D, Fabris C, Cussigh A, Fontanini E, et al. (2010) Vitamin D receptor gene polymorphisms and hepatocellular carcinoma in alcoholic cirrhosis. World J Gastroenterol 16: 3016–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Virtamo J, Pietinen P, Huttunen JK, Korhonen P, Malila N, et al. (2003) Incidence of cancer and mortality following alpha-tocopherol and beta-carotene supplementation: a postintervention follow-up. JAMA 290: 476–485. [DOI] [PubMed] [Google Scholar]
- 20. Bouillon R, Auwerx J, Dekeyser L, Fevery J, Lissens W, et al. (1984) Serum vitamin D metabolites and their binding protein in patients with liver cirrhosis. J Clin Endocrinol Metab 59: 86–89. [DOI] [PubMed] [Google Scholar]
- 21. Petta S, Camma C, Scazzone C, Tripodo C, Di Marco V, et al. (2010) Low vitamin D serum level is related to severe fibrosis and low responsiveness to interferon-based therapy in genotype 1 chronic hepatitis C. Hepatology. 51: 1158–1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linkage disequilibrium of SNPs in CYP2R1 , GC , and DHCR7 investigated in the present study.
(DOC)
Primers for SNP genotyping assays.
(DOC)
Summary of associations between SNPs in CYP2R1 , GC , and DHCR7 , and HCV-related hepatocellular carcinoma development, considering the SCCS as case-control study.
(DOC)