Abstract
An HIV-1 infection progresses in most human individuals sooner or later into AIDS, a devastating disease that kills more than a million people worldwide on an annual basis. Nonetheless, certain HIV-1-infected persons appear to act as long-term non-progressors, and elite control is associated with the presence of particular MHC class I allotypes such as HLA-B*27 or -B*57. The HIV-1 pandemic in humans arose from the cross-species transmission of SIVcpz originating from chimpanzees. Chimpanzees, however, appear to be relatively resistant to developing AIDS after HIV-1/SIVcpz infection. Mounting evidence illustrates that, in the distant past, chimpanzees experienced a selective sweep resulting in a severe reduction of their MHC class I repertoire. This was most likely caused by an HIV-1/SIV-like retrovirus, suggesting that chimpanzees may have experienced long-lasting host-virus relationships with SIV-like viruses. Hence, if natural selection is allowed to follow its course, prospects for the human population may look grim, thus underscoring the desperate need for an effective vaccine.
Keywords: AIDS, Chimpanzee, HIV-1, HLA, Human, MHC, Repertoire reduction, SIVcpz, Zoonosis
Review
AIDS: facts and figures
Acquired immunodeficiency syndrome (AIDS) is caused by the human immunodeficiency virus type 1 (HIV-1) and poses a major threat to human health. At present, over thirty million individuals are infected worldwide, and more than twenty five million people have died from AIDS since the beginning of the pandemic (UNAIDS 2011). Based on genetic characteristics, the virus strains can be clustered into different groups designated M, N, O and P. HIV-1 group M is responsible for the vast majority of infections recorded in human individuals globally [1,2]. Thanks to the currently available anti-retroviral (AR) drugs, the number of people dying yearly from AIDS reached a plateau around 2004, and the number has even begun to decline (UNAIDS 2011). However, it is important to mention that highly active anti-retroviral therapy (HAART) does not eradicate HIV-1 [3], and several studies have demonstrated that discontinuation of HAART may result in a rapid viral rebound [4-7]. Moreover, AR therapy carries with it high costs, resulting in only a limited number of HIV-1-infected individuals having access to it. Especially in developing countries, where most of the infected individuals live, AIDS is still a prominent health problem. Another worry is that sooner or later the virus may develop resistance to these drugs [8], and the spread of drug-resistant HIV-1 strains would be devastating. Therefore, a series of low-cost drugs and, more optimistically, an efficacious vaccine able to protect against infection are urgently needed.
HIV-1 in a nutshell: its genome and infection routes
The genome of HIV-1 is relatively small, approximately 10 kilobases, and encodes a limited number of gene products (Figure 1). This retrovirus belongs to the group of lentiviruses. Like most retroviruses, the HIV-1 genome contains the three functional proteins Gag, Pol, and Env, which are essential for the construction of the virus particle [9]. In addition, lentiviral genomes encode a number of regulatory (Tat and Rev) and accessory genes (Vif, Vpr, Vpu, and Nef). The Tat and Rev proteins are essential for replication of the virus. The accessory proteins are attached to viral messenger RNA, but they are not required for replication of the virus in vitro. However, in vivo these proteins are necessary for the replication and virulence of the virus [10].
Figure 1.
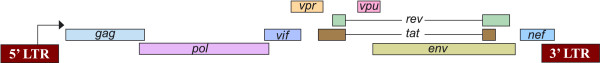
Schematic representation of the HIV-1 genome and its gene products. The arrow indicates the transcription initiation site.
HIV-1 possesses specific characteristics that hamper or have hampered the development of broadly efficacious and efficient anti-retroviral drugs and vaccines. On the one hand, the virus can cause a persistent infection and hide in an inactive form in cellular reservoirs. On the other hand, it can replicate quickly, and − depending on the copy number present in one individual − may produce 109 to 1010 particles per day [11,12]. Moreover, the genome of HIV-1 encodes an error-prone reverse transcriptase characterized by a high mutation rate (approximately 3 × 10-5 per nucleotide/replication cycle) [13]. This, in combination with a relatively small genome size and the high viral production rate within a given individual, may ultimately result in the establishment of a virus swarm. There is also the risk of emergence of new recombinant strains as multiple HIV-1 viruses, which are prone to exchange genetic material, infect a single cell. Hence, a high mutation rate in concert with recombination provides the biological ability for HIV-1 to escape the immune recognition/control of its host.
HIV-1 enters the human body via mucosal sites, blood-blood contact, or breast milk [14]. After entry, the virus either directly infects CD4+ T cells, is recruited by dendritic cells (DC), or is taken up by macrophages. The latter two cell types act as a kind of Trojan horses, and transport HIV-1 through different layers of epithelial cells to the lymphoid tissues, where they can transfer the virus to CD4+ T cells [15,16]. DCs can either internalize HIV-1 into endosomes, and the cell-surface molecule that is used by the virus for entry into the cell is the C-type lectin receptor DC-SIGN [17], or they can recruit HIV-1 virions to their endosomal compartment, where the virions accumulate at regions that come into contact with neighboring T cells (referred to as the infectious synapse) [18]. Macrophages may phagocytize HIV-1, and can either function as a reservoir for long-term persistence of the virus or can be responsible for the transport and dissemination of the virus. The “primary” non-syncytium-inducing (NSI) HIV-1 viruses mainly infect CD4+ memory T cells that express CC-chemokine receptor-5 (CCR5). Later in the course of the infection, the NSI strains can switch to a syncytium-inducing (SI) phenotype, which can infect T cells by using the C-X-C-chemokine receptor-4 (CXCR4) as co-receptor. This switch is associated with a loss of sensitivity to chemokines (like RANTES, and MIP-1α and β), a rapid decrease in CD4+ T cells, and progression towards AIDS [19]. Moreover, an infection of memory CD4+ T cells that revert back to a resting state ensues as well, resulting in a dormant stage of the virus that is undetectable by the host’s immune system [20]. In essence, HIV-1 infection and its subsequent manipulations take place at the heart of the immune system.
Zoonotic infections: what may happen if we open Pandora’s box
More than 40 African non-human primate species are infected naturally with various simian immunodeficiency virus (SIV) strains, but progression towards AIDS is rarely observed [21,22]. From among these natural primate hosts, the SIV infection has been most thoroughly studied in three species, sooty mangabeys, African green monkeys, and mandrills, and has led to the insight that non-pathogenicity need not be linked to effective immune control. Apparently, many of the species have co-evolved with SIV infections, and have found ways to manage/control it. These non-human primates, however, are potential reservoirs for viruses that could be spread among related species. One example of such a zoonotic transmission is provided by sooty mangabeys (Cercocebus atys). This species, infected naturally with a SIV strain (SIVsm), is considered to represent the origin of the HIV-2 infection in humans, causing an AIDS-like disease [23]. The infection in humans is mostly confined to the West African territory, and indeed overlaps with the natural habitat of sooty mangabeys.
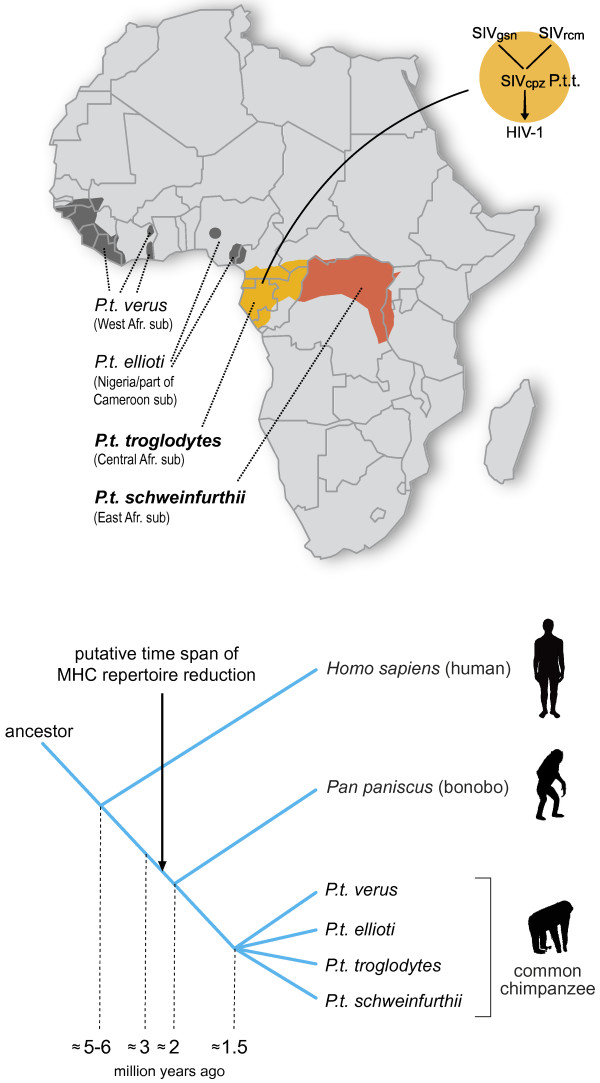
Chimpanzees (Pan troglodytes), humankind’s closest living relative, can also be infected with SIV [24]. Based on the geographic distribution as well as on morphological and genetic data, chimpanzees have been divided into four different subspecies/populations sharing a common ancestor approximately 1.5 million years ago [25-27], and incidences of natural SIVcpz infection have been recorded in contemporary animals from at least two of the four subspecies (Figure 2). These SIVcpz-strains appear to have a mosaic genome, consisting of gene segments from different types of SIV strains obtained from distinct Old World monkey species [28]. This suggests that chimpanzees have acquired their types of SIV infections by predation on Old World monkeys sharing the same habitat. In turn, the different HIV-1 groups M, N, O and P, have arisen from at least four separate introductions of SIV into the human population. The initiator of the human pandemic (HIV-1 group M) is the SIVcpz strain derived from chimpanzees of the Central-African subspecies (Pan troglodytes troglodytes) [29] (Figure 2), and the HIV-1 group N also arose from an SIVcpz strain from this species. The HIV-1 groups O and P most likely originated from gorillas (SIVgor) [30,31], in which there is an indication that P.t.t. animals are the source of SIVgor[32].
Figure 2.
Map of the African continent, highlighting habitats of the four different chimpanzee subspecies. Populations that show evidence of contemporary natural infections with SIVcpz strains are Pan troglodytes troglodytes (light orange) and P.t. schweinfurthii (dark orange). Superimposed is a diagram illustrating the cross-species transmission events that led to the emergence of HIV-1 group M, the initiator of the human pandemic [28]. The lower panel in the figure illustrates the speciation events in the homo-pan lineage. The arrow indicates the putative time span of the MHC repertoire reduction in the pan lineage.
Chimpanzees and AIDS
Apart from humans, chimpanzees are one of the non-human primate species that is susceptible to infection with HIV-1. Recently, it has been shown that pig-tailed macaques can also be infected through a modified HIV-1 strain that only differs in the vif gene [33,34]. In the past, a substantial number of West-African chimpanzees (P.t.verus) were experimentally infected with different HIV-1 strains, but most of these animals seem to be relatively resistant to developing AIDS [35,36]. These experiments were conducted before it was known that HIV-1 was derived from a zoonotic transmission of a virus that has its roots in a chimpanzee reservoir. In the late 1980s and early 1990s, the first chimpanzees to be infected naturally with an HIV-1-related lentivirus were discovered [37-39]. Additional natural SIVcpz infections in captive and wild-living chimpanzees were described later [40,41]. Some of these chimpanzees were kept under close surveillance for long periods, but signs of an AIDS-like disease were not seen. Moreover, transmission of one of the viruses to West-African chimpanzees (animals in which natural infection with SIVcpz is not observed) also did not result in the progression to AIDS [42]. Recently, population analyses have shown that wild-ranging East-African chimpanzees (P.t. schweinfurthii) experience natural infections with SIVcpz. For one of the populations, evidence of AIDS-like symptoms has been documented [43], and AIDS-related symptoms have also been described for a naturally infected Central-African chimpanzee [44]. In addition to these observations of AIDS in SIVcpz-related infections, there is one case documenting that chimpanzees can develop AIDS after experimental infection with HIV-1 [45]. This chimpanzee of West-African origin was infected with three different HIV-1 isolates. The animal eventually developed AIDS, and subsequent analysis showed that a recombinant HIV-1 strain had emerged. Three other chimpanzees that were afterwards infected with this recombinant strain all developed signs of AIDS-like disease [46]. This set of experiments shows that viruses that have been edited in the human population, and that are re-introduced into their original host species, can cause AIDS. Thus, escape from immune control in chimpanzees is possible in exceptional cases, even within a species considered to be relatively resistant.
Control of AIDS development in humans: an introduction to the MHC
In HIV-1-infected human cohorts, certain individuals do not progress towards AIDS. Some of these long-term non-progressors (LTNP)/elite controllers (ECs) have been infected for more than two decades [47], and their situation appears to resemble that of HIV-1-infected chimpanzees in terms of their relative resistance to developing AIDS. The LTNP/EC status in humans is strongly associated with the presence of particular major histocompatibility complex (MHC) class I molecules, such as HLA-B*27:05 and -B*57:01 [48-54].
The MHC plays a central role in the induction of adaptive immune responses, and encodes two clusters of cell-surface proteins. In humans, these molecules are designated HLA-A, -B, and -C (class I cluster), and -DP, -DQ, and -DR (class II cluster), and almost all these genes display abundant levels of polymorphism [55]. MHC class I antigens are expressed on virtually all nucleated cells, and generally present peptides of 8-10 amino acids in length. One of the biological functions of MHC class I molecules is to sample degraded peptide fragments from pathogens that are able to establish intracellular infections. In most cases, such peptides are derived from viruses trying to manipulate the intracellular machinery of the host cell in favor of their own replication purposes. MHC class II molecules, on the other hand, display differential tissue distribution and usually bind substantially longer peptides. They sample degraded peptide fragments from pathogens that are establishing extra-cellular infections, or that have life stages that are taking place outside of the host cell. In the case of an intracellular infection, the immune system will try to kill the target cell by inducing apoptosis-mediated lysis by CD8+ cytotoxic T cells (CTL). Extra-cellular infections are usually eliminated by antibodies that are generated by B cells under the control of CD4+ T helper cells.
In order to initiate an immune response, each individual will present a range of antigenic peptides as dictated by their unique HLA repertoire. Thus, MHC polymorphism warrants that many individuals within a given population have the capacity to generate unique adaptive immune responses to eliminate infections. In other words, MHC polymorphism minimizes the chance that a given pathogen will eliminate an entire population. Several pathogens have evolved different ways to interfere with the MHC class I and II antigen presentation pathways in order to avoid immune recognition [56-59]. Indeed, it has been suggested that in human populations exposed to HIV-1 there is enrichment for particular HLA specificities [60,61].
The MHC and resistance to the development of AIDS in chimpanzees
Grosso modo, humans and chimpanzees share the same MHC class I and II loci. However, chimpanzees possess an additional oligomorphic locus designated Patr-AL (A-like), which is characterized by a differential haplotype distribution and low expression levels [62,63]. An overview of the reported number of alleles for each locus is provided (Table 1). With regard to lineages, in comparison to humans, chimpanzees have a reduced MHC class I repertoire. The very first indications that chimpanzees may lack the evolutionary equivalents of particular HLA lineages became evident in serological studies [64]. In humans, the HLA-A locus alleles are divided into two lineages comprising six families (Table 2) [65,66], and the experiments of van Rood et al.[64] showed that chimpanzees lack the HLA-A2 related serotypes. Subsequent molecular studies suggested that chimpanzees might only possess the equivalents of the HLA-A1/A3/A11/A30 family [67]. Although the number of animals tested in this initial study was relatively small, the outcome was unexpected. Humans and chimpanzees are each other’s closest living relatives, and they shared a common ancestor about 5 million years ago [68]. Thus, the most logical explanation would be that humans and chimpanzees share for the A and B loci the same lineages, since they were inherited in a trans-species mode of evolution [69].
Table 1.
Number of alleles detected for Mhc class I and II loci in humans (HLA) and chimpanzees (Patr)
| Locus | HLA | Patr | |
|---|---|---|---|
| |
A |
2188 |
33 |
|
Mhc class I |
B |
2862 |
57 |
| |
C |
1746 |
31 |
| |
E |
11 |
2 |
| |
F |
22 |
1 |
| |
G |
50 |
1 |
| |
AL |
0 |
5 |
| |
DPA1 |
36 |
3 |
| |
DPB1 |
159 |
29 |
|
Mhc class II |
DQA1 |
49 |
7 |
| |
DQB1 |
193 |
10 |
| |
DRA |
7 |
2 |
| |
DRB |
1386 |
79 |
| #Individuals analyzed | >> | ≈100 |
Data were retrieved from the databases http://www.ebi.ac.uk/imgt/hla[70] and http://www.ebi.ac.uk/ipd/mhc/nhp[71].
Table 2.
Subdivision of the HLA-A locus members into lineages and families
| Lineage | Family |
|---|---|
| |
HLA-A2 |
|
A2 |
HLA-A10 |
| |
HLA-A19 |
| |
HLA-A1/A3/A11/A30 |
|
A3 |
HLA-A9 |
| HLA-A80 |
As a follow-up to McAdam’s study [67], the MHC class I repertoire of a pedigreed West-African chimpanzee colony comprising more than 30 wild-caught founder animals was analyzed. Again, only orthologs of the HLA-A1/A3/A11/A30 family were encountered [72]. Moreover, MHC analyses of West-African chimpanzees by other research teams, and of small numbers of Central- and East-African chimpanzees, also provided data indicating only the presence of HLA-A1/A3/A11/A30 family orthologs [72-76]. Although the results suggest that there was a repertoire reduction at the MHC class I region in chimpanzees, conclusions in this direction should be arrived at cautiously. The presence of one particular omnipresent HLA-A-like family in chimpanzees can, in theory, also be explained by convergent evolution. Additionally, the imbalance in sample size between the number of chimpanzees and humans analyzed for their MHC repertoire can hamper an accurate interpretation of the data. For instance, one can argue that particular alleles and/or lineages were missed in chimpanzees due to a limited sample size. In addition, the MHC class I and II gene products of different species/populations may experience different sorts of natural selection, depending on their habitat and on the pathogens that are present. Another viable explanation could be that there was an expansion of the HLA class I lineages. To answer such questions, we designed a study that investigated the influence of selection operating on the classical MHC class I loci. This involved the comparison of intron variation of chimpanzee and human MHC class I alleles. The different intron 2 variations observed in both species are clustering into various lineages, of which some are shared (Figures 3 and 4). On the basis of two well-defined populations, the MHC class I intron 2 data regarding twenty-five randomly chosen human Caucasian individuals [77] were compared with the data regarding our twenty-five P.t.v. chimpanzees. The χ2 statistics showed that the intron 2 variation found in humans is 2.56 times higher (confidence interval (CI) 95% is 0.87-7.55, P = 0.07) for the MHC-A locus, and 2.64 times higher (CI 95% is 1.20-5.82, P = 0.01) for the B locus [78]. The human cohort used, however, was not typed for the HLA-C locus, and therefore we have reanalyzed the data using a different human cohort that is typed at high-resolution level for HLA-A, -B, and -C (random panel IHB). To determine for all three loci whether the number of intron 2 alleles detected in humans is significantly higher as compared to that in chimpanzees, the difference in unique number of alleles (∆ne) was calculated by using bootstrap analyses. Additionally, for each locus, the ratio of the number of unique alleles in humans divided by the number of unique alleles in chimpanzees was calculated. The ∆ne and the ratios of unique alleles were considered statistically significant if their confidence interval did not enclose 0 or 1, respectively. The analyses revealed that all three loci show a statistical significant difference for the ∆ne and ratio in humans versus chimpanzees (Table 3). Thus, the approach confirms that chimpanzees indeed experienced a selective sweep targeting the MHC class I repertoire. The repertoire condensation was claimed to predate the (sub)speciation of chimpanzees, as the same intron lineages are shared between the different populations of chimpanzees, and was calibrated to have happened approximately 2-3 million years ago [78] (Figure 2). This suggests that in the distant past, ancestors of the contemporary chimpanzee populations must have been in close contact with each other, and that a plague may have affected the entire species. The fact that chimpanzees experienced a repertoire reduction in the MHC class I region, and the knowledge that most chimpanzees infected experimentally with HIV-1 are relatively resistant to developing AIDS, resulted in the hypothesis that the ancient repertoire reduction may have been caused by an HIV-1/SIVcpz-like or a closely related retrovirus [78].
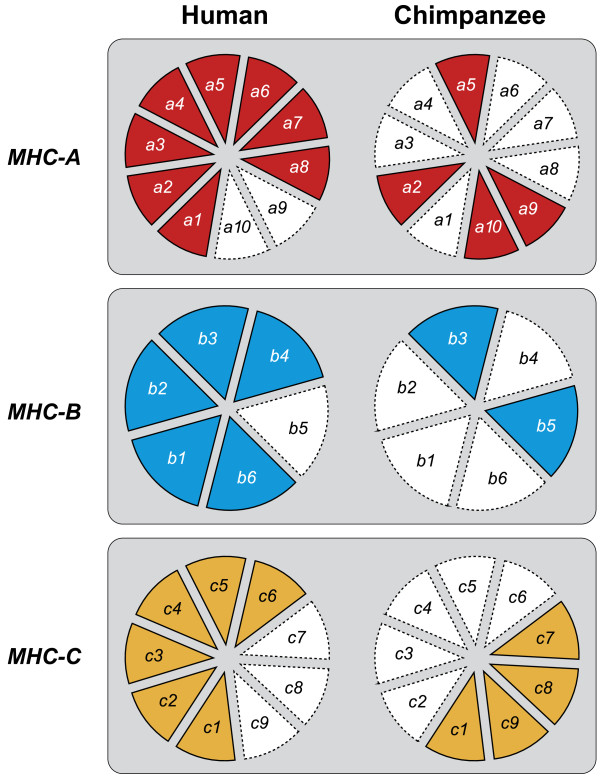
Figure 3.
Pie charts showing the presence/absence of MHC class I intron 2 lineages in humans and chimpanzees. A colored section in a pie indicates the presence of a particular intron 2 lineage in that species; red for MHC-A, blue for MHC-B, and orange for MHC-C.
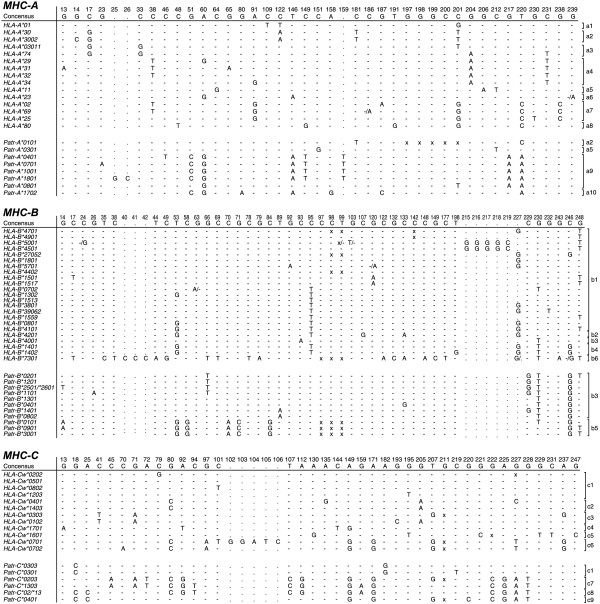
Figure 4.
MHC class I intron 2 sequences in humans (HLA) and chimpanzees (Patr). Only the polymorphic nucleotide positions are indicated. Identity to the consensus sequence (depicted at the top) is indicated by dashes. Substitutions and inserts are depicted by the conventional one-letter code; deletions are marked “x”. For instance, “-/A” indicates that differences in a particular sequence have been reported in the literature. The brackets indicate the division of the intron 2 alleles into lineages, and this is based on phylogenetic analysis. (The figure is adapted from De Groot N.G. et al., PNAS 99, 11748-11753, 2002).
Table 3.
Difference in the number of unique alleles (∆ne (95% CI)) and the ratio of unique alleles (95% CI) in humans as compared to chimpanzees for the different MHC class I loci
| Locus | Δne (95% CI) | Ratio (95% CI) |
|---|---|---|
|
Mhc-A |
4.00 (2.00 - 7.00) |
1.80 (1.33 - 2.40) |
|
Mhc-B |
5.00 (2.00 - 8.00) |
1.63 (1.22 - 2.17) |
| Mhc-C | 6.00 (3.00 - 8.00) | 2.25 (1.60 - 3.67) |
For the statistical analysis, 50 haplotypes were re-sampled 100,000 times and with replacement, separately for both species. The ∆ne was calculated by subtracting the ne of the chimpanzee from the ne of the human, and the ratio of unique alleles in the two species was calculated by dividing the ne of the human by the ne of the chimpanzee. The median was defined as the 50,000th value and the lower and upper confidence limits were defined as the 250th and 99,750th values emanating from the bootstrapping. Calculations were performed using the R statistical package (version 2.15.3).
A successive comparative genomic approach, using microsatellite markers located in the MHC region and markers mapping at a variety of other chromosomes, revealed that both chimpanzees and humans show similar amounts of variation for most of the non-MHC markers. Nonetheless, a significant lower allelic variation for the majority of the markers mapping in the chimpanzee MHC region was documented [79]. These data support the claim that chimpanzees possess a reduced MHC repertoire. Subsequent multi-locus demographic analyses highlighted the fact that chimpanzees experienced a selective sweep that mainly targeted the chromosomal segment carrying the MHC class I region [79].
The evolution of chimpanzee MIC genes: further substantiation of selection
Activated CD4+ memory T cells are selectively and rapidly depleted after an HIV-1/SIV infection [80,81]. This cell type is present in abundance in the intestine, but other mucosal surfaces (lung, vagina) that are frequently exposed to environmental pathogens also contain a high percentage of activated lymphocytes [82]. In humans and macaques in particular, the intestine is the prominent site of CD4+ T-cell depletion during the first few weeks after exposure to HIV-1/SIV [81,83]. The major histocompatibility complex class I chain-related gene (MIC) molecules are highly expressed on the gastrointestinal epithelium [84], and the genes encoding these molecules map to chromosome 6 within the MHC region. In humans, seven genes (MICA to MICG) have been distinguished, though only the MICA and -B genes produce functional transcripts and display high levels of polymorphism [70]. The molecules are thought to be involved in signaling stress to the immune system.
Chimpanzees appear to have only one functional MIC gene (localized near the Patr-B locus), which has an intermediate character resembling in part the human MICA and -B genes. A large ancestral deletion of 95 kb resulted in the birth of a hybrid Patr-MICA/B gene [85]. We have investigated MIC gene polymorphism in chimpanzees, and found that the Patr-MICA/B fusion gene controls only one lineage showing moderate allelic variation [86]. Thus, as compared to humans, chimpanzees have a reduced MIC gene repertoire, which is consistent with the selective sweep observed for the Patr-B locus. Moreover, the hybrid MICA/B gene appears to represent a fixed entity in all chimpanzee (sub)species. This is peculiar, as most other primate species possess haplotypes that carry both MICA and -B genes. Hence, somewhere during chimpanzee evolution a haplotype with a hybrid MICA/B gene was generated, and all other haplotypes carrying MICA and -B genes have gone missing. As the MICA/B gene maps next to the Patr-B gene, it is possible that selection on particular B alleles favored the preferential selection of this hybrid MIC gene by way of a piggyback effect. However, it cannot be excluded that the hybrid MICA/B gene itself, or the combination of this gene with particular Patr-B alleles, provided a selective advantage.
In humans, MIC is the ligand for the NKG2D receptor, which is expressed on natural killer (NK) cells, γδ T cells, and particular CD8+ αβ T cells [87]. A cellular “stress” signal, triggered for instance by a viral infection or malignant transformation, may upregulate the expression of MIC, which can ultimately lead to an immune response. The Patr-MICA/B fusion molecule is recognized by human Vδ1 γδ T cells specific for MICA and B, suggesting a conserved recognition site [88]. In addition, the NKG2D receptor of humans and chimpanzees is also highly similar [89]. Whether MIC plays a role in NK effector responses against HIV-1/SIVcpz has yet to be proven, as does the functional role of the Patr-MICA/B gene in viral infections [86].
Signs of selection beyond the MHC in chimpanzees
HIV-1 targets CD4+ T cells by the use of the CD4 receptor and different co-receptors. Orthologs of the human CD4 receptor, and of the CCR5 and CXCR4 co-receptors, are present in chimpanzees. In humans, several genetic CCR5 modifications are associated with resistance/susceptibility to HIV-1 infection [90]. The CCR5 variant that possesses a 32-base pair deletion (CCR5-∆32) conferring nearly complete resistance to HIV-1 infection in homozygous individuals is present in approximately 1% of the Caucasian population [91]. In chimpanzees, no 32-base pair deletion in the CCR5 gene has been observed, and the relative resistance of chimpanzees to developing AIDS after an HIV-1 infection cannot be attributed to this genetic modification [92-94]. Variations in the human 5′ cis-regulatory region of CCR5 (5′CCR5) were found to be associated with different transcription levels that influence HIV-1 entry and may affect disease progression [95]. Hence, the human 5′CCR5 haplotype that shows the lowest promoter activity resulting in control of AIDS development is the most common haplotype in chimpanzees [96]. By assuming a wide range of demographic histories, Wooding et al.[96] demonstrated that the human 5′CCR5 promotor region experienced balancing selection, and that, in contrast, the chimpanzee equivalent was affected by a selective sweep. This suggests that if the chimpanzee 5′CCR5 promotor region is linked to functional variants that influence progression to AIDS, it may contribute to the relative resistance of chimpanzees to developing AIDS after an HIV-1/SIVcpz infection. Corresponding to this finding, MacFie et al.[97] demonstrated differential patterns of diversity for the HIV-related loci CCR5, CXCR4, and CX3CR1 in three chimpanzee subspecies (West- and Central-African, and Nigeria-Cameroon) [97]. For the CCR5 locus, they showed that it has low levels of diversity for all three subspecies, and this appeared to be tightly centered, as flanking loci displayed normal variation in all subspecies. The results suggest that the CCR5 locus experienced a selective sweep and that this may have predated subspeciation. For the Central-African chimpanzees, natural infections with SIVcpz are documented [39], and a selective sweep at the CCR5 locus may be related to recent co-evolution with SIVcpz. For the other two subspecies, natural infections with SIVcpz are not documented in the contemporary living animals [44,98]. The fact that these subspecies do show evidence for selection at CCR5 could indicate that they were infected with SIVcpz-like virus strains in the past, and that infections are rare or absent at present. Alternatively, the entire ancestral chimpanzee population may have been infected with an SIVcpz-like virus, resulting in a selection event at that stage for which the signature still can be measured. Depending on novel infections with SIVcpz strains, particular selection forces are still operative.
Different cellular restriction factors, such as tripartite motif protein 5 alpha (TRIM5α) and apolipoprotein B mRNA-editing enzyme 3G (APOBEC3G), can target the intracellular replication of HIV-1 [99]. The APOBEC3G gene underwent strong positive selection, and during primate evolution this gene experienced several episodes of adaptations [100]. Polymorphisms in the human APOBEC3G gene have been documented, but its relationship with control of viral replication needs further investigation [101]. The chimpanzee APOBEC3DE, one of the other seven members of the APOBEC family, has potent antiretroviral activity against HIV-1. This is thought to be driven by an ancient lentiviral selective pressure dating back approximately 2 to 6 million years [102], which is consistent with our hypothesis that chimpanzees experienced a selective sweep caused by HIV-1/SIVcpz-like or a closely related retrovirus prior to chimpanzee (sub)speciation [78].
In addition, chimpanzees experienced selection on the fourth component (C4) gene of the complement system that maps in the MHC class III region [103]. Most humans possess two copies of the gene, designated C4A and C4B, and both genes are known to display size polymorphism due to the insertion of a complete endogenous retrovirus of 6.3 kb [104]. The long version of the C4A gene is found in humans, orangutans, and a variety of Old World monkeys [103,105]. However, chimpanzees only possess the equivalents of the short versions of the C4A and C4B genes. Since the presence of the long C4A gene predates human and Old World monkey speciation, the equivalent of this gene has most likely been lost during chimpanzee evolution.
Therefore, different research angles provide evidence that chimpanzees, as compared to humans, have experienced selection operating on the MHC and other genomic regions during their evolution. Chimpanzees shared a common ancestor with the bonobo (Pan paniscus) approximately 2 million years ago. A recent comparison of the bonobo and chimpanzee genome revealed that in chimpanzees, the MHC region in particular has experienced positive selection [106].
Ancient selective sweep in chimpanzees most likely caused by an SIV-like retrovirus: supporting data from functional studies
Control of AIDS development in HIV-1-infected human cohorts is strongly associated with the presence of HLA-B*27:05 and -B*57:01 [48,52-54,60], and adaptive immune responses to the HIV-1 Gag protein are thought to play an important role in the control of viral replication [107,108]. Hence, the Gag protein was taken as a model system to measure immune responses to this protein in chimpanzees. More specifically, we investigated whether the selective sweep had resulted in the preferential selection of Patr class I allotypes, and, if so, whether these allotypes could target similar HIV-1/SIV Gag regions as do HLA molecules associated with control of AIDS development. Therefore, the peptide-binding motifs of four Patr class I molecules, occurring at high frequency in a thoroughly characterized West-African chimpanzee population, were determined [109]. The obtained motifs were used to scan the HIV-1/SIVcpz Gag proteins for potential CTL epitopes, and the relevant peptides were subsequently tested in binding studies. Two of the studied Patr molecules have peptide-binding motifs that resemble those of HLA-B*27 or -B*57, and can target similar areas of the HIV-1/SIVcpz Gag protein (Figure 5). In addition, the two other studied allotypes, although divergent in their peptide-binding anchors, also appear to target the conserved areas of the HIV/SIVcpz proteome similar to the AIDS-controlling HLA-B*27 and -B*57 molecules. Thus, particular human and chimpanzee allotypes share similar qualitative functional characteristics.
Figure 5.
Human (HLA) and chimpanzee (Patr) peptide-binding motifs of relevant MHC class I molecules, and the Gag protein regions that are potentially targeted. P2 and PΩ (carboxyl-terminus) represent the anchor-binding positions of the MHC class I molecules; amino acids that are preferred on these positions are indicated by the conventional one-letter code, whereas tolerated amino acids are indicated between brackets. Patr-B*01:01 has a peptide-binding motif that resembles that of HLA-B*57:01, and Patr-B*03:01 has a peptide-binding motif that resembles that of HLA-B*27:05. Parts of the Gag consensus sequences of HIV-1 (HBX2) and SIVcpz are given (http://www.hiv.lanl.gov). A lower-case letter in the SIVcpz consensus indicates a variable position. The HLA-B*27 and -B*57 CTL epitopes are indicated by red arrows. For the respective Patr class I molecules, blue arrows indicate the potential Gag epitopes that are tested in peptide-binding studies. Based on IC50 (μM) values determined in peptide-binding competition assays, high (1) or intermediate (2) binding affinities of the peptides to their respective MHC class I molecule are indicated [109].
Recently, an in silico approach comparing human and chimpanzee MHC class I alleles illustrated that, with regard to the peptide-binding repertoire, the Patr-A molecules show signs of a selective sweep [110]. The Patr-A allotypes, in general, appear to have promiscuous binding profiles, and as a consequence they can target conserved areas of the Gag protein. The chimpanzee-specific Patr-AL molecule has a peptide-binding motif similar to HLA-A*02 [111]. However, as compared to the classical MHC class I loci, Patr-AL has distinct characteristics (low expression level and present only on approximately 50% of the chimpanzee MHC haplotypes), and further investigation is needed as to whether this molecule contributes significantly to adaptive immune responses.
In the founder population that we analyzed thoroughly, 94% of the chimpanzees possess at least one of the four Patr class I allotypes studied (Figure 5). Moreover, many chimpanzees were observed to express multiple allotypes, which are able to bind peptides derived from various conserved Gag regions. This has led us to suggest that chimpanzees may have developed a “double-lock” strategy to respond to an HIV-1/SIVcpz infection, meaning that the chance of the virus escaping by mutations has been severely diminished. As only four Patr allotypes have been studied at this point, this quantitative aspect of the immune response might become more prominent if more peptide-binding motifs become available. Evaluation of the immune response data of three different HIV-1-infected chimpanzees revealed that the animals displayed broadly reactive CTL responses to conserved epitopes of the Gag protein [109,112,113]. Thus, the functional characteristics of the chimpanzee MHC class I repertoire suggest that the ancient selective sweep was caused by a lentiviral pandemic. The effect of this sweep can still be measured, as most extant chimpanzee populations appear to have a reduced MHC class I repertoire. However, most animals do possess (multiple) allotypes with functional characteristics similar to the AIDS-controlling HLA-B*27/B*57 molecules in humans.
There is evidence that the selective sweep or subsequent selection processes must have been more prominent in West-African chimpanzees than in other chimpanzee populations [96,97,114]. Population separation may have influenced this. Moreover, chimpanzees and their ancestors were most likely infected through predating on different monkey species that are infected with disparate types of SIV strains. Thus, depending on the monkey species predated upon, and their respective SIV infection, repertoires could have been edited in slightly different manners [109]. Predation may result in ongoing new infections. As a consequence, new recombinant SIVcpz strains may be generated occasionally [115], and some of these strains may develop pathogenic characteristics. Thus, non-human primates are challenged by SIV infections for long periods of time, and must have developed ways to control the development of AIDS [22].
In rhesus macaque SIV models, currently used widely to study immunopathogenesis as well as to modulate the immunological responses induced by HIV-1 vaccine and vaccine components, EC is correlated with immune responses primarily directed towards the proteins Vif and Nef [116,117]. The macaque class I allotype Mamu-B*008, which is involved in EC, has a peptide-binding motif that resembles HLA-B*27, and only three CD8+ T-cell epitopes were shown to be responsible for the T-cell responses controlling replication of the pathogenic SIVmac239 in these ECs [118]. Two of the epitopes, Vif RL9 and Nef RL10, are conserved, and similar motifs are present in SIVcpz and HIV-1 (Figure 6). Modeling the peptide binding of the proteins Vif and Nef to HLA-B*27:05 and the four studied Patr class I allotypes showed that they can target these conserved regions of Vif RL9 and Nef RL10 as well (Figure 6). This may suggest that, in different species, evolutionarily unrelated MHC class I molecules possessing similar peptide binding motifs are important for control of the lentiviral infection.
Figure 6.
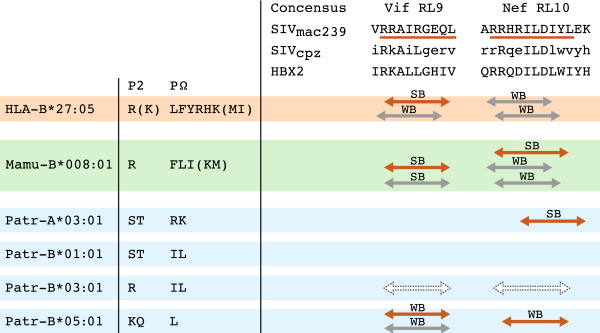
Conserved regions around the Mamu-B*008 epitopes Vif RL9 and Nef RL10 that can be targeted by HLA and Patr class I molecules. P2 and PΩ (carboxyl-terminus) represent the anchor-binding positions of the MHC class I molecules; amino acids that are preferred on these position are indicated by the conventional one-letter code, whereas amino acids indicated between brackets are tolerated. The epitopes Vif RL9 and Nef RL10 in SIVmac239 are indicated by a red line [118]. For HIV-1 (HBX2) and SIVcpz the consensus sequences surrounding these Vif and Nef epitopes are given (http://www.hiv.lanl.gov). A lower-case letter in the SIVcpz consensus indicates a variable position. A red arrow indicates that the respective MHC class I molecule is predicted to target the SIVmac239 consensus sequence. A grey arrow indicates prediction of the MHC molecule to target the SIVcpz/HBX2 consensus sequence. SB stands for solid binding (affinity < 100 nM); WB stands for weak binding (affinity between 100 to 700 nM). Binding affinity of the peptides was predicted using the NetMHCpan algorithm [119], except for Patr-B*03:01 (white dotted arrows), for which binding was assumed based on agreement in the anchor binding positions.
Conclusions
The selective sweep in chimpanzees: a mirror of humankind’s future?
A remaining question concerns how many chimpanzees died during the pandemic that may have been caused by an HIV-1/SIVcpz-like retrovirus. This is a difficult question to answer, but it is likely that the number must have been huge. Natural history has provided a few examples in which the introduction of a “novel” virus into a naïve population resulted in mass mortality. For instance, contact between European individuals and the native population of America led to the death of millions of Amerindians as a result of infection with measles, small pox, and other viruses [120]. The deliberate infection of rabbits with the myxoma virus exterminated approximately 99% of the rabbit population in Australia [121]. Natural infections in animals can also have strong effects on population size, as is illustrated by the rinderpest epidemic in African buffalo [122], and by the decimation of seals by morbilli and influenza viruses [123,124].
Modern humans, who have their cradle in Africa, have existed for about 150,000-200,000 years [125], and display an impressive amount of MHC class I polymorphism [126]. Recent cohort studies have demonstrated that particularly those individuals equipped with HLA-B*27 and/or -B*57 are able to control an HIV-1 infection [49,50]. Nevertheless, in these individuals one also sees that an escape by the virus may occur, although mostly with a cost to viral fitness [127-129]. Furthermore, there is an additional effect on viral control caused by a phenomenon such as heterozygous advantage [130,131]. Chimpanzees may reflect the latter stages of selection. First, the Patr molecules have the ability to respond to an HIV-1/SIVcpz infection. Second, a quantitative aspect seem to be operative in chimpanzees, as each individual may possess several Patr molecules that can respond to different conserved parts of the HIV-1/SIVcpz proteome, termed “double lock” strategy.
The selective sweep in chimpanzees resulted in the preferential selection of Patr class I allotypes that can target conserved areas of Gag similar to those targeted by HLA-B*27/B*57. In humans, these allotypes also seem to predispose for the immune-mediated pathologies ankylosing spondylitis and psoriasis, respectively. Evidence for the presence of these immune-mediated pathologies in chimpanzees is very low [132,133]. In humans, the exact role of HLA-B*27 in ankylosing spondylitis is also not known. Moreover, chimpanzee MHC class I molecules can target similar conserved motifs such as HLA-B*27/B*57, but this does not tell us whether the structure of the chimpanzee MHC class I molecules are comparable to HLA-B*27/B*57, as these molecules belong to different ancestral lineages [72,112].
In general, a virus survives best if it is able to replicate and disseminate itself within a population without killing all of its hosts. Such a host/virus state of equilibrium is, for instance, reached in different non-human primate species that are infected naturally with SIV strains. For HIV-1 and its human hosts, the battle seems to be in full swing, and without proper treatment the human population may be hit hard by this pandemic. For humans up until now, control of HIV-1 replication, without treatment, is significantly associated with the MHC region, in particular with the MHC class I molecules HLA-B*27:05 and -B*57:01, which can target conserved regions of the HIV-1 Gag protein. The observations adduced in this review have important consequences for vaccine design, as different HLA specificities may target different sections of the HIV-1 proteome. One major implication is that many human individuals may not possess HLA class I molecules that have the capacity to bind conserved HIV-1 epitopes that are associated with control of AIDS development. A truly protective vaccine has not yet been reported. Therefore, until that moment arrives, prevention of the infection itself will be, for the individual, one of the most important goals as regards surviving the current HIV-1 pandemic.
Competing interests
The authors declare that they do not have competing interests.
Authors’ contributions
NG and RB wrote the manuscript. Both authors have given their final approval of the manuscript.
Contributor Information
Natasja G de Groot, Email: groot@bprc.nl.
Ronald E Bontrop, Email: bontrop@bprc.nl.
Acknowledgements
The authors would like to thank D Devine for editing the manuscript, H van Westbroek for preparing the figures, W Verduyn and Dr. F Claas for supplying the HLA-data, Dr. EJ Remarque for the statistical analyses, and Drs. MN Carrington, G Koopman, and EJ Verschoor for their suggestions and critical reading of the manuscript.
References
- Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Thomson MM, Perez-Alvarez L, Najera R. Molecular epidemiology of HIV-1 genetic forms and its significance for vaccine development and therapy. Lancet Infect Dis. 2002;2:461–471. doi: 10.1016/S1473-3099(02)00343-2. [DOI] [PubMed] [Google Scholar]
- Chun TW, Fauci AS. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS. 2012;26:1261–1268. doi: 10.1097/QAD.0b013e328353f3f1. [DOI] [PubMed] [Google Scholar]
- Chun TW, Davey RT Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, Fauci AS. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. 2000;6:757–761. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- Davey RT Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia F, Plana M, Vidal C, Cruceta A, O’Brien WA, Pantaleo G, Pumarola T, Gallart T, Miro JM, Gatell JM. Dynamics of viral load rebound and immunological changes after stopping effective antiretroviral therapy. AIDS. 1999;13:F79–F86. doi: 10.1097/00002030-199907300-00002. [DOI] [PubMed] [Google Scholar]
- Harrigan PR, Whaley M, Montaner JS. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS. 1999;13:F59–F62. doi: 10.1097/00002030-199905280-00001. [DOI] [PubMed] [Google Scholar]
- Friedland GH, Williams A. Attaining higher goals in HIV treatment: the central importance of adherence. AIDS. 1999;13(Suppl 1):S61–S72. [PubMed] [Google Scholar]
- Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- Subbramanian RA, Cohen EA. Molecular biology of the human immunodeficiency virus accessory proteins. J Virol. 1994;68:6831–6835. doi: 10.1128/jvi.68.11.6831-6835.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD. Perspectives series: host/pathogen interactions. Dynamics of HIV-1 replication in vivo. J Clin Invest. 1997;99:2565–2567. doi: 10.1172/JCI119443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JA. HIV and the Pathogenesis of AIDS. Washington, DC: American Society of Microbiology; 2007. [Google Scholar]
- Bomsel M, David V. Mucosal gatekeepers: selecting HIV viruses for early infection. Nat Med. 2002;8:114–116. doi: 10.1038/nm0202-114. [DOI] [PubMed] [Google Scholar]
- Pope M, Haase AT. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat Med. 2003;9:847–852. doi: 10.1038/nm0703-847. [DOI] [PubMed] [Google Scholar]
- van Kooyk Y, Geijtenbeek TB. DC-SIGN: escape mechanism for pathogens. Nat Rev Immunol. 2003;3:697–709. doi: 10.1038/nri1182. [DOI] [PubMed] [Google Scholar]
- McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Churchill M, Crowe SM, Cunningham AL, Gabuzda D. Pathogenesis of macrophage tropic HIV-1. Curr HIV Res. 2005;3:53–60. doi: 10.2174/1570162052772951. [DOI] [PubMed] [Google Scholar]
- Pierson T, McArthur J, Siliciano RF. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- Pandrea I, Apetrei C, Gordon S, Barbercheck J, Dufour J, Bohm R, Sumpter B, Roques P, Marx PA, Hirsch VM. Paucity of CD4 + CCR5+ T cells is a typical feature of natural SIV hosts. Blood. 2007;109:1069–1076. doi: 10.1182/blood-2006-05-024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodora DL, Allan JS, Apetrei C, Brenchley JM, Douek DC, Else JG, Estes JD, Hahn BH, Hirsch VM, Kaur A. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat Med. 2009;15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Fruteau C, Noe R, Peeters M, Brookfield JF. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Cote d’Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–12527. doi: 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Rodenburg CM, Kamenya S, Bibollet-Ruche F, Gao F, Bailes E, Meleth S, Soong SJ, Kilby JM, Moldoveanu Z. SIVcpz in wild chimpanzees. Science. 2002;295:465. doi: 10.1126/science.295.5554.465. [DOI] [PubMed] [Google Scholar]
- Bowden R, MacFie TS, Myers S, Hellenthal G, Nerrienet E, Bontrop RE, Freeman C, Donnelly P, Mundy NI. Genomic tools for evolution and conservation in the chimpanzee: Pan troglodytes ellioti is a genetically distinct population. PLoS Genet. 2012;8:e1002504. doi: 10.1371/journal.pgen.1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Wills C, Gerloff U, Tautz D, Morin PA, Boesch C, Fruth B, Hohmann G, Ryder OA, Woodruff DS. Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc Natl Acad Sci USA. 1999;96:5077–5082. doi: 10.1073/pnas.96.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, Moore JJ, Chakraborty R, Jin L, Goodall J, Woodruff DS. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol. 2009;83:1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Ambrose Z, Chung NP, Piatak M Jr, Yuan F, Trubey CM, Coalter V, Kiser R, Schneider D, Smedley J. A macaque model of HIV-1 infection. Proc Natl Acad Sci USA. 2009;106:4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thippeshappa R, Polacino P, Yu Kimata MT, Siwak EB, Anderson D, Wang W, Sherwood L, Arora R, Wen M, Zhou P. Vif substitution enables persistent infection of pig-tailed macaques by human immunodeficiency virus type 1. J Virol. 2011;85:3767–3779. doi: 10.1128/JVI.02438-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz N, Sauter D, Usmani SM, Fritz JV, Goffinet C, Heigele A, Geyer M, Bibollet-Ruche F, Learn GH, Fackler OT. Reacquisition of Nef-mediated tetherin antagonism in a single in vivo passage of HIV-1 through its original chimpanzee host. Cell Host Microbe. 2012;12:373–380. doi: 10.1016/j.chom.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney JL. AIDS: a disease of impaired Th-cell renewal? Immunol Today. 1995;16:515–520. doi: 10.1016/0167-5699(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- Peeters M, Fransen K, Delaporte E, Van den Haesevelde M, Gershy-Damet GM, Kestens L, van der Groen G, Piot P. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS. 1992;6:447–451. doi: 10.1097/00002030-199205000-00002. [DOI] [PubMed] [Google Scholar]
- Peeters M, Honore C, Huet T, Bedjabaga L, Ossari S, Bussi P, Cooper RW, Delaporte E. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS. 1989;3:625–630. doi: 10.1097/00002030-198910000-00001. [DOI] [PubMed] [Google Scholar]
- Nerrienet E, Santiago ML, Foupouapouognigni Y, Bailes E, Mundy NI, Njinku B, Kfutwah A, Muller-Trutwin MC, Barre-Sinoussi F, Shaw GM. Simian immunodeficiency virus infection in wild-caught chimpanzees from cameroon. J Virol. 2005;79:1312–1319. doi: 10.1128/JVI.79.2.1312-1319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago ML, Lukasik M, Kamenya S, Li Y, Bibollet-Ruche F, Bailes E, Muller MN, Emery M, Goldenberg DA, Lwanga JS. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii) J Virol. 2003;77:7545–7562. doi: 10.1128/JVI.77.13.7545-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney JL, Rutjens E, Verschoor EJ, Niphuis H, ten Haaft P, Rouse S, McClure H, Balla-Jhagjhoorsingh S, Bogers W, Salas M. Transmission of simian immunodeficiency virus SIVcpz and the evolution of infection in the presence and absence of concurrent human immunodeficiency virus type 1 infection in chimpanzees. J Virol. 2006;80:7208–7218. doi: 10.1128/JVI.00382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Nerrienet E, LeBreton M, Bibila GT, Foupouapouognigni Y, Rousset D, Nana A, Djoko CF, Tamoufe U, Aghokeng AF. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology. 2011;8:4. doi: 10.1186/1742-4690-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre FJ, Saucier M, Anderson DC, Klumpp SA, O’Neil SP, Brown CR 2nd, Hart CE, Guenthner PC, Swenson RB, McClure HM. Development of AIDS in a chimpanzee infected with human immunodeficiency virus type 1. J Virol. 1997;71:4086–4091. doi: 10.1128/jvi.71.5.4086-4091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre FJ, de Rosayro J, Nidtha S, O’Neil SP, Gibson TR, Evans-Strickfaden T, Hart CE, McClure HM. Rapid CD4(+) T-cell loss induced by human immunodeficiency virus type 1(NC) in uninfected and previously infected chimpanzees. J Virol. 2001;75:1533–1539. doi: 10.1128/JVI.75.3.1533-1539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, Chaix ML, Lambotte O, Avettand-Fenoel V, Le Clerc S, de Senneville LD. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS genome wide association 01 study. PLoS One. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips RE, McMichael AJ. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Hum Retroviruses. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O’Brien SJ, Rinaldo C. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK. An update to HLA nomenclature, 2010. Bone Marrow Transplant. 2010;45:846–848. doi: 10.1038/bmt.2010.79. [DOI] [PubMed] [Google Scholar]
- Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol. 2009;9:503–513. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- Harding CV, Boom WH. Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat Rev Microbiol. 2010;8:296–307. doi: 10.1038/nrmicro2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst D, Verweij MC, Davison AJ, Ressing ME, Wiertz EJ. Viral evasion of T cell immunity: ancient mechanisms offering new applications. Curr Opin Immunol. 2011;23:96–103. doi: 10.1016/j.coi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Jackson SE, Mason GM, Wills MR. Human cytomegalovirus immunity and immune evasion. Virus Res. 2011;157:151–160. doi: 10.1016/j.virusres.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Walker BD. HIV and HLA Class I: an evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Daniuk CA, Diallo TO, Capina RE, Kimani J, Wachihi C, Kimani M, Bielawny T, Peterson T, Mendoza MG. For protection from HIV-1 infection, more might not be better: a systematic analysis of HIV Gag epitopes of two alleles associated with different outcomes of HIV-1 infection. J Virol. 2012;86:1166–1180. doi: 10.1128/JVI.05721-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams EJ, Cooper S, Parham P. A novel, nonclassical MHC class I molecule specific to the common chimpanzee. J Immunol. 2001;167:3858–3869. doi: 10.4049/jimmunol.167.7.3858. [DOI] [PubMed] [Google Scholar]
- Geller R, Adams EJ, Guethlein LA, Little AM, Madrigal JA, Parham P. Linkage of Patr-AL to Patr-A and- B in the major histocompatibility complex of the common chimpanzee (Pan troglodytes) Immunogenetics. 2002;54:212–215. doi: 10.1007/s00251-002-0452-x. [DOI] [PubMed] [Google Scholar]
- van Rood JJ, van Leeuwen A, Balner H. HL-A and ChL-A: similarities and differences. Transplant Proc. 1972;4:55–62. [PubMed] [Google Scholar]
- Domena JD, Hildebrand WH, Bias WB, Parham P. A sixth family of HLA-A alleles defined by HLA-A*8001. Tissue Antigens. 1993;42:156–159. doi: 10.1111/j.1399-0039.1993.tb02186.x. [DOI] [PubMed] [Google Scholar]
- Kato K, Trapani JA, Allopenna J, Dupont B, Yang SY. Molecular analysis of the serologically defined HLA-Aw19 antigens. A genetically distinct family of HLA-A antigens comprising A29, A31, A32, and Aw33, but probably not A30. J Immunol. 1989;143:3371–3378. [PubMed] [Google Scholar]
- McAdam SN, Boyson JE, Liu X, Garber TL, Hughes AL, Bontrop RE, Watkins DI. Chimpanzee MHC class I A locus alleles are related to only one of the six families of human A locus alleles. J Immunol. 1995;154:6421–6429. [PubMed] [Google Scholar]
- Fujiyama A, Watanabe H, Toyoda A, Taylor TD, Itoh T, Tsai SF, Park HS, Yaspo ML, Lehrach H, Chen Z. Construction and analysis of a human-chimpanzee comparative clone map. Science. 2002;295:131–134. doi: 10.1126/science.1065199. [DOI] [PubMed] [Google Scholar]
- Klein J. Origin of major histocompatibility complex polymorphism: the trans-species hypothesis. Hum Immunol. 1987;19:155–162. doi: 10.1016/0198-8859(87)90066-8. [DOI] [PubMed] [Google Scholar]
- Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–D1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Robinson J, Blancher A, Lafont BA, Marsh SG, O’Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE. Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Arguello R, Watkins DI, Doxiadis GG, Madrigal JA, Bontrop RE. Major histocompatibility complex class I diversity in a West African chimpanzee population: implications for HIV research. Immunogenetics. 2000;51:398–409. doi: 10.1007/s002510050638. [DOI] [PubMed] [Google Scholar]
- Adams EJ, Cooper S, Thomson G, Parham P. Common chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes. Immunogenetics. 2000;51:410–424. doi: 10.1007/s002510050639. [DOI] [PubMed] [Google Scholar]
- Caggiari L, Simula MP, Marzotto A, Shiina M, Rehermann B, De Re V. Identification of novel chimpanzee MHC class I and II alleles using an improved sequence-based typing strategy. Hum Immunol. 2006;67:63–72. doi: 10.1016/j.humimm.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Hughes AL, Ghim SH, Letvin NL, Watkins DI. Two more chimpanzee Patr-A locus alleles related to the HLA-A1/A3/A11 family. Immunogenetics. 1993;38:238–240. doi: 10.1007/BF00211524. [DOI] [PubMed] [Google Scholar]
- Kowalski H, Erickson AL, Cooper S, Domena JD, Parham P, Walker CM. Patr-A and B, the orthologues of HLA-A and B, present hepatitis C virus epitopes to CD8+ cytotoxic T cells from two chronically infected chimpanzees. J Exp Med. 1996;183:1761–1775. doi: 10.1084/jem.183.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, van Bree S, Gijbels Y, van Veen A, van Rood JJ, Class FH. Comparison of the humoral and cytotoxic T-lymphocyte response to individual HLA class I alloantigens in highly immunized patients. Hum Immunol. 1991;30:156–161. doi: 10.1016/0198-8859(91)90029-9. [DOI] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Doxiadis GG, Balla-Jhagjhoorsingh SS, Heeney JL, van Rood JJ, Gagneux P, Bontrop RE. Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci USA. 2002;99:11748–11753. doi: 10.1073/pnas.182420799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Heijmans CM, de Groot N, Otting N, de Vos-Rouweller AJ, Remarque EJ, Bonhomme M, Doxiadis GG, Crouau-Roy B, Bontrop RE. Pinpointing a selective sweep to the chimpanzee MHC class I region by comparative genomics. Mol Ecol. 2008;17:2074–2088. doi: 10.1111/j.1365-294X.2008.03716.x. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, Lackner AA. Getting to the guts of HIV pathogenesis. J Exp Med. 2004;200:697–700. doi: 10.1084/jem.20041464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci USA. 2003;100:7708–7713. doi: 10.1073/pnas.1230533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Garcia CA, Verschoor EJ, Doxiadis GG, Marsh SG, Otting N, Bontrop RE. Reduced MIC gene repertoire variation in West African chimpanzees as compared to humans. Mol Biol Evol. 2005;22:1375–1385. doi: 10.1093/molbev/msi127. [DOI] [PubMed] [Google Scholar]
- Gleimer M, Parham P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity. 2003;19:469–477. doi: 10.1016/S1074-7613(03)00272-3. [DOI] [PubMed] [Google Scholar]
- Steinle A, Groh V, Spies T. Diversification, expression, and gamma delta T cell recognition of evolutionarily distant members of the MIC family of major histocompatibility complex class I-related molecules. Proc Natl Acad Sci USA. 1998;95:12510–12515. doi: 10.1073/pnas.95.21.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum BP, Flodin LR, Muir DG, Rajalingam R, Khakoo SI, Cleland S, Guethlein LA, Uhrberg M, Parham P. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168:240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao XH, Merrill G. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Benton PA, Lee DR, Kennedy RC. Sequence comparisons of non-human primate HIV-1 coreceptor homologues. Mol Immunol. 1998;35:95–101. doi: 10.1016/S0161-5890(98)00016-9. [DOI] [PubMed] [Google Scholar]
- Pretet JL, Zerbib AC, Girard M, Guillet JG, Butor C. Chimpanzee CXCR4 and CCR5 act as coreceptors for HIV type 1. AIDS Res Hum Retroviruses. 1997;13:1583–1587. doi: 10.1089/aid.1997.13.1583. [DOI] [PubMed] [Google Scholar]
- Voevodin A, Samilchuk E, Dashti S. A survey for 32 nucleotide deletion in the CCR-5 chemokine receptor gene (deltaccr-5) conferring resistance to human immunodeficiency virus type 1 in different ethnic groups and in chimpanzees. J Med Virol. 1998;55:147–151. doi: 10.1002/(SICI)1096-9071(199806)55:2<147::AID-JMV11>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Watkins WS, Wooding S, Stone AC, Jorde LB, Weiss RB, Ahuja SK. A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci USA. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Stone AC, Dunn DM, Mummidi S, Jorde LB, Weiss RK, Ahuja S, Bamshad MJ. Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5. Am J Hum Genet. 2005;76:291–301. doi: 10.1086/427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFie TS, Nerrienet E, de Groot NG, Bontrop RE, Mundy NI. Patterns of diversity in HIV-related loci among subspecies of chimpanzee: concordance at CCR5 and differences at CXCR4 and CX3CR1. Mol Biol Evol. 2009;26:719–727. doi: 10.1093/molbev/msp016. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Strebel K, Luban J, Jeang KT. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobieszczyk ME, Lingappa JR, McElrath MJ. Host genetic polymorphisms associated with innate immune factors and HIV-1. Curr Opin HIV AIDS. 2011;6:427–434. doi: 10.1097/COH.0b013e3283497155. [DOI] [PubMed] [Google Scholar]
- Duggal NK, Malik HS, Emerman M. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J Virol. 2011;85:11361–11371. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontrop RE, Broos LA, Otting N, Jonker MJ. Polymorphism of C4 and CYP21 genes in various primate species. Tissue Antigens. 1991;37:145–151. doi: 10.1111/j.1399-0039.1991.tb01862.x. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Strachan T, Anderson M, Campbell RD, Collier S, Lako M. Identification of a novel family of human endogenous retroviruses and characterization of one family member, HERV-K(C4), located in the complement C4 gene cluster. Nucleic Acids Res. 1994;22:5211–5217. doi: 10.1093/nar/22.24.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider PM, Witzel-Schlomp K, Steinhauer C, Stradmann-Bellinghausen B, Rittner C. Rapid detection of the ERV-K(C4) retroviral insertion reveals further structural polymorphism of the complement C4 genes in old world primates. Exp Clin Immunogenet. 2001;18:130–134. doi: 10.1159/000049192. [DOI] [PubMed] [Google Scholar]
- Prufer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R. The bonobo genome compared with the chimpanzee and human genomes. Nature. 2012;486:527–531. doi: 10.1038/nature11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghans JA, Molgaard A, de Boer RJ, Kesmir C. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS One. 2007;2:e920. doi: 10.1371/journal.pone.0000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland M, Heckerman D, Deng W, Rousseau CM, Coovadia H, Bishop K, Goulder PJ, Walker BD, Brander C, Mullins JI. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One. 2008;3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Heijmans CM, Zoet YM, de Ru AH, Verreck FA, van Veelen PA, Drijfhout JW, Doxiadis GG, Remarque EJ, Doxiadis II. AIDS-protective HLA-B*27/B*57 and chimpanzee MHC class I molecules target analogous conserved areas of HIV-1/SIVcpz. Proc Natl Acad Sci USA. 2010;107:15175–15180. doi: 10.1073/pnas.1009136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom HW, Hoof I, Bontrop RE, Kesmir C. A comparative analysis of viral peptides presented by contemporary human and chimpanzee MHC class I molecules. J Immunol. 2011;187:5995–6001. doi: 10.4049/jimmunol.1102236. [DOI] [PubMed] [Google Scholar]
- Gleimer M, Wahl AR, Hickman HD, Abi-Rached L, Norman PJ, Guethlein LA, Hammond JA, Draghi M, Adams EJ, Juo S. Although divergent in residues of the peptide binding site, conserved chimpanzee Patr-AL and polymorphic human HLA-A*02 have overlapping peptide-binding repertoires. J Immunol. 2011;186:1575–1588. doi: 10.4049/jimmunol.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla-Jhagjhoorsingh SS, Koopman G, Mooij P, Haaksma TG, Teeuwsen VJ, Bontrop RE, Heeney JL. Conserved CTL epitopes shared between HIV-infected human long-term survivors and chimpanzees. J Immunol. 1999;162:2308–2314. [PubMed] [Google Scholar]
- Balla-Jhagjhoorsingh SS, Mooij P, ten Haaft PJ, Bogers WM, Teeuwsen VJ, Koopman G, Heeney JL. Protection from secondary human immunodeficiency virus type 1 infection in chimpanzees suggests the importance of antigenic boosting and a possible role for cytotoxic T cells. J Infect Dis. 2001;184:136–143. doi: 10.1086/322019. [DOI] [PubMed] [Google Scholar]
- Hvilsom C, Carlsen F, Siegismund HR, Corbet S, Nerrienet E, Fomsgaard A. Genetic subspecies diversity of the chimpanzee CD4 virus-receptor gene. Genomics. 2008;92:322–328. doi: 10.1016/j.ygeno.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Leitner T, Dazza MC, Ekwalanga M, Apetrei C, Saragosti S. Sequence diversity among chimpanzee simian immunodeficiency viruses (SIVcpz) suggests that SIVcpzPts was derived from SIVcpzPtt through additional recombination events. AIDS Res Hum Retroviruses. 2007;23:1114–1118. doi: 10.1089/aid.2007.0071. [DOI] [PubMed] [Google Scholar]
- Mothe BR, Sidney J, Dzuris JL, Liebl ME, Fuenger S, Watkins DI, Sette A. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol. 2002;169:210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- Valentine LE, Loffredo JT, Bean AT, Leon EJ, MacNair CE, Beal DR, Piaskowski SM, Klimentidis YC, Lank SM, Wiseman RW. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J Virol. 2009;83:11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ. Environmental and social influences on emerging infectious diseases: past, present and future. Philos Trans R Soc Lond B Biol Sci. 2004;359:1049–1058. doi: 10.1098/rstb.2004.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers L, Inglis B, Labudovic A, Janssens PA, van Leeuwen BH, Kerr PJ. Virulence and pathogenesis of the MSW and MSD strains of Californian myxoma virus in European rabbits with genetic resistance to myxomatosis compared to rabbits with no genetic resistance. Virology. 2006;348:72–83. doi: 10.1016/j.virol.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Wenink PW, Groen AF, Roelke-Parker ME, Prins HH. African buffalo maintain high genetic diversity in the major histocompatibility complex in spite of historically known population bottlenecks. Mol Ecol. 1998;7:1315–1322. doi: 10.1046/j.1365-294x.1998.00463.x. [DOI] [PubMed] [Google Scholar]
- Domingo M, Visa J, Pumarola M, Marco AJ, Ferrer L, Rabanal R, Kennedy S. Pathologic and immunocytochemical studies of morbillivirus infection in striped dolphins (Stenella coeruleoalba) Vet Pathol. 1992;29:1–10. doi: 10.1177/030098589202900101. [DOI] [PubMed] [Google Scholar]
- Geraci JR, St Aubin DJ, Barker IK, Webster RG, Hinshaw VS, Bean WJ, Ruhnke HL, Prescott JH, Early G, Baker AS. Mass mortality of harbor seals: pneumonia associated with influenza A virus. Science. 1982;215:1129–1131. doi: 10.1126/science.7063847. [DOI] [PubMed] [Google Scholar]
- Mellars P. Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model. Proc Natl Acad Sci USA. 2006;103:9381–9386. doi: 10.1073/pnas.0510792103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J Virol. 2009;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, Tebit DM, Zhao H, Avila S, Lobritz MA. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 2009;5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- Matthews PC, Listgarten J, Carlson JM, Payne R, Huang KH, Frater J, Goedhals D, Steyn D, van Vuuren C, Paioni P. Co-operative additive effects between HLA alleles in control of HIV-1. PLoS One. 2012;7:e47799. doi: 10.1371/journal.pone.0047799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biella U, Haustein UF, Seifert S, Adler J, Schuppel KF, Eulenberger K. [Psoriasis in a female chimpanzee] Hautarzt. 1991;42:322–323. [PubMed] [Google Scholar]
- Taurog JD. Animal models of spondyloarthritis. Adv Exp Med Biol. 2009;649:245–254. doi: 10.1007/978-1-4419-0298-6_18. [DOI] [PubMed] [Google Scholar]