Abstract
Purpose of review
With the advent of whole-transcriptome sequencing, or RNA-seq, we now know that alternative splicing is a generalized phenomenon, with nearly all multi-exonic genes subject to alternative splicing. In this review we highlight recent studies examining alternative splicing as a modulator of cellular cholesterol homeostasis, and as an underlying mechanism of dyslipidemia.
Recent findings
A number of key genes involved in cholesterol metabolism are known to undergo functionally relevant alternative splicing. Recently, we have identified coordinated changes in alternative splicing in multiple genes in response to alteration in cellular sterol content. We and others have implicated several splicing factors as regulators of lipid metabolism. Furthermore, a number of cis-acting human gene variants that modulate alternative splicing have been implicated in a variety of human metabolic diseases.
Summary
Alternative splicing is of importance in various types of genetically influenced dyslipidemias, and in the regulation of cellular cholesterol metabolism.
Keywords: PTBP1, HMGCR, LDLR, statin, SFRS10
Introduction
Alternative splicing is the process by which a single gene produces more than one transcript, and is considered to be the principal mechanism of expanding diversity within the eukaryotic proteome. The most common forms of alternative splicing include exon skipping, alternative 5′ or 3′ splice-site selection, intron retention and mutually exclusive exon utilization [1]. Molecular analyses during the past two decades have demonstrated that alternative splicing influences binding properties, intracellular localization of both transcripts as well as the proteins they encode, enzymatic activity, protein stability and post-translational modifications [2].
As recently as 20 years ago, alternative splicing was thought to be a relatively rare phenomenon, with only 5% of human genes subject to alternative splicing [3]. However, with the development of new detection technologies, this value has been repeatedly revised upwards. Analyses based on expressed sequence tags (ESTs) increased estimates of alternative splicing to 40–60% [4, 5], with a later jump to 74% using exon-junction microarrays [6]. With the advent of whole transcriptome sequencing, or RNA-seq, we now know that nearly every multi-exonic gene undergoes some form of alternative splicing, demonstrating that this is a universal phenomenon [7, 8]. Furthermore, the large number of RNA-seq datasets on a wide variety of primary human tissues currently being generated through The Common Fund’s Genotype-Tissue Expression (GTEx) program will likely reveal a plethora of yet unidentified tissue-specific and rare splice variants. With further improvements in sequencing technology coupled with decreasing costs, transcript quantification platforms that include information on both transcript levels as well as structure will replace the gene expression array as the standard technology.
While it has been known for several years that many key regulators of cholesterol metabolism undergo functionally relevant alternative splicing [9–12], the wider extent of this process has recently become evident. The objective of this paper is to provide an overview of the role of alternative splicing in cellular cholesterol homeostasis and plasma lipid metabolism.
The functional effects of alternative splicing
Alternative splicing events are often divided into one of two classes, 1) “functional” or “productive alternative splicing”, by which transcripts with biological functions are created, and 2) splice variants with no known functional effect, called “unproductive” [13], “erroneous” [14], or “aberrant alternative splicing” [15]”. Often these unproductive splicing events either encode defective proteins, or stimulate one of the many mechanisms that prevent mRNA translation, such as nonsense-mediated decay [16], non-stop decay [17] or no-go decay [18]. With 60% of the alternatively spliced transcripts characterized in the ENCODE pilot project lacking an annotated protein coding sequence, it is now clear that alternative splicing is not simply a mechanism for increasing protein diversity [19, 20].
There has been debate about whether these unproductive splicing events are derived from errors in the splicing machinery [15, 21], or represent targeted changes that influence cellular processes [22]. Proposed functions for these variants include roles in gene expression regulation or gene evolution [14]. Indeed, it has been shown through evolutionary conservation studies of the DNA polymerase β transcript that, regardless of whether the specific RNA transcripts are functional, alternative splicing yielding unproductive splice variants may have adaptive significance [13]. Furthermore, given evidence of widespread coupling of alternative spicing and nonsense-mediated decay in humans, regulated unproductive splicing may be an under appreciated means of regulating protein expression [23].
Alternative splicing as a regulatory mechanism
Although it has been suggested by some that unproductive alternative splicing is not regulated [15], recently we reported evidence that alternative splicing may be a generalized mechanism for regulating genes in the cholesterol biosynthesis and uptake pathways [10]. 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR), the gene that encodes the rate limiting enzyme of the cholesterol biosynthesis pathway, undergoes alternative splicing of exon 13 to generate a second transcript called designated HMGCR13(−). Although the open reading frame is maintained past the exon 12–14 splice junction, the resulting HMGCR protein cannot catalyze the conversion of HMG-CoA into mevalonate, and thus exon 13 skipping could be considered a form of “unproductive” alternative splicing. Using a variety of cellular models, we found that while sterol depletion up-regulated overall expression levels of both the canonical full-length HMGCR transcript, HMGCR13(+), as well as HMGCR13(−), the relative degree on induction of the 13(−) variant was smaller than that of the 13(+) variant, causing an overall reduction in the proportion of alternatively spliced transcripts. This effect was reversed by add-back of either LDL-C or 25-hydroxycholesterol. Similar sterol induced changes in the relative number of alternatively spliced transcripts were observed in other genes encoding enzymes in the cholesterol biosynthetic pathway, including HMG-CoA synthase 1 (HMGCS1) and mevalonate kinase (MVK) as well as key regulators of cholesterol uptake, namely the low density lipoprotein receptor (LDLR) and proprotein convertase subtilisin/kexin type 9 (PCSK9). Similarly, cholesterol feeding of African Green monkeys increased the relative levels of alternatively spliced variants of MVK, LDLR and PCSK9. Furthermore, inter-individual variation in sterol-induced increases in expression of an LDLR splice variant lacking exon 12, LDLR12(−), was directly correlated with degree of change in hepatic cholesterol ester content. Since the splice variants studied were all either known or predicted to attenuate or abolish protein activity, reductions in alternative splicing under conditions of sterol depletion would be expected to increase activity of these two pathways, while increased alternative splicing observed under conditions of sterol excess would decrease both cholesterol synthesis and uptake. Thus, these changes were consistent with the expected directionality necessary to achieve cholesterol homeostasis, and suggested that cellular sterol levels may directly regulate alternative splicing.
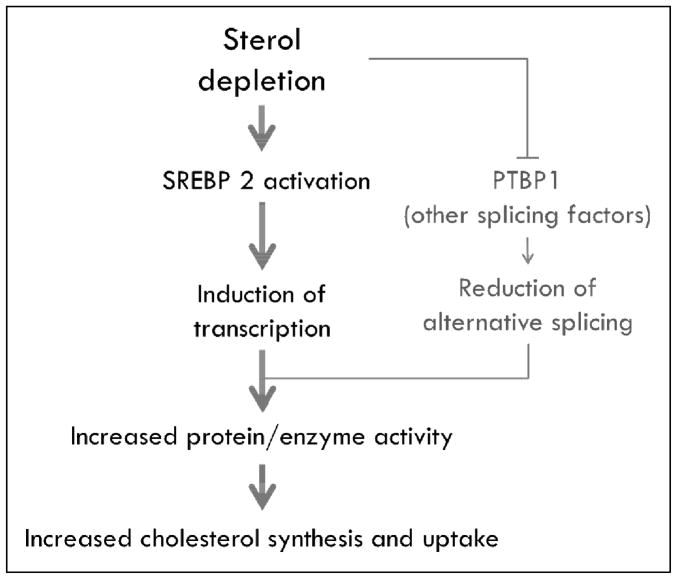
These results strongly suggest that alternative splicing is a novel and generalized mechanism of regulating the cholesterol biosynthesis and uptake pathways, and that it functions in conjunction with SREBP-induced increases in transcription (Figure 1). Although it may be argued that sterol-induced changes in alternative splicing are quite modest in comparison to SREBP2-mediated regulation of transcription, since we observed changes in alternative splicing within 15 minutes of sterol depletion, whereas SREBP-mediated transcriptional regulation did not appear until 4–6 hours after sterol depletion, we have proposed that this may be a mechanism to rapidly modulate or fine tune the effects of SREBP-mediated transcriptional regulation in response to changes in intracellular sterol content.
Figure 1.
Proposed model of alternative splicing as a regulatory mechanism of cellular cholesterol homeostasis.
Splicing factors implicated in cholesterol metabolism
The process of alternative splicing is regulated in part by interactions between RNA-binding proteins (also called splicing factors) and specific sequences contained within the pre-mRNA. There are two major classes of splicing factors, serine-arginine (SR) proteins and heteronuclear riboprotein (hnRNP) proteins. These two classes tend to work in opposition to one another with SR proteins generally promoting splicing by recruiting components of the spliceosome to the exon (leading to exon inclusion, or canonical splicing), while hnRNP proteins generally repress splicing by interfering with spliceosome binding (leading to complete or partial exon skipping, or alternative splicing) [1]. To date, only a few splicing factors have been shown to modulate splicing of genes involved in lipid metabolism: polypyrimidine tract binding protein 1 (PTBP1), Transformer-2alpha (TRA2A) and Transformer-2beta (TRA2B).
Polypyrimidine tract binding protein 1 (PTBP1 also known as hnRNP I) is a member of the hnRNP family. PTBP1 is found mainly in the nucleus, where it is thought to mediate repression of splicing of a number of pre-mRNAs; however, it is also found in the cytoplasm where it has been shown to bind both the 3′ and 5′ UTRs of mRNA to regulate mRNA localization, stability, and translational initiation [24]. While intracellular localization of PTBP1 has been shown to be influenced by glucose [25], recently we have also found that PTBP1 transcript levels are regulated by sterols, with sterol depletion decreasing PTBP1 transcript levels by almost 50%, an effect that is reversed with either LDLC or 25-hydroxycholesterol add-back [10]. We have shown that PTBP1 modulates alternative splicing of several genes involved in cholesterol biosynthesis and uptake including LDLR, MVK, HMGCS1 and PSCK9 [10]. PTBP1 knock-down in HepG2 cells reduced the relative levels of alternatively spliced variants of LDLR, MVK, HMGCS1 and PSCK9 but did not appear to regulate HMGCR exon 13 alternative splicing. Notably, PTBP1 knockdown either attenuated or ameliorated sterol-induced changes in alternative splicing of HMGCS1, MVK, LDLR and PCSK9, consistent with the likelihood that these changes are mediated at least in part by sterol regulation of PTBP1.
PTBP1 has also been implicated in the production on both omega-3 and omega-6 polyunsaturated fatty acids. Notably, Reardon et al reported that PTBP1 regulates alternative splicing of both fatty acid desaturase 2 (FADS2), the gene that encodes the first and rate limiting enzyme in the biosynthesis of long chain polyunsaturated fatty acids, as well as fatty acid desaturase (FADS3). In addition, they found that PTBP1 knock-down reduced both omega-3 and omega-6 fatty acids in HepG2 cells, but did not affect either monounsaturated or saturated fatty acids. Since the most dramatic effects (50% reduction) were observed with reduction in eicosapentaenoic acid, PTBP1 knock-down modulated the ratio of omega-3 to omega-6 fatty acids [26].
Transformer-2 alpha and beta (TRA2A and TRA2B) belong to the SR-like protein family of splicing factors, and are the human homologs of the Drosophila transformer -2 (TRA-2) protein, one of the most well studied splicing regulators in Drosophila [27]. Both TRA2A and TRA2B have been implicated in human cholesterol metabolism. TRA2A has been shown to target the scavenger receptor class B, member 1 (SCARB1) gene, which, through alternative splicing of exon 12, encodes both the scavenger receptor class B type I and II (SR-BI and SR-BII) proteins [12, 28]. Although both SR-BI and SR-II bind high density lipoprotein (HDL) particles and serve as the major receptor mediating reverse cholesterol transport from HDL to the liver, the two isoforms differ in their intracellular localization and efficiency in promoting cholesterol efflux. SCARB1 alternative splicing is regulated by estrogen, and similar to what we observed with sterol regulation of PTBP1, estrogen-induced changes in SCARB1 alternative splicing appear to be mediated by estrogen regulation of TRA2A [12].
Pihlajamaki et al reported that genes involved in RNA splicing were down-regulated in liver and muscle of insulin-resistant obese humans, as well as a mouse model of diet-induced obesity [29]. Knock-down of one of these genes, TRA2B (also known as SFRS10) in HepG2 cells was shown to increase expression levels of lipogenic genes such as SREBP1c, FASN, ACC1 and DGAT2, leading to a 60% increase in lipogenesis. These effects were confirmed in vivo, as heterozygous Sfrs10+/− mice had increased expression of lipogenic and triacylglycerol (TAG) synthesis genes, leading to elevated hepatic VLDL secretion as well as a 3-fold increase in plasma VLDL. This effect appeared to be attributed in part to altered expression of lipin 1 (LPIN1), a key regulator of lipoprotein metabolism. Using mini-gene constructs as well as knockdown in HepG2 cells, SFRS10 was shown to modulate the relative expression levels of the LPIN1 α versus β splice variants, with no effect on overall LPIN1 levels. Notably, SFRS10-induced changes in expression of lipogenic genes, as well as measures of cellular lipogenesis, TAG and lysophosphatidic acid, were reversed with LPIN1β knock-down. Very recently, the physiological relevance of this finding has been called into question as Brosch et al were unable to replicate the reduced SFRS10 expression levels in an independent cohort of lean versus obese human subjects [30]. However, given the complexities of accurately quantifying SFRS10 due to the expression of numerous splice variants, and the fact that different quantitation methods were employed by the two groups [31], further studies are required to determine the true relationship between SFRS10 and human metabolic disease.
Alternative splicing and human genetic variation in cholesterol metabolism and statin response
Originally, the relationships between alternative splicing and cholesterol metabolism were identified through human genetics. For example, a number of studies demonstrated that a SNP within HMGCR intron 13, rs3846662, or other SNPs in tight linkage disequilibrium, are associated with variation in endogenous levels of plasma cholesterol [32, 33]. Associations between rs3846662 and statin response have also been reported in populations treated with a number of statins including simvastatin, pravastatin, rosuvastatin and atorvastatin, indicating a relationship with statin efficacy as a class effect versus a specific statin drug [34–37]. These relationships have been attributed to alternative splicing of HMGCR since rs3846662 was shown to regulate exon 13 skipping [33,38], and inter-individual variation in the magnitude of statin-induced change in exon 13 skipping was directly correlated with variation in in vivo plasma LDL-C reduction with statin treatment [38]. Furthermore, loss of exon 13 abolishes the catalytic activity of the resulting HMGCR isoform [33, 39], while enrichment of the HMGCR13(−) splice variant reduces statin sensitivity of the HMGCR enzyme [38, 40].
There are numerous examples in the literature of other variants (both common and rare) in cis-elements causing aberrant splicing in a number of cholesterol related diseases. The most well-known example is familial hypercholesterolemia, in which many of the mutations identified within the low density lipoprotein receptor (LDLR) have been shown to cause skipping of various exons in the transcript, thus attenuating or abolishing LDLR activity [9, 11, 41]. Very recently, mutation analysis found that the molecular basis for a form of primary hereditary hypertriglyceridemia was a single base mutation in the glycerol-3-phosphate dehydrogenase 1 (GPD1) gene causing irregular splicing and generation of a truncated pathogenic GPD1 isoform [42]. Similarly, cases of hyperalphalipoproteinemia have been attributed to SNPs causing aberrant splicing of cholesterol ester transfer protein (CETP) in multiple independent populations [43, 44]. Splicing mutations in microsomal triglyceride transfer protein (MTTP) have been identified in cases of abetalipoproteinemia [45, 46], while splicing mutations in 7-dehydrocholesterol reductase (DHCR7) were found in individuals with Smith-Lemli-Optiz syndrome [47].
Remaining questions and challenges
Although we and others have started to define the relationship between alternative splicing and its effects on cellular cholesterol metabolism as well as its role in human lipid disorders, there are still a number of questions remaining. For example, while sterol-induced changes in alternative splicing are mediated by sterol regulated splicing factors; the molecular mechanism for how sterols regulate expression and activity of these splicing factors remains unknown. In addition, since functional studies are usually limited to testing for effects of alternative splicing on a specific catalytic activity or known function of the canonical protein isoform, it is possible that expression of alternatively spliced variants may affect cellular processes beyond cholesterol metabolism. For example, mevalonate kinase, the enzyme immediately following HMGCR in the cholesterol biosynthesis pathway, has also been shown to act as a mRNA binding protein whereby it specifically binds the luteinizing hormone receptor (LHR) mRNA to suppresses its translation [48]. While splice variants that are not translated are often assumed to be non-functional, both the prevalence of expression of non-coding transcripts as well as recent findings for specific regulatory roles of non-coding transcripts challenge this assumption [49].
Conclusion
With recent advances in sequencing technology, we now know that alternative splicing is a universal phenomenon that impacts the whole transcriptome, and thus potentially every cellular process. Numerous key regulatory genes involved in cholesterol metabolism are known to undergo functionally relevant changes in alternative splicing, and these changes appear to be part of an orchestrated mechanism of increasing or decreasing activity of the cholesterol biosynthesis and uptake pathways in conjunction with SREBP-mediated transcriptional regulation in response to changes in intracellular sterol content. A number of splicing factors have been shown to not only modulate this regulation, but also to directly affect plasma lipid metabolism suggesting the possibility that there may be both cis-acting and trans-acting gene variants underlying various dyslipidemias. Overall these studies validate the physiological relevance of alternative splicing as a mechanism that influences cholesterol homeostasis, and highlight the likelihood that our current understanding represents only a glimpse of the extent of the role of alternative splicing in lipid metabolism.
Key points.
Alternative splicing is a generalized phenomenon.
Expression of splice variants, whether they are translated into active proteins or not, can impact cholesterol metabolism.
Alternative splicing is a coordinated and generalized mechanism of regulating genes in the cholesterol biosynthesis and uptake pathways.
Splicing factors can modulate cholesterol metabolism by altering expression of splice variants.
One of the major challenges to the field is functionalizing the impact alternative spliced transcripts.
Acknowledgments
This work was funded by NIH NHLBI R01 104133 and U19 HL069757.
Abbreviations
- PTBP1
polypyrimidine tract binding protein 1
- TRA2A
Transformer-2alpha
- TRA2B
Transformer-2beta
- SFRS10
arginine/serine rich 10
- ESTs
expressed sequence tags
- HMGCR
3-hydroxy-3-methylglutaryl coenzyme A reductase
- HMGCR13(−)
HMGCR transcript lacking exon 13
- HMGCR13(+)
HMGCR transcript with exon 13
- LDL
low density lipoprotein
- HMGCS1
3-hydroxy-3-methylglutaryl coenzyme A synthase 1
- MVK
mevalonate kinase
- LDLR
low density lipoprotein receptor
- PCSK9
proprotein convertase subtilisin/kexin type 9
- LDLR12(−)
LDLR splice variant lacking exon 12
- SREBP
sterol response element binding protein
- hnRNP
heterogenous nuclear ribonuclear protein
- SR
serine-arginine
- FADS2
fatty acid desaturase 2
- FADS3
fatty acid desaturase 3
- UTR
untranslated region
- FASN
fatty acid synthase
- ACC1
acetyl-CoA carboxylase 1
- DGAT2
diacylglycerol O-acyltransferase 2
- LPIN1
lipin 1
- SNP
single nucleotide polymorphism
- GPD1
glycerol-3-phosphate dehydrogenase 1
- CETP
cholesterol ester transfer protein
- MTTP
microsomal triglyceride transfer protein
- DHCR7
7-dehydrocholesterol reductase
Footnotes
Conflict of Interest:
None
References and recommended reading
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelemen O, Convertini P, Zhang Z, et al. Function of alternative splicing. Gene. 2012 doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–15. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Kim B, Shin Y, et al. ECgene: an alternative splicing database update. Nucleic Acids Res. 2007;35:D99–103. doi: 10.1093/nar/gkl992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–9. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JM, Castle J, Garrett-Engele P, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holla OL, Nakken S, Mattingsdal M, et al. Effects of intronic mutations in the LDLR gene on pre-mRNA splicing: Comparison of wet-lab and bioinformatics analyses. Mol Genet Metab. 2009;96:245–252. doi: 10.1016/j.ymgme.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 10*.Medina MW, Gao F, Naidoo D, et al. Coordinately regulated alternative splicing of genes involved in cholesterol biosynthesis and uptake. PLoS One. 2011;6:e19420. doi: 10.1371/journal.pone.0019420. This is the first demonstration that key regulatory genes involved in cellular cholesterol homeostasis undergo regulated alternative splicing in response to changes in cellular sterol content. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tveten K, Ranheim T, Berge KE, et al. Analysis of alternatively spliced isoforms of human LDL receptor mRNA. Clin Chim Acta. 2006;373:151–7. doi: 10.1016/j.cca.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Moor AN, Merkler KA, et al. Regulation of alternative splicing of liver scavenger receptor class B gene by estrogen and the involved regulatory splicing factors. Endocrinology. 2007;148:5295–304. doi: 10.1210/en.2007-0376. [DOI] [PubMed] [Google Scholar]
- 13.Skandalis A, Frampton M, Seger J, Richards MH. The adaptive significance of unproductive alternative splicing in primates. RNA. 2010;16:2014–22. doi: 10.1261/rna.2127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu SN, Hertel KJ. Spliceosomes walk the line: splicing errors and their impact on cellular function. RNA Biol. 2009;6:526–30. doi: 10.4161/rna.6.5.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 17.Frischmeyer PA, van Hoof A, O’Donnell K, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–61. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 18.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–4. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mudge JM, Frankish A, Fernandez-Banet J, et al. The origins, evolution, and functional potential of alternative splicing in vertebrates. Mol Biol Evol. 2011;28:2949–59. doi: 10.1093/molbev/msr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tress ML, Martelli PL, Frankish A, et al. The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci U S A. 2007;104:5495–500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Xin D, Wang P, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–93. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–92. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–7. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 25.Knoch KP, Meisterfeld R, Kersting S, et al. cAMP-dependent phosphorylation of PTB1 promotes the expression of insulin secretory granule proteins in beta cells. Cell Metab. 2006;3:123–34. doi: 10.1016/j.cmet.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 26**.Reardon HT, Park WJ, Zhang J, et al. The polypyrimidine tract binding protein regulates desaturase alternative splicing and PUFA composition. J Lipid Res. 2011;52:2279–86. doi: 10.1194/jlr.M019653. One of the first demonstrations that modulation of a splicing factor, in this case PTBP1, can modulate cellular lipid metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci U S A. 1996;93:9004–9. doi: 10.1073/pnas.93.17.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webb NR, Connell PM, Graf GA, et al. SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. J Biol Chem. 1998;273:15241–8. doi: 10.1074/jbc.273.24.15241. [DOI] [PubMed] [Google Scholar]
- 29**.Pihlajamaki J, Lerin C, Itkonen P, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011;14:208–18. doi: 10.1016/j.cmet.2011.06.007. This study describes the identification of SFRS10, a splicing factor, associated with obesity and dyslipidemia, and specifically was the first study to demonstrate the metabolic impact of a splicing factor using an rodent model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brosch M, von Schonfels W, Ahrens M, et al. SFRS10--a splicing factor gene reduced in human obesity? Cell Metab. 2012;15:265–6. doi: 10.1016/j.cmet.2012.02.002. author reply 267–9. [DOI] [PubMed] [Google Scholar]
- 31.Pihlajamaki J, Lerin C, Kaminska D, et al. Response to Brosch et al. Cell Metab. 2012;15:267–269. doi: 10.1016/j.cmet.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkhardt R, Kenny EE, Lowe JK, et al. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler Thromb Vasc Biol. 2008;28:2078–84. doi: 10.1161/ATVBAHA.108.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung JY, Cho SK, Oh ES, et al. Effect of HMGCR variant alleles on low-density lipoprotein cholesterol-lowering response to atorvastatin in healthy Korean subjects. J Clin Pharmacol. 2012;52:339–46. doi: 10.1177/0091270011398239. [DOI] [PubMed] [Google Scholar]
- 35.Poduri A, Khullar M, Bahl A, et al. Common variants of HMGCR, CETP, APOAI, ABCB1, CYP3A4, and CYP7A1 genes as predictors of lipid-lowering response to atorvastatin therapy. DNA Cell Biol. 2010;29:629–37. doi: 10.1089/dna.2009.1008. [DOI] [PubMed] [Google Scholar]
- 36.Chasman DI, Posada D, Subrahmanyan L, et al. Pharmacogenetic study of statin therapy and cholesterol reduction. Jama. 2004;291:2821–7. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 37.Krauss RM, Mangravite LM, Smith JD, et al. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117:1537–44. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 38.Medina MW, Gao F, Ruan W, et al. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–62. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clendening JW, Pandyra A, Boutros PC, et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci U S A. 2010;107:15051–6. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipkin SM, Chao EC, Moreno V, et al. Genetic variation in 3-hydroxy-3-methylglutaryl CoA reductase modifies the chemopreventive activity of statins for colorectal cancer. Cancer Prev Res (Phila) 2010;3:597–603. doi: 10.1158/1940-6207.CAPR-10-0007. [DOI] [PubMed] [Google Scholar]
- 41.Al-Khateeb A, Zahri MK, Mohamed MS, et al. Analysis of sequence variations in low-density lipoprotein receptor gene among Malaysian patients with familial hypercholesterolemia. BMC Med Genet. 2011;12:40. doi: 10.1186/1471-2350-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basel-Vanagaite L, Zevit N, Har Zahav A, et al. Transient infantile hypertriglyceridemia, fatty liver, and hepatic fibrosis caused by mutated GPD1, encoding glycerol-3-phosphate dehydrogenase 1. Am J Hum Genet. 2012;90:49–60. doi: 10.1016/j.ajhg.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohtani R, Inazu A, Noji Y, et al. Novel mutations of cholesteryl ester transfer protein (CETP) gene in Japanese hyperalphalipoproteinemic subjects. Clin Chim Acta. 2012;413:537–43. doi: 10.1016/j.cca.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 44.Cefalu AB, Noto D, Magnolo L, et al. Novel mutations of CETP gene in Italian subjects with hyperalphalipoproteinemia. Atherosclerosis. 2009;204:202–7. doi: 10.1016/j.atherosclerosis.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 45.Pons V, Rolland C, Nauze M, et al. A severe form of abetalipoproteinemia caused by new splicing mutations of microsomal triglyceride transfer protein (MTTP) Hum Mutat. 2011;32:751–9. doi: 10.1002/humu.21494. [DOI] [PubMed] [Google Scholar]
- 46.Di Filippo M, Crehalet H, Samson-Bouma ME, et al. Molecular and functional analysis of two new MTTP gene mutations in an atypical case of abetalipoproteinemia. J Lipid Res. 2012;53:548–55. doi: 10.1194/jlr.M020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koo G, Conley SK, Wassif CA, Porter FD. Discordant phenotype and sterol biochemistry in Smith-Lemli-Opitz syndrome. Am J Med Genet A. 2010;152A:2094–8. doi: 10.1002/ajmg.a.33540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair AK, Young MA, Menon KM. Regulation of luteinizing hormone receptor mRNA expression by mevalonate kinase--role of the catalytic center in mRNA recognition. FEBS J. 2008;275:3397–407. doi: 10.1111/j.1742-4658.2008.06490.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]