Abstract
Objective
A sexual dimorphism exists in body fat distribution; females deposit relatively more fat in subcutaneous/inguinal depots whereas males deposit more fat in the intra-abdominal/gonadal depot. Our objective was to systematically document depot- and sex-related differences in the accumulation of adipose tissue and gene expression, comparing differentially expressed genes in diet-induced obese mice with mice maintained on a chow diet.
Research Design and Methods
We used a microarray approach to determine whether there are sexual dimorphisms in gene expression in age-matched male, female or ovariectomized female (OVX) C57/BL6 mice maintained on a high-fat (HF) diet. We then compared expression of validated genes between the sexes on a chow diet.
Results
After exposure to a high fat diet for 12 weeks, females gained less weight than males. The microarray analyses indicate in intra-abdominal/gonadal adipose tissue in females 1642 genes differ by at least twofold between the depots, whereas 706 genes differ in subcutaneous/inguinal adipose tissue when compared with males. Only 138 genes are commonly regulated in both sexes and adipose tissue depots. Inflammatory genes (cytokine–cytokine receptor interactions and acute-phase protein synthesis) are upregulated in males when compared with females, and there is a partial reversal after OVX, where OVX adipose tissue gene expression is more ′male-like′. This pattern is not observed in mice maintained on chow. Histology of male gonadal white adipose tissue (GWAT) shows more crown-like structures than females, indicative of inflammation and adipose tissue remodeling. In addition, genes related to insulin signaling and lipid synthesis are higher in females than males, regardless of dietary exposure.
Conclusions
These data suggest that male and female adipose tissue differ between the sexes regardless of diet. Moreover, HF diet exposure elicits a much greater inflammatory response in males when compared with females. This data set underscores the importance of analyzing depot-, sex- and steroid-dependent regulation of adipose tissue distribution and function.
Keywords: high-fat diet, inflammation, fat partitioning, gender dimorphism, mouse, microarray
Introduction
There are sex-based differences in total body fat distribution and content, with women having higher levels of adiposity deposited in the subcutaneous depot compared with men.1–5 After menopause or surgical removal of the ovaries, the fat deposition in women shifts toward a more central/male-like pattern of adipose distribution.6–10 These sex differences in the distribution of body fat are statistically associated with differential risks to various chronic diseases.11,12 Epidemiological and clinical studies show that the amount of thigh/ subcutaneous fat tissue is associated with a lower metabolic risk profile (lower triglycerides and higher high-density lipoprotein cholesterol, glucose and insulin levels13) in both men and women.
Sex differences in fat distribution are manifested by differences in the number and size of fat cells in lower/truncal subcutaneous fat depots. Specifically, women have smaller visceral fat cells independent of obesity. 14–18 Sex- and depot-related differences in the uptake and release of metabolic fuels (fatty acids, glycerol and lactate) or adipokines are thought to mediate the association of fat distribution and metabolic risk, but the molecular mechanisms involved are not well understood.18 Adipose tissue of females produces more leptin, antiinflammatory genes and insulin-sensitizing adipokines, such as adiponectin and omentin, independent of fat cell size or fat mass.19–21 Isolated adipocytes from subcutaneous depots of premenopausal females and young rats are also more sensitive to insulin when compared with males.22–27 The enhanced uptake and storage of fatty acids in lower body/truncal fat depots in premenopausal females is because of their relatively higher lipoprotein lipase activity and expression of fatty acid transporters.17
Sex differences in adipocyte metabolism and depot differences may at least partially be mediated by differences in sex steroids and/or expression of steroid receptors such as the estrogen and androgen receptors.18,28–31 Numerous depot differences in gene expression between subcutaneous and abdominal adipose tissues of men or women have been documented.18,32 For rodents, sex differences between inguinal/subcutaneous (inguinal white adipose tissue (IWAT)) depots may be related to the fact that IWAT in female rodents contains mammary glands. Sex differences in gonadal/intra-abdominal (gonadal white adipose tissue (GWAT)) may be because of the close proximity to reproductive tissue. Despite these obvious differences, only a few previous studies have attempted to systematically compare male and female adipose tissue gene expression33,34 to more fully understand whether sex-based differences in male and female adipose tissues are related to differences in function or sex steroids.
Recent studies by Strissel et al.35 show that high-fat-diet (HF diet)-induced obesity causes inflammation and remodeling of GWAT adipose tissue of male C57/BL6 mice, with very little alteration in the IWAT. No previous studies have compared regional differences in the inflammatory response and remodeling after exposure to an HF diet in adipose tissues between males and females. We specifically assessed whole adipose tissue rather than isolated adipocytes because of potential changes in gene expression and inflammation induced by the isolation procedure. The objective of this study was to systematically document depot- and sex-related differences in the accumulation of adipose tissue and gene expression using diet-induced obese mice as a model. We compared these findings with those of a select panel of validated differentially regulated genes in mice consuming a chow diet. The influence of female sex steroids was assessed by comparing females and ovariectomized (OVX) animals. Our results reveal that depot-related differences in gene expression are common in males and females when maintained on a chow diet, but that females are resistant to adipose tissue inflammation associated with HF-diet-induced obesity.
Materials and methods
Animals
The study was performed using 21 male, 21 female and 21 OVX female C57/BL6 mice fed from day 21 (after weaning) for 12 weeks on an HF diet (4.7 Kcalg−1 and 45% calories from fat; Research Diets, Inc., New Brunswick, NJ, USA). An additional cohort of 10 male, 10 female and 10 OVX C57/ BL6 mice were weaned and maintained for 12 weeks on a chow diet (Harlan Teklad, Madison, WI, USA, 2916). Mice were single caged and maintained at a controlled temperature (22°C) and a 12 h light-dark cycle (light on from 0800 to 2000h). Daily food intake and body weight were monitored. All procedures were approved by the animal care and use committee of the University of Cincinnati and the University of Texas Southwestern Medical Center.
Ovariectomy (OVX)
OVX or sham surgery was performed in anesthetized female mice by making bilateral dorsal incisions through the skin, such that the ovary and oviduct could be rapidly removed. In the sham operation, the ovary and oviduct were visualized but left intact before the incisions were sutured. The success of the OVX procedure was confirmed by measuring uterine weights. To equate for any interaction between gene expression profiles and surgical stress, all mice (male, female and OVX) were exposed to a similar surgical paradigm. We represent our groups as males (sham operated), females (sham operated) and OVX.
Nuclear magnetic resonance/body adiposity methods
Body fat was estimated using two procedures. During an ongoing experiment, body fat was assessed using nuclear magnetic resonance (EchoMRI; EchoMedical Systems, Houston, TX, USA). Unanesthetized mice were placed in a restraint tube and inserted into the nuclear magnetic resonance machine. Upon killing, individual fat pads were weighed.
Tissue collection
After 12 weeks of exposure to the diet, animals were killed during the first 2 h of the beginning of the light cycle after a 12-h fast. All females were killed in the proestrus phase of their estrus cycle. Two different WAT depots—GWAT and IWAT—were rapidly dissected and the orientation of the fat pad was maintained. Half of the fat pad was rapidly frozen for microarray analysis, and the other half of the fat pad was rapidly frozen for validation of target genes. From the other side of the animal, the same fat pads were removed and processed for tissue morphology. Where indicated, samples were rapidly frozen in liquid nitrogen and stored at −70°C for RNA analysis.
Plasma analysis
Cardiac puncture was used to extract plasma. Blood was also collected, stored at room temperature for 1 h and overnight at 4°C, and was then centrifuged at 1000g for 10 min to collect the serum, which was stored for 5 days at −20 °C until analysis. Plasma leptin was analyzed using a rat leptin radioimmunoassay kit (Linco Research, St Charles, MO, USA). This assay is able to detect leptin in 100 µl samples of plasma with intra- and interassay coefficients of variation of 4.6–5.7% for leptin. We also assayed the plasma using the Luminex Multi Analysis (Luminex Corporation, Austin, TX, USA) system for the simultaneous measurement of up to 20 different hormones, peptides or cytokines in mouse plasma (cytokine panel) in a single reaction vessel based on latex microbeads dyed with two fluorophores.
RNA isolation
GWAT/IWAT were homogenized in 800 µl Trizol reagent (Invitrogen, Carlsbad, CA, USA), and total cellular RNA was isolated according to the specifications of the manufacturer. Total RNA was further purified using the RNeasy Mini kit (Qiagen, Valencia, CA, USA). The quality and concentration of the RNA was determined by measuring the absorbance at 260 and 280nm, and RNA integrity was confirmed by bioanalysis (Agilent 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA).
Microarray analysis
For the microarray analysis, seven independent pooled samples were analyzed from GWAT and IWAT fat pads. Each sample comprised a pool of tissue from three animals (for pooled samples, reverse transcription was performed on each sample individually and equal amounts of complementary DNA (cDNA) were pooled); thus, samples were obtained from a total of 21 mice. GWAT/IWAT adipose RNA pools were generated from the following groups of mice: (1) males, (2) females and (3) OVX. To identify genes that were differentially expressed in the two WAT fat pads, comparisons were made between normalized signal intensity from the control group (males) and experimental groups (female and OVX) from each experiment. Each total RNA sample was processed according to protocols recommended by the manufacturers. In brief, total RNA is reverse-transcribed with an oligo-dT primer and double-stranded cDNA is generated. The cDNA serves as a template for the in vitro transcription reaction that produces amplified amounts of biotin-labeled antisense mRNA. This biotinylated RNA is referred to as labeled cRNA. After fragmentation, the cRNA is hybridized onto oligonucleotide micrarrays (Mouse Genome 430 2.0; 40 000 mouse probe sets). A GeneChip Operating System absolute expression analysis was performed for each Gene-Chip genome array hybridization. After the initial analysis, the absolute analysis was re-run using global scaling to an average target intensity of 350. The scaling allows for the direct comparison of hybridization values from the different targets analyzed in this project (and with any additional GeneChip sample assays using the same array type). For each analysis, scaled or unscaled, AMC Project Report Version 12 (06/27/07) GeneChip Operating System parameters α1 and α2 are set to 0.05 and 0.065, respectively. These parameters establish the point at which a probe set is called present (P), marginal (M) or undetectable (A). This call is based on the detection of the P-value of the probe set as determined using the software.
Analysis of gene expression
For the microarray data analysis, normalized expression values from the Affymetrix identifier were imported into the online software server Genesifter (VizX Labs, Inc., Seattle, WA, USA). For the comparisons of the microarray data sets, multiple t-tests were used to identify genes with at least a twofold differences in gene expression (with Benjamini and Hockberg correction; P < 0.05) and at least an expression level of 100. Samples from at least one of the groups had to have a 100% present call (7 out of 7) according to Affymetric MAS 5.0.
Real-time reverse transcriptase-PCR
Real-time reverse transcriptase-PCR was performed on GWAT/IWAT as described previously.36 RNA samples (1 µg) were reverse transcribed using random hexamer primers (Promega, Madison, WI, USA). The reverse transcriptase product was then optimized for each primer/probe set through a titration PCR analysis. The reaction mixture (10µl) consisted of 5 µl of TaqMan Universal PCR Master Mix, 300nM specific target gene primers, 80nM 18S rRNA gene primers, 250nM specific probe and 2 µl cDNA. Amplification was performed using the ABI/Prism 7700 Sequences Detector System (Applied Biosystems, Foster City, CA, USA) with 2 min at 50 °C, 10 min at 95 °C and then 45 cycles each at 95 °C for 15 s and 60 °C for 60s. Primer/probe sets were purchased from Applied Biosystems (see Supplementary Table 1 that lists probe set catalog numbers).
Inflammatory cytokines and receptors superarray
Reverse transcription was performed on 1 µg of RNA using TaqMan Reverse Transcription Kit (Applied Biosystems). For each reaction, cDNA synthesis was performed using 1 µg of RNA in a reaction containing 2 µl of 10 × reverse transcriptase buffer, 4 µl of 25 mM MgCl2, 3 µl of 10 mM deoxynucleo-tide triphosphates, 1 µl of 50 µM random hexamers, 0.5 µl of RNase inhibitor, 1 µl of MultiScribe Reverse Transcriptase, q.s. to 20 µl with nuclease-free water. Reverse transcriptase reactions were performed on an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany) programmed for 25 °C for 10 min, 37 °C for l h and 95 °C for 5 min. Samples were diluted with 80 µl nuclease-free water and stored at 4 °C until quantitative PCR (qPCR) was performed. qPCR was performed according to the manufacturer’s directions (RT2Pro-filer PCR Array User Manual, v1.5) using RT2 Profiler PCR Array mouse Inflammatory Cytokines and Receptors array (Cat no. PAMM-011) on an ABI 7300 Real-Time PCR System (Applied Biosystems). For each 96-well plate, a master mix was made containing 1225 µl Power SYBR Green PCR Master Mix (Applied Biosystems), 98 µl diluted cDNA synthesis reaction and 1127 µl double-distilled water. The master mix was vortexed and 21 µl was added to each well. qPCR reactions were 95 °C for 10 min, and 40 cycles of 95 °C for 15s followed by 60 °C for l min. A dissociation (melting) curve was performed according to the manufacturer’s instructions and only samples with one peak at temperatures of > 80°C were accepted. Auto Ct values were calculated using 7300 RQ Study Software v.1.3 (Applied Biosystems), which was verified and adjusted as necessary. Gene expression values are expressed as ΔCt for each set of duplicates. The fold-change for each gene was calculated as 2(−ΔΔCt), in which ΔΔCt = ΔCt (female group)−ΔCt (male group).
Adipose tissue morphology
Fat was dissected, fixed, embedded in paraffin and sectioned.37 Sections were stained with hematoxylin and eosin or with Gomori trichrome. Digital images were acquired with an Olympus DX51 light microscope. For each mouse, morpho-metric data were obtained from 500 adipocytes from three or more sections that were cut at least 50 µm apart. Fat cell area/ size was determined using ImageJ software (NIH). Immuno-histochemistry was performed using VectaStain kits (Vector Labs). Antibodies were rat anti-mouse F4/80 (Serotec). Negative controls were nonimmune immunoglobulin G and peptide-neutralized primary antibody.
Adipocyte death
Dead/dying adipocytes were identified by the presence of crown-like structures. These structures have been shown to correspond to perilipin-negative lipid droplets surrounded by macrophage (MΦ) crowns.37 The frequency of adipocyte death was calculated from micrographs as (number of dead adipocytes/number of total adipocytes) × 100.
Statistical analysis
Data are expressed as means±s.e.m. (n = 6–8). Two-way analysis of variance was used to determine the significance of the effect of sex on the measured parameters. Single differences between male, female and OVX were assessed using Student’s t-test. Simple correlations were assessed using Pearson’s correlation coefficients. Threshold significance was defined at P < 0.05. For the Kegg pathway analysis, the % change represents the percentage of genes that met the differential expression criteria of the analysis compared with the total number of genes in this pathway. The % represents the percentage of genes that were above the set detection limits in the adipose tissue compared with the total number of genes in this pathway. The z-score represents an analysis of the observed number of genes in this pathway compared with the number of genes that would be expected to change considering the overall analysis. A positive z-score indicates that the pathway has a greater number of genes meeting the criteria than would be expected by chance.38
Results
Nuclear magnetic resonance/body adiposity
Body and fat pad weight are summarized in Table 1. Under HF diet, both male and OVX mice gained more weight than intact females (P < 0.05, two-way analysis of variance).
Table 1.
Body weight, body adiposity, lean tissue and plasma analysis
| Male | Female | OVX | |
|---|---|---|---|
| Final body weight (g) | 42.25±0.67 | 32.54±1.14* | 36.25±1.15 |
| Body weight change (g) | 9.02±0.29 | 5.8± 0.37* | 9.61±0.61 |
| Body fat (g) | 14.98±0.36 | 10.8±1.06* | 14.55±1.0 |
| Lean mass (g) | 24.9±0.33 | 20.06±0.24 | 20.26±0.32 |
| Inguinal fat pad (g) | 1.5±0.06 | 0.92±0.08* | 1.22±0.12 |
| Retroperitoneal fat pad (g) | 0.4±0.04 | 0.30±0.04 | 0.28±0.02 |
| Gonadal fat pad (g) | 1.66±0.16 | 0.86±0.08* | 1.26±0.12 |
| Mesenteric fat pad (g) | 2.16±0.12 | 1.08±0.12 | 1.48±0.16 |
| % Inguinal fat pad (depot/BW) | 3.55±0.06 | 2.82±0.10 | 3.37±0.11 |
| % Retroperitoneal fat pad (depot/BW) | 0.95±0.05 | 1.35±0.04 | 0.77±0.03 |
| % Gonadal fat pad (Depot/BW) | 3.93±0.09 | 2.64±0.11 | 3.48±0.12 |
| % Mesenteric fat pad (Depot/BW) | 2.53±0.13 | 3.32±0.13 | 4.08±1.15 |
| Inguinal/intra-abdominal | 0.35 | 0.41 | 0.40 |
| Cumulative food intake (g) | 162.35±1.17 | 157.1±2.47 | 164.77±2.7 |
| Leptin | 78.42±17.53 | 34.37±7.68* | 55.05±12.3 |
| TNF-α | 12.6±4.0 | 10.9±7.0 | 19.5±10.0 |
| IL-1β | 39.7±3.0 | 18.2±7.0 | 55.2±7.0 |
| IL-6 | 11.9±12.0 | 16.4±16.0 | 16.3±19.0 |
| MCP-1 | 87.0±8.0 | 61.1±16.0 | 77.9±16.0 |
| GmCSF | 130.8±12.0 | 153.7±18.0 | 149.8±18.0 |
Abbreviations: GmCSF, granulocyte-macrophage colony-stimulating factor; HF, high-fat; IL, interleukin; MCP-1, monocyte chemotactic protein-1; OVX, ovariectomy; TNF-α, tumor necrosis factor-α.
Statistically different from control, males (P < 0.05). Body weight (g), carcass fat(g), lean mass (g), tissue weights (g), plasma leptin, TNF-α, IL-1β, IL-6, MCP-1 and GmCSF of male, female or OVX mice maintained on HF diet for 12 weeks (n = 21/group).
Plasma analysis
Measured plasma leptin levels reflect total adiposity on HF diet, with males having significantly higher plasma levels of leptin than females. Removal of the ovaries in females increases plasma leptin levels, which is a reflection of the increase in total body adiposity (Table 1). The remaining hormones, measured by the Luminex (tumor necrosis factor-α, interleukin (IL)-1β, IL-6, monocyte chemotactic protein-1 and granulocyte-macrophage colony-stimulating factor), do not achieve statistical differences across the groups on HF diet.
Sex-dependent differences in gene expression: microarray analysis
A scatter plot depicting comparisons of male versus female GWAT and IWAT on HF diet is depicted in Supplementary Figures. We show here that 1642 probe sets amplify differentially expressed RNA between the two sexes (Supplementary Figure 2 and Supplementary Table 5); more than two-thirds of the differently expressed probe sets amplify RNAs that are significantly lower in female GWAT than in male GWAT. Analysis of male IWAT versus female IWAT found that only 706 amplification probe sets are different between the two sexes. In contrast to GWAT, more than two-thirds of the probe sets corresponding to differentially regulated genes are higher in females. Although both fat pads have a relatively large number of sex-dependent differences, only a small number of these probe sets that amplify differentially expressed messages are commonly regulated in GWAT and IWAT (8 and 20%, respectively; Supplementary Figure 2). There are also 129 probe sets that correspond to genes that are regulated in opposite directions; that is, higher in male GWAT than in IWAT, and higher in female IWAT than in GWAT. Examples of these include keratin 8, 13 and 18, prolactin receptor, cadherin 1 and lipocalin 2.
It should be noted that the parameters used for the microarray analysis are considered conservative, requiring an expression level of 100 and a twofold differential expression. If the analysis was widened to include all probe sets with a significant 1.5 + fold change (P < 0.05 using Student’s t-test) in expression and 100% present call in at least one group (but no limit on expression values), this would result in a large increase in the number of differentially expressed genes. For GWAT there are 4063 probe sets that amplify differentially expressed genes between male and female and it was 2025 in IWAT. In addition, we denote statistical significance only for those genes that we confirmed using PCR. We directly compared the GWAT expression of these validated genes with those of mice maintained on chow.
Ovarian-dependent differences in gene expression
A scatter plot depicting comparisons of female versus OVX female GWAT and IWAT on HF diet is represented in Supplementary Figure 1. When compared with differences in gene expression between male and female animals (Supplementary Figure 1a and b), there are very few differences in gene expression between female and OVX females (Supplementary Figure 1c and d) in either the GWAT or IWAT on HF diet. Indeed, there are only 118 probe sets that amplify RNA with differential expression in the GWAT and 28 probe sets with differential expression in IWAT between the two groups (Supplementary Figure 3). However, 75% of the probe sets corresponding to differentially expressed genes in GWAT, and 79% in IWAT, are in common with genes differentially expressed between male and female fat pads, indicating that these are likely steroid-regulated genes. Of the probe sets corresponding to differentially expressed genes between the female and OVX females, only five are commonly regulated in the two fat pads.
When using the less stringent parameters outlined above, there is again a large increase in the number of probe sets that amplify differentially expressed genes. This modification in analysis parameters increases the number of probe sets corresponding to differentially expressed genes in the GWAT to 517 and in IWAT to 205.
Kegg pathway analysis
To begin to identify functional differences in fat depots between male and females, we performed a Kegg Pathway analysis. Table 2 lists pathways identified by this analysis for the different comparisons (a pathway analysis was performed using Panther v 6.1 on the 448 genes that were differentially expressed across the clusters in Table 2, see Supplementary Table 6). For the male GWAT versus female GWAT comparison, the cytokine signaling pathway shows the greatest number of genes (26 genes) with differential expression (see Supplementary Table 2). With the less stringent analysis, this number increases to 50 genes with differential expression. As expected, there is also a significant difference in numerous genes involved in the insulin signaling pathway (Supplementary Table 3), with 10 differentially expressed genes (25 with the less stringent analysis). For the Kegg Pathway analysis of the male IWAT versus female IWAT comparison, only four genes were identified from the cytokine signaling and three from the insulin signaling pathway (see Supplementary Table 2 and Supplementary Table 3, respectively). However, using the less stringent analysis, 21 cytokine signaling and 13 insulin signaling pathways were identified. Neither of these pathways was highlighted by the Kegg pathway analysis for female versus OVX female, for either of the fat pads. The z-scores for genes that were up- and downregulated independent of each other have been calculated using methods previously reported.38 A z-score of ≥ 2 is considered to be significant. Table 2 shows only Kegg pathways with a z-score of > 2.
Table 2.
Kegg pathway analysis of microarray data
| Kegg pathway | Up | Down | % Changeda | % Presentb | Z-score up | Z-score down |
|---|---|---|---|---|---|---|
| Male GWAT versus female GWAT | ||||||
| Cytokine–cytokine receptor interaction | 2 | 24 | 13 | 54 | −1.14 | 2.92 |
| CAMs | 3 | 17 | 19 | 54 | 0.46 | 3.61 |
| Leukocyte transendothelial migration | 2 | 17 | 24 | 80 | 0.09 | 4.61 |
| B-cell receptor signaling pathway | 0 | 14 | 34 | 90 | −1.04 | 5.99 |
| Natural killer cell-mediated cytotoxicity | 0 | 13 | 18 | 64 | −1.36 | 3.19 |
| Complement and coagulation cascades | 2 | 9 | 27 | 56 | 1.05 | 3.38 |
| Fc epsilon RI signaling pathway | 0 | 10 | 26 | 67 | −1.10 | 3.30 |
| Hematopoietic cell lineage | 0 | 10 | 16 | 47 | −1.20 | 2.71 |
| Insulin signaling pathway | 5 | 5 | 13 | 89 | 2.15 | −0.66 |
| Type 2 diabetes mellitus | 5 | 4 | 21 | 54 | 4.49 | 0.70 |
| Male IWAT versus female IWAT | ||||||
| CAMs | 12 | 0 | 11 | 59 | 6.33 | −0.99 |
| Tight junction | 8 | 0 | 9 | 76 | 4.22 | −0.91 |
| Cell communication | 5 | 2 | 10 | 50 | 2.87 | 1.87 |
| Leukocyte transendothelial migration | 7 | 0 | 9 | 86 | 3.68 | −0.89 |
| Female GWAT versus OVX GWAT | ||||||
| Natural killer cell-mediated cytotoxicity | 5 | 0 | 7 | 58 | 5.75 | −0.31 |
| Leukocyte transendothelial migration | 5 | 0 | 6 | 70 | 6.01 | −0.30 |
| Female IWAF versus OVX IWAF | ||||||
| None |
Abbreviations: CAM, cell adhesion molecule; GWAT gonadal white adipose tissue; IWAT, inguinal white adipose tissue; OVX, ovariectomy.
The percentage of genes that met the differential expression criteria of the analysis compared with the total number of genes in this pathway.
The percentage of genes that were above the set detection limits in the adipose tissue compared with the total number of genes in this pathway. The z-score represents an analysis of the observed number of genes in this pathway compared with the number of genes that would be expected to change considering the overall analysis. A positive z-score indicates that the pathway has a greater number of genes meeting the criteria that would be expected by chance.38 Up- or downregulated genes in males, relative to females, or OVX in Kegg pathway analysis: male gonadal fat (GWAT) is compared with microarray data from female GWAT; male inguinal fat (IWAT) is compared with microarray data from female IWAT; female GWAT is compared with microarray data from OVX GWAT; female IWAT is compared with microarray data from OVX IWAT.
qPCR confirmation of microarray results
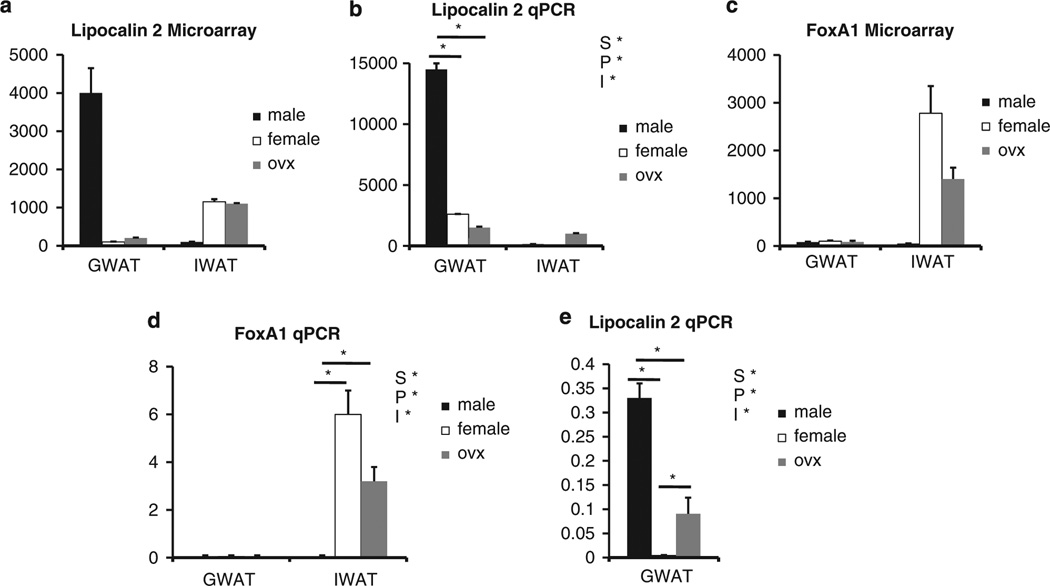
To initially test the consistency of the microarray results, we chose a selection of genes with large sex-dependent or fat pad-dependent differences. Figure 1 illustrates comparisons between the microarray and qPCR results for lipocalin 2 and FoxA1 (forkhead box A1). In all instances, for both sex and fat pad, there is strong agreement between the microarray analysis and qPCR confirmation. We found lipocalin 2 highly expressed in male GWAT relative to females and OVX regardless of diet (Figure 1a – d). It should be noted that although FoxA1 is highly expressed in only the IWAT of female and OVX females (Figure 1), this is likely because of expression in mammary gland epithelium, which is present specifically in IWAT of females, rather than expression in adipocytes.
Figure 1.
Validation of microarray results with qPCR. (a) Differences in lipocalin 2 microarray gene expression between males, females and OVX in gonadal fat (GWAT) and inguinal fat (IWAT). (b) qPCR of lipocalin 2 mRNA in the three groups on HF diet. (c) Differences in FoxA1 microarray gene expression between males, females and OVX. (d) qPCR of FoxA1 mRNA in the three groups. (e) qPCR of GWAT lipocalin 2 mRNA in the three groups on chow (Note: S* = P<0.05 compared between the sexes; P* = P<0.05 compared between the fat pad; and I* = P<0.05 independent of the sexes).
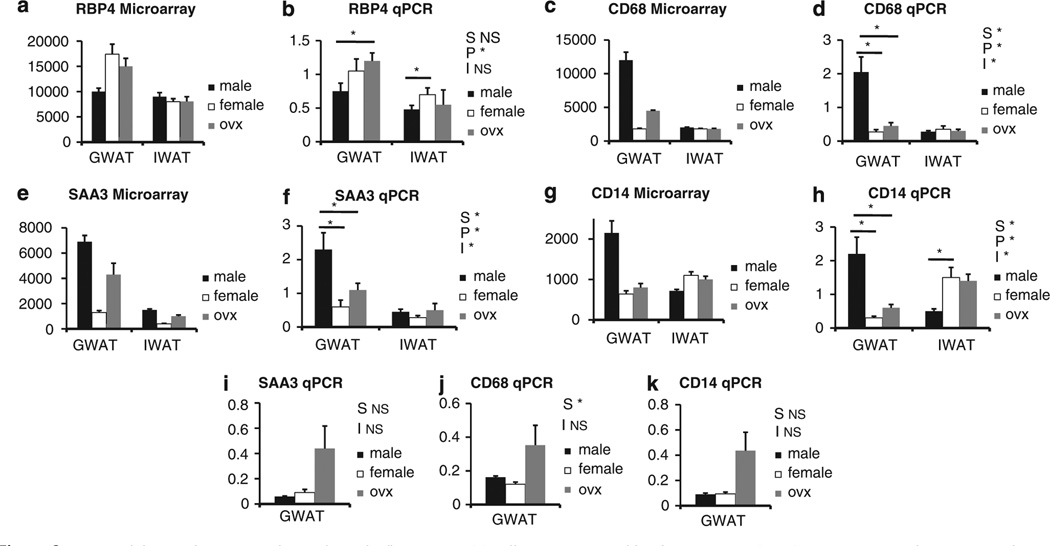
To confirm changes in the cytokine signaling pathway, we used a SuperArray qPCR platform (see Supplementary Table 4). The qPCR analysis for the male GWAT versus female GWAT confirmed many of the differences identified by microarray analysis, as well as identified a few additional genes that did not show up on the microarray results. These additional genes are likely due to the increased sensitivity of the qPCR technique. This was especially apparent for the analysis of the male IWAT versus female IWAT comparison, in which numerous genes not identified by microarray analysis were shown to be significantly different by qPCR (see Supplementary Table 4). Although a comparison of the GWAT versus IWAT was not performed, it is apparent from the average Ct values that expression of cytokines is higher in GWAT when compared with IWAT for both sexes. Again, the increased sensitivity of the qPCR technique has shown that cytokine expression in both GWAT and IWAT from the male is higher than the female; however, the relative level of expression of these genes is much lower in IWAT. Furthermore, we performed standard qPCR for additional genes associated with inflammation, macrophage infiltration and activation. Figure 2 shows a strong agreement between the microarray and qPCR results for retinol-binding protein 4 (panels a and b), CD68 (panels c and d), serum amyloid A3 (SAA3; panels e and f) and CD14 (panels g and h). The one discrepancy between the two techniques was for SAA3, in which the microarray results indicated that OVX resulted in a near male pattern of expression in GWAT, whereas the qPCR showed only a trend toward a change in expression between female versus OVX female. Genes that are expressed in adipocytes involved in the acute-phase response and insulin resistance, including SAA3, is higher in male than female GWAT, consistent with the other inflammatory markers. SAA3, CD68 and CD14 (Figure 2 i,j,k) were similarly expressed regardless of sex when the mice were maintained on chow. In addition, we found that after OVX there was a trend for increased SAA3, CD68 and CD14 that did not reach significance, but suggests a potential anti-inflammatory role of estrogen (Figure 2 i,j,k). Furthermore, these genes were significantly elevated in males after HF diet exposure.
Figure 2.
qPCR validation of microarray for markers of inflammation. (a) Differences in retinol binding protein 4 (RBP4) gene expression by microarray between males, females and OVX in gonadal fat (GWAT) and inguinal fat (IWAT). (b) qPCR of RBP4 mRNA on HF diet. (c) Differences in CD68 gene expression by microarray. (d) qPCR of CD68 mRNA on HF diet. (e) Differences in SAA3 gene expression by microarray. (f) qPCR of SAA3 mRNA on HF diet. (g) Differences in CD14 gene expression by microarray. (h) qPCR of CD14 mRNA on HF diet. (i) qPCR of GWAT SAA3 mRNA on chow. (j) qPCR of GWAT CD68 on chow. (k) qPCR of GWAT CD14 mRNA on chow (Note: S* = P<0.05 compared between the sexes; P* = P<0.05 compared between the fat pad; and I* = P < 0.05 independent of the sexes).
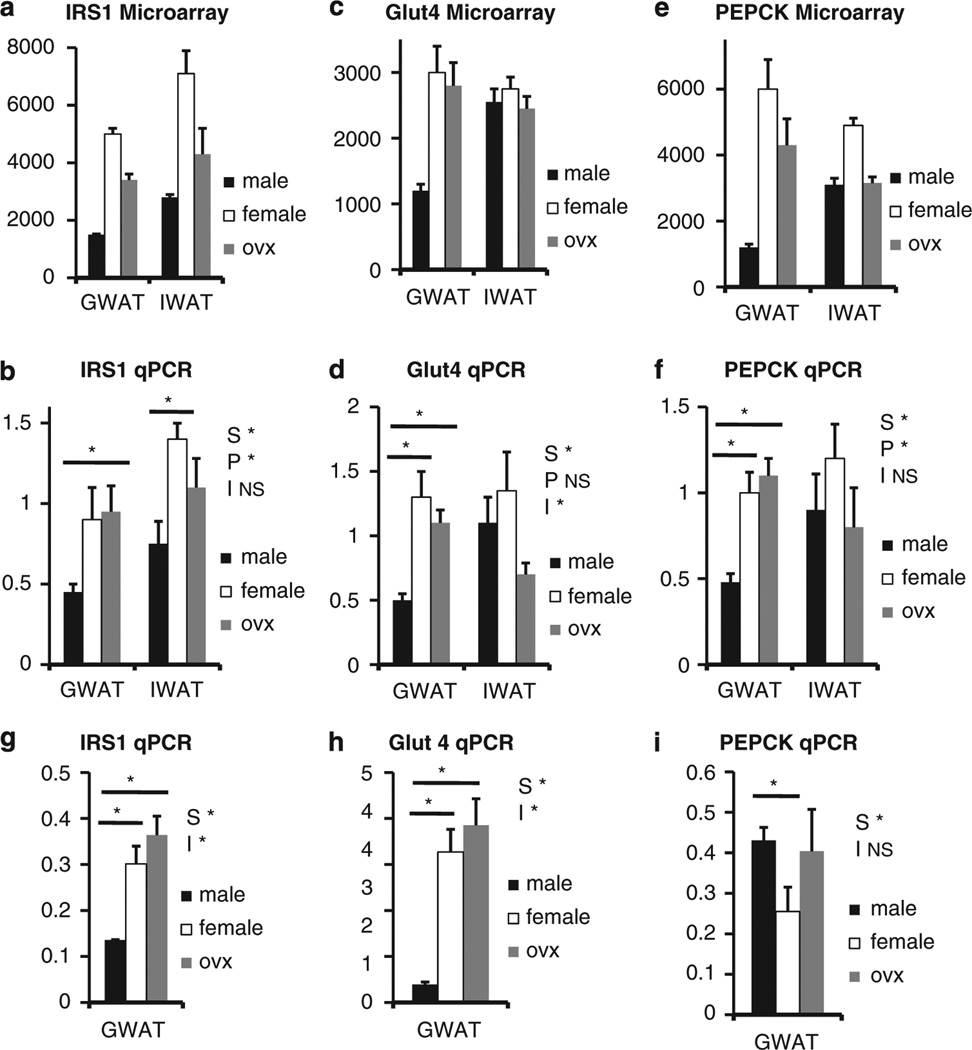
Finally, Figure 3 shows a comparison of microarray and qPCR results for several genes involved in regulation of insulin sensitivity. Again, there is strong agreement between the two techniques for the insulin receptor substrate 1 (panels a and b), the glucose transporter 4 (panels c and d) and phosphoenolpyruvate carboxykinase (PEPCK) (panels e and f). We found a similar pattern of expression for insulin receptor substrate 1 and glucose transporter 4 (Figure 3 g,h) expression in chow-fed animals. However, phosphoenolpyruvate carboxykinase did not show any sexual dimorphism until animals were exposed to HF diet.
Figure 3.
qPCR validation of microarray for markers of the insulin signaling pathway. (a) Differences in insulin receptor substrate 1 (IRS1) gene expression by microarray between males, females and OVX in gonadal fat (GWAT) and inguinal fat (IWAT). (b) qPCR of IRS1 mRNA on HF diet. (c) Differences in glucose transporter 4 (Glut4) gene expression by microarray. (d) qPCR of Glut4 mRNA on HF diet. (e) Differences in phosphoenolpyruvate carboxykinase (PEPCK) gene expression by microarray. (f) qPCR of PEPCK mRNA on HF diet. (g) qPCR of GWAT IRS1 mRNA on chow. (h) qPCR of GWAT Glut4 mRNA on chow. (i) qPCR of GWAT PEPCK mRNA on chow (Note: S* = P<0.05 compared between the sexes; P* = P<0.05 compared between the fat pad; and I* = P < 0.05 independent of the sexes).
Adipose tissue morphology
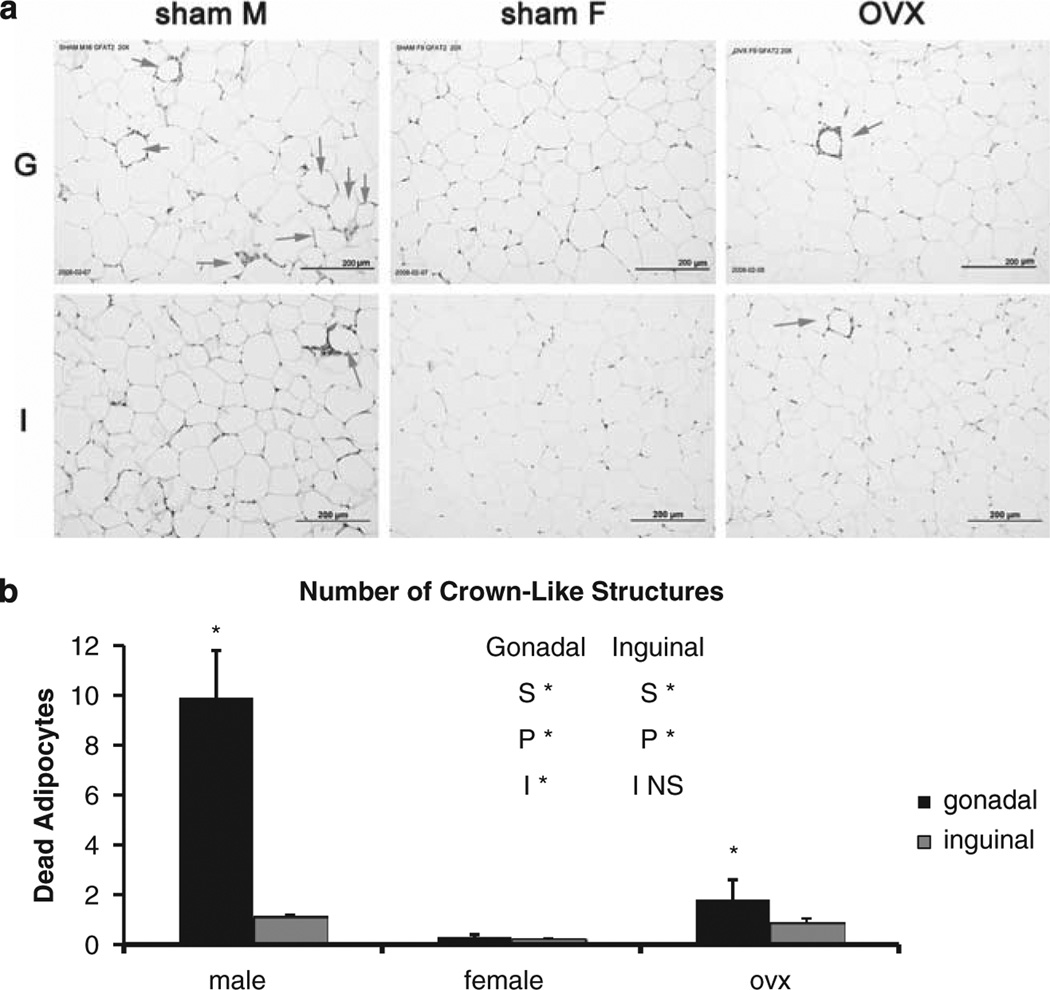
Adipose tissue morphology was assessed for crown-like structures, which are clusters/rings of macrophages that have been recruited to the periphery of a dead or dying adipocyte.35 The male GWAT adipose tissue contains more crown-like structures than females, consistent with their increased expression of inflammatory genes (Figure 4). Furthermore, OVX increases the number of crown-like structures in GWAT compared with females, suggesting an anti-inflammatory role for female sex hormones. Fat cell area/size was determined, and in GWAT, there was a significant increase in OVX fat cell size relative to males or females (male: 5985.75±357.45; female: 5508.5±4778.74; and OVX: 12858.15±4778.74). For IWAT, the females had statistically smaller fat cells when compared with the males or the OVX (male: 5220.01±535.76; female: 3365.91 ± 583.93; and OVX: 4996.61 ± 572.59).
Figure 4.
(a) Adipose tissue morphology. Histology (hematoxylin and eosin-stained sections) of male, female or OVX gonadal (G) or inguinal (I) showing crown-like structures (arrows, at week 12 because of numerous crown-like structures). (b) Quantification of adipocyte death determined from multiple histological sections (> 500 cells) of 6–8 mice per group per time point. Bars identified by * are significantly different (P<0.05) by Tukey′s procedure.
Discussion
After exposure to a high fat diet, we found that age-matched males gained weight and body fat, and increased their percentage of adiposity more than females. In addition, there is a trend toward relatively more fat deposited in intraabdominal depots of the males when compared with the females. Analysis of micro- and qPCR arrays suggests a depot difference in inflammatory pathways (IWAT≪GWAT) in both sexes, and a significant upregulation of inflammatory genes in the males relative to the females that does not occur when the mice are maintained on chow. Removal of the ovaries (OVX) caused a significant increase in percentage of adiposity and a trend toward relatively more fat deposited in the intra-abdominal depots when compared with intact females. Although OVX increases expression levels of inflammatory genes in GWAT compared with the intact females, OVX does not reach the level of expression observed in the males, even though this group had similar levels of adiposity. Consistent with the gene expression profiles when compared with the females, males have more crown-like structures in both depots, and we found an intermediate number in the OVX. These findings suggest that after exposure to an HF diet, male adipose tissue becomes more inflamed when compared with female and OVX GWAT. In the presence of the HF diet, male GWAT re-models more than female adipose tissue. Our findings suggest that ovarian hormones are only partially responsible for these differences.
Despite the higher level of total body fat, female humans and rodents are more insulin sensitive than males. Women have improved glucose tolerance and increased insulin sensitivity when compared with men,33,39,40 and are more resistant to fatty acid-induced insulin resistance.25,41–43 Insulin resistance has been associated with consumption of HF diets and induction of a low-grade inflammation.35 Female mice are less prone to diet-induced insulin resistance,25,44–46 and many genetically induced forms of insulin resistance have a milder phenotype in females compared with males.25,34,44,45 In this study we extend these findings and by analyses of GWAT of mice fed on HF and chow diet, we find that males show clear evidence of inflammation, with an upregulation of genes encoding cytokines-cytokine receptor interactions, natural killer cell-mediated cytotoxicity, leukocyte transendothelial migration and complement and coagulaton cascades (Kegg pathways analyses), compared with females when placed on the HF diet. PCR analysis by Superarray confirmed the significantly higher expression of genes involved in cytokine signaling in males when compared with females on HF diet. The acute-phase reactant, SAA3, is higher in males than females, but only when the mice are maintained on the HF diet. These findings are consistent with a greater level of inflammation and activation of the innate immune system after exposure to the HF diet in the males.
Recently, Greenberg and colleagues35,37 showed that after consumption of the HF diet, there is re-modeling of male adipose tissue and increased infiltration of macrophages as evidenced by higher levels of crown-like structures and this is associated with insulin resistance. In this study we extend those findings and show increased crown-like structures in male GWAT when compared with female GWAT and these data are consistent with the microarray findings of elevations in inflammatory markers. More importantly we report that the inflammation/macrophage infiltration is higher in GWAT of males when compared with females. In addition, CD68 and CD14, which are markers of macrophages, are upregulated in male when compared with female GWAT. Only when males are maintained on the HF diet show a slight increase in the remodeling of GWAT in the OVX females with increased crownlike structures relative to the females, but this did not reach the level of the males. The increased inflammation/macrophage infiltration in male GWAT compared with females is consistent with males being more prone to diseases associated with obesity and inflammation when compared with females.
Males have lower insulin receptor substrate 1 and glucose transporter 4 expression in GWAT (microarray result confirmed using qPCR) than females regardless of dietary exposure. These findings are consistent with a recently published paper by Macotela et al.25 and consistent with earlier reports comparing male and female adipose tissue in rats,39 showing increased expression of genes related to insulin signaling in female compared with male GWAT rodents fed standard chow. Interestingly, removal of ovarian hormones in the OVX partially reverses gene expression for SAA3, regardless of dietary exposure. Phosphoenolpyruvate carboxykinase, an inflammation gene that is rate determining for glyceroneogenesis and the main pathway of fatty acid esterification47 in adipocytes, is higher in the female GWAT despite similarly sized cells. In addition, the adipocyte secretory protein, retinol-binding protein 4, is higher in females than males and higher in GWAT than IWAT in both sexes. Taken together with the parallel changes in glucose transporter 4, we predict that triglyceride turnover, that is, lipolysis and re-esterification of fatty acids, is higher in females regardless of diet. These data are consistent with Macotela et al.,25 in which they found that adipocytes from females have higher mRNA/protein levels of several genes involved in glucose and lipid metabolism; however, they found more lipogenic enzymes and genes in lipogenic pathways increased in females when compared with males than what we report in this study. In addition, these data are consistent with those of Koutsari et al.,48 in which they found in humans, greater lipogenic activity in IWAT in women when compared with men.
Macotela et al.,25 found that GWAT adipocytes from male mice were 60% larger than adipocytes from the analogous depot in females (P < 0.05). Additionlly, IWAT adipocytes from males were only 20% bigger than females. After OVX, GWAT and IWAT adipocytes increased in size by 70 and 50%, respectively (P < 0.05) in their study.25 In this study we confirm their data, and show a significant increase in adipocyte size in OVX relative to males and females in GWAT. Importantly, our data differ from those of Macotela et al.25 in that we found no difference between male and female adipocyte size in the GWAT. These data suggest that after exposure to the HF diet, female adipocytes expand (get larger) to store excess lipid in the presence of a HF diet. We found that females did have significantly smaller adipocytes in the IWAT than males and OVX, and these data are consistent with previous findings.25 Our data further suggest that the HF diet is having the greatest effect on GWAT adipose tissue in males.
Some genes are oppositely regulated between fat pads in males and females. One of these genes, lipocalin 2, which binds to and transports small hydrophobic molecules such as retinol, fatty acids, steroids and thyroid hormone,49 is significantly elevated in males when compared with females, regardless of diet. Lipocalin 2 is highly expressed by fat cells in vivo and in vitro. Expression of lipocalin 2 is elevated by agents that promote insulin resistance and is reduced by thiazolidinediones. Lipocalin 2 serum levels are elevated in multiple rodent models of obesity, and forced reduction of lipocalin 2 in 3T3-L1 adipocytes improves insulin action.50 However, the role of lipocalin 2 in adipose tissue is unclear, because it has been reported to have proapoptotic effects on both neutrophils and leukocytes.51
Conclusions
There are major sex-based differences in gene expression profiles between male and female mice, and these differences are amplified after exposure to the HF diet. Specifically, despite no difference in adipocyte size, gene expression profiles in GWAT seem to differ most significantly between the sexes, and this difference is enhanced after exposure to the HF diet. Interestingly, removal of the ovaries does not completely reverse or alter gene expression profiles, despite increased adipocyte size and weight gain in OVX. The increased expression of genes in the insulin signaling cascade observed in adipose tissue from females may account for their lower level of insulin resistance and diabetes risk when compared with males, and this gene expression trend is maintained after exposure to the HF diet. The increased expression of genes associated with inflammation and cytokines in the males after exposure to HF diets suggests that male adipose tissue is more inflamed than females, potentially leading to a greater incidence of insulin resistance and diabetes risk when compared with females. Our descriptive data show sex- and depot-dependent variations in patterns of gene expression in age-matched male and female C57/BL6 mice on HF and chow diets. These data further suggest that the metabolic profile of male and female adipose tissue differs based on the location of the adipose tissue, the presence of sex hormones and dietary fatty acid exposure. This data set underscores the importance of analyzing depot-, sex- and steroid- dependent regulation of adipose tissue distribution and function.
Supplementary Material
Acknowledgements
We thank the ISIS network for their funding and support of the research. We also thank the members of the ISIS group: Jeffrey Chang, Jennifer Lovejoy, Nori Geary, Joel Elmquist, Philipp Scherer, Randy Seeley, Richard Simerly and Steve Smith. For technical assistance, we thank Sarah Williams, salary support RR00163 (KLG), as well as Jody Caldwell and Kathi Smith.
Footnotes
Conflict of interest The authors declare no conflict of interest.
Supplementary Information accompanies the paper on International Journal of Obesity website (http://www.nature.com/ijo)
References
- 1.Dua A, Hennes MI, Hoffman RG, Maas DL, Krakower GR, Sonnenberg GE, et al. Leptin: a significant indicator of total body fat but not of visceral fat and insulin insensitivity in African-American women. Diabetes. 1996;45:1635–1637. doi: 10.2337/diab.45.11.1635. [DOI] [PubMed] [Google Scholar]
- 2.Havel PJ, Kasim-Karakas S, Dubuc GR, Mueller W, Phinney SD. Gender differences in plasma leptin concentrations. Nat Med. 1996;2:949–950. doi: 10.1038/nm0996-949b. [DOI] [PubMed] [Google Scholar]
- 3.Kotani K, Tokunaga K, Fujioka S, Kobatake T, Keno Y, Yoshida S, et al. Sexual dimorphism of age-related changes in whole-body fat distribution in the obese. Int J Obes Relat Metab Disord. 1994;18 207–202. [PubMed] [Google Scholar]
- 4.Legato MJ. Gender-specific physiology: how real is it? How important is it? Int J Fertil Womens Med. 1997;42:19–29. [PubMed] [Google Scholar]
- 5.Wajchenberg BL. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr Rev. 2000;21:697–738. doi: 10.1210/edrv.21.6.0415. [DOI] [PubMed] [Google Scholar]
- 6.Clegg DJ, Brown LM, Woods SC, Benoit SC. Gonadal hormones determine sensitivity to central leptin and insulin. Diabetes. 2006;55:978–987. doi: 10.2337/diabetes.55.04.06.db05-1339. [DOI] [PubMed] [Google Scholar]
- 7.Gambacciani M, Ciaponi M, Cappagli B, Piaggesi L, De Simone L, Orlandi R, et al. Body weight, body fat distribution, and hormonal replacement therapy in early postmenopausal women. J Clin Endocrinol Metab. 1997;82:414–417. doi: 10.1210/jcem.82.2.3735. [DOI] [PubMed] [Google Scholar]
- 8.Haarbo J, Hansen BF, Christiansen C. Hormone replacement therapy prevents coronary artery disease in ovariectomized cholesterol-fed rabbits. Apmis. 1991;99:721–727. doi: 10.1111/j.1699-0463.1991.tb01250.x. [DOI] [PubMed] [Google Scholar]
- 9.Haarbo J, Marslew U, Gotfredsen A, Christiansen C. Postmenopausal hormone replacement therapy prevents central distribution of body fat after menopause. Metabolism. 1991;40:1323–1326. doi: 10.1016/0026-0495(91)90037-w. [DOI] [PubMed] [Google Scholar]
- 10.Lovejoy JC, Sainsbury A. Sex differences in obesity and the regulation of energy homeostasis. Obes Rev. 2009;10:154–167. doi: 10.1111/j.1467-789X.2008.00529.x. [DOI] [PubMed] [Google Scholar]
- 11.Despres JP. The insulin-resistance-dyslipidemic syndrome of visceral obesity: effect on patients’ risk. Obes Res. 1998;6(Suppl 1):8S–17S. doi: 10.1002/j.1550-8528.1998.tb00683.x. [DOI] [PubMed] [Google Scholar]
- 12.Lamarche B. Abdominal obesity and its metabolic complications: implications for the risk of ischaemic heart disease. Coron Artery Dis. 1998;9:473–481. doi: 10.1097/00019501-199809080-00002. [DOI] [PubMed] [Google Scholar]
- 13.Van Pelt RE, Jankowski CM, Gozansky WS, Schwartz RS, Kohrt WM. Lower-body adiposity and metabolic protection in postmenopausal women. J Clin Endocrinol Metab. 2005;90:4573–4578. doi: 10.1210/jc.2004-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edens NK, Fried SK, Kral JG, Hirsch J, Leibel RL. In vitro lipid synthesis in human adipose tissue from three abdominal sites. Am J Physiol. 1993;265:E374–E379. doi: 10.1152/ajpendo.1993.265.3.E374. [DOI] [PubMed] [Google Scholar]
- 15.Fried SK, Leibel RL, Edens NK, Kral JG. Lipolysis in intraabdominal adipose tissues of obese women and men. Obes Res. 1993;1:443–448. doi: 10.1002/j.1550-8528.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 16.Jensen MD. Health consequences of fat distribution. Horm Res. 1997;48(Suppl 5):88–92. doi: 10.1159/000191335. [DOI] [PubMed] [Google Scholar]
- 17.Shadid S, Koutsari C, Jensen MD. Direct free fatty acid uptake into human adipocytes in vivo: relation to body fat distribution. Diabetes. 2007;56:1369–1375. doi: 10.2337/db06-1680. [DOI] [PubMed] [Google Scholar]
- 18.Tchernof A, Belanger C, Morisset AS, Richard C, Mailloux J, Laberge P, et al. Regional differences in adipose tissue metabolism in women: minor effect of obesity and body fat distribution. Diabetes. 2006;55:1353–1360. doi: 10.2337/db05-1439. [DOI] [PubMed] [Google Scholar]
- 19.D’Eon TM, Souza SC, Aronovitz M, Obin MS, Fried SK, Greenberg AS. Estrogen regulation of adiposity and fuel partitioning. Evidence of genomic and non-genomic regulation of lipogenic and oxidative pathways. J Biol Chem. 2005;280:35983–35991. doi: 10.1074/jbc.M507339200. [DOI] [PubMed] [Google Scholar]
- 20.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 21.Guerre-Millo M. Adiponectin: an update. Diabetes Metab. 2008;34:12–18. doi: 10.1016/j.diabet.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Arner P. Differences in lipolysis between human subcutaneous and omental adipose tissues. Ann Med. 1995;27:435–438. doi: 10.3109/07853899709002451. [DOI] [PubMed] [Google Scholar]
- 23.Bolinder J, Engfeldt P, Ostman J, Arner P. Site differences in insulin receptor binding and insulin action in subcutaneous fat of obese females. J Clin Endocrinol Metab. 1983;57:455–461. doi: 10.1210/jcem-57-3-455. [DOI] [PubMed] [Google Scholar]
- 24.Hellmer J, Marcus C, Sonnenfeld T, Arner P. Mechanisms for differences in lipolysis between human subcutaneous and omental fat cells. J Clin Endocrinol Metab. 1992;75:15–20. doi: 10.1210/jcem.75.1.1320047. [DOI] [PubMed] [Google Scholar]
- 25.Macotela Y, Boucher J, Tran TT, Kahn CR. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes. 2009;58:803–812. doi: 10.2337/db08-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostman J, Arner P, Engfeldt P, Kager L. Regional differences in the control of lipolysis in human adipose tissue. Metabolism. 1979;28:1198–1205. doi: 10.1016/0026-0495(79)90131-8. [DOI] [PubMed] [Google Scholar]
- 27.Tordjman J, Guerre-Millo M, Clement K. Adipose tissue inflammation and liver pathology in human obesity. Diabetes Metab. 2008;34:658–663. doi: 10.1016/S1262-3636(08)74601-9. [DOI] [PubMed] [Google Scholar]
- 28.Dicker A, Ryden M, Naslund E, Muehlen IE, Wiren M, Lafontan M, et al. Effect of testosterone on lipolysis in human pre-adipocytes from different fat depots. Diabetologia. 2004;47:420–428. doi: 10.1007/s00125-003-1324-0. [DOI] [PubMed] [Google Scholar]
- 29.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, et al. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 30.Hoffstedt J, Arner P, Hellers G, Lonnqvist F. Variation in adrenergic regulation of lipolysis between omental and subcutaneous adipocytes from obese and non-obese men. J Lipid Res. 1997;38:795–804. [PubMed] [Google Scholar]
- 31.Nilsson M, Dahlman I, Ryden M, Nordstrom EA, Gustafsson JA, Arner P, et al. Oestrogen receptor alpha gene expression levels are reduced in obese compared to normal weight females. Int J Obes (Lond) 2007;31:900–907. doi: 10.1038/sj.ijo.0803528. [DOI] [PubMed] [Google Scholar]
- 32.Boivin A, Brochu G, Marceau S, Marceau P, Hould FS, Tchernof A. Regional differences in adipose tissue metabolism in obese men. Metabolism. 2007;56:533–540. doi: 10.1016/j.metabol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Boyns DR, Crossley JN, Abrams ME, Jarrett RJ, Keen H. Oral glucose tolerance and related factors in a normal population sample. I Blood sugar, plasma insulin, glyceride, and cholesterol measurements and the effects of age and sex. Br Med J. 1969;1:595–598. doi: 10.1136/bmj.1.5644.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li AC, Brown KK, Silvestre MJ, Willson TM, Palinski W, Glass CK. Peroxisome proliferator-activated receptor gamma ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2000;106:523–531. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strissel KJ, Stancheva Z, Miyoshi H, Perfield JW, 2nd, DeFuria J, Jick Z, et al. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes. 2007;56:2910–2918. doi: 10.2337/db07-0767. [DOI] [PubMed] [Google Scholar]
- 36.Xiao XQ, Grove KL, Smith MS. Metabolic adaptations in skeletal muscle during lactation: complementary deoxyribonucleic acid microarray and real-time polymerase chain reaction analysis of gene expression. Endocrinology. 2004;145:5344–5354. doi: 10.1210/en.2004-0721. [DOI] [PubMed] [Google Scholar]
- 37.Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46:2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Doniger S, Hofmann T, Yeh J. Predicting CNS permeability of drug molecules: comparison of neural network and support vector machine algorithms. J Comput Biol. 2002;9:849–864. doi: 10.1089/10665270260518317. [DOI] [PubMed] [Google Scholar]
- 39.Guerre-Millo M, Leturque A, Girard J, Lavau M. Increased insulin sensitivity and responsiveness of glucose metabolism in adipocytes from female versus male rats. J Clin Invest. 1985;76:109–116. doi: 10.1172/JCI111932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yki-Jarvinen H. Sex and insulin sensitivity. Metabolism. 1984;33:1011–1015. doi: 10.1016/0026-0495(84)90229-4. [DOI] [PubMed] [Google Scholar]
- 41.Frias JP, Macaraeg GB, Ofrecio J, Yu JG, Olefsky JM, Kruszynska YT. Decreased susceptibility to fatty acid-induced peripheral tissue insulin resistance in women. Diabetes. 2001;50:1344–1350. doi: 10.2337/diabetes.50.6.1344. [DOI] [PubMed] [Google Scholar]
- 42.Hevener A, Rei D, Janez A, Olefsky J. Female rats do not exhibit free fatty acid-induced insulin resistance. Diabetes. 2002;51:1907–1912. doi: 10.2337/diabetes.51.6.1907. [DOI] [PubMed] [Google Scholar]
- 43.Soeters MR, Sauerwein HP, Groener JE, Aerts JM, Ackermans MT, Glatz JF, et al. Gender-related differences in the metabolic response to fasting. J Clin Endocrinol Metab. 2007;92:3646–3652. doi: 10.1210/jc.2007-0552. [DOI] [PubMed] [Google Scholar]
- 44.Corsetti JP, Sparks JD, Peterson RG, Smith RL, Sparks CE. Effect of dietary fat on the development of non-insulin dependent diabetes mellitus in obese Zucker diabetic fatty male and female rats. Atherosclerosis. 2000;148:231–241. doi: 10.1016/s0021-9150(99)00265-8. [DOI] [PubMed] [Google Scholar]
- 45.Trevaskis JL, Meyer EA, Galgani JE, Butler AA. Counterintuitive effects of double-heterozygous null melanocortin-4 receptor and leptin genes on diet-induced obesity and insulin resistance in C57BL/6J mice. Endocrinology. 2008;149:174–184. doi: 10.1210/en.2007-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zierath JR, Houseknecht KL, Gnudi L, Kahn BB. High-fat feeding impairs insulin-stimulated GLUT4 recruitment via an early insulin-signaling defect. Diabetes. 1997;46:215–223. doi: 10.2337/diab.46.2.215. [DOI] [PubMed] [Google Scholar]
- 47.Geisler RW, Hansen RJ. Effects of insulin on the adaptation of glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in rat adipose tissue. Biochim Biophys Acta. 1972;279:139–145. doi: 10.1016/0304-4165(72)90248-6. [DOI] [PubMed] [Google Scholar]
- 48.Koutsari C, Snozek CL, Jensen MD. Plasma NEFA storage in adipose tissue in the postprandial state: sex-related and regional differences. Diabetologia. 2008;51:2041–2048. doi: 10.1007/s00125-008-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kjeldsen L, Cowland JB, Borregaard N. Human neutrophil gelatinase-associated lipocalin and homologous proteins in rat and mouse. Biochim Biophys Acta. 2000;1482:272–283. doi: 10.1016/s0167-4838(00)00152-7. [DOI] [PubMed] [Google Scholar]
- 50.Yan QW, Yang Q, Mody N, Graham TE, Hsu CH, Xu Z, et al. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes. 2007;56:2533–2540. doi: 10.2337/db07-0007. [DOI] [PubMed] [Google Scholar]
- 51.Kehrer JP. Lipocalin-2: pro- or anti-apoptotic? Cell Biol Toxicol. 2009 doi: 10.1007/s10565-009-9119-9. [DOI] [PubMed] [Google Scholar]
- 52.van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.