Abstract
The efficacy of a newly created software package for predictive modeling of developmental gene regulatory networks (GRNs) has recently been demonstrated (Peter et al., 2012). The program GeNeTool computes spatial gene expression patterns based on GRN interactions and thereby allows the direct comparison of predicted and observed spatial expression patterns. GeNeTool also permits in silico exploration of both cis- and trans- perturbations of GRN interactions. Here, we present this program, review briefly its major features and applications, and provide a detailed and accessible tutorial.
Key words: artificial life, gene networks, genetic analysis, recognition of genes, regulatory elements
In developmental genomic regulatory interactions among genes, encoding transcription factors and signaling molecules constitute gene regulatory networks (GRNs), and these networks generate spatially restricted gene expression in developmental time. The major function of developmental GRN circuitry is to control the occurrence of such Boolean patterns of gene expression in space, as visually accessed by whole mount in situ hybridization. In every well-known embryonic system, zygotically expressed regulatory genes are either on or off in the given territories of the embryo at each point in time. GRNs also control the progressive subdivision of these regulatory state territories, generally by determining signal presentation. Thus GRNs hold the key to causal understanding of development, since the fates and functions expressed by the cells of every lineage arising in the embryo depend on their progression of regulatory states. It is essential to be able to model predictively proposed GRNs in order to test their sufficiency, and also to determine the consequences of programmed rewiring of their circuitry. The GeNeTool software was initially built and utilized to examine the sea urchin embryo GRNs in these ways (Peter et al., 2012), but it will have applications in a broad range of developmental and synthetic systems. Given initial conditions, this software permits a dynamic reconstruction of changing spatial regulatory states according to the regulatory logic relationships resident in GRN structure, producing an automaton—a process that runs by itself on internally computed inputs in silico.
GeNeTool is designed to simultaneously compute progressive regulatory gene expression in separate spatial domains over developmental time based on experimentally or theoretically inferred GRN circuitry. The program generates Boolean spatial gene expression patterns on the basis of inputs extracted from experimentally established or synthetically constructed GRNs. Input information that the program is fed from the starting GRNs includes the repertoires of genes included in the GRN, the identities of the regulatory genes to which each gene in the system responds (positively or negatively) according to its genomically encoded cis-regulatory system, and the logic transactions these cis-regulatory systems execute. The input identities and the logic transactions are encoded in GeNeTool as a set of vector equations, which are expressed in Boolean logic such that the output of a gene is 1 if and only if all the necessary input conditions are true, otherwise it's 0.
In successive time intervals, the GeNeTool software computes the output of each gene, sweeping across all spatial domains considered. Thus for all spatial domains defined in the model, the complete set of vector equations is used to compute the genes expressed based on the regulatory inputs in each respective domain. That is, for each gene the output at each temporal step is the predicted consequence of the inputs to which the gene is exposed, but these inputs are themselves the outputs of other genes within the system at prior times. The dynamics are controlled by statements also included in the vector equations that provide the intervals (in real time or merely as a sequence of steps) between the activation of a gene and the activation of its direct target genes. GeNeTool includes fields in the vector equations in which a variety of other input information that is utilized to determine whether the output at a given time and place is 1 or 0, including presence or absence of signaling where this is relevant; location in the spatial domains of the system; and again where relevant, choice of alternative regulatory modules (i.e., in silico, alternative vector equations).
GeNeTool permits the use of the computations of spatial and temporal gene expression, which it performs in many ways. It generates direct graphic comparisons between observed and computed expression patterns, and it generates comparisons revealing the deviations between expression patterns computed under “normal” vs. perturbed conditions. Every kind of perturbation can be modeled in GeNeTool including gene product knockdowns, cis-regulatory target site mutations, relative temporal alterations, alteration of the relative positions of spatial domains in the developing system (i.e., of signal availability), and alterations of GRN circuit design.
GeNeTool accommodates many essential features general to the processes of development. The program incorporates and utilizes for its computations input information on all the following: (i) all genes in the system about which there is specific spatial regulatory information; (ii) initial inputs, for example, localized maternal factors; (iii) cis-regulatory results indicating how combinatorial regulatory inputs interact and what these inputs are, and any results that indicate the Boolean logic functions of the cis-regulatory modules controlling each gene; (iv) data on use of different cis-regulatory modules at different times; (v) results of all the perturbation data on which the interaction maps of the GRNs are based; (vi) the progressive spatial geometry of the embryonic territories with respect to one another; (vii) signaling interactions, that is, source and targets; (viii) digitized representation of observed spatial expression patterns, which it reserves for automated comparison with the computed spatial patterns of spatial gene expression.
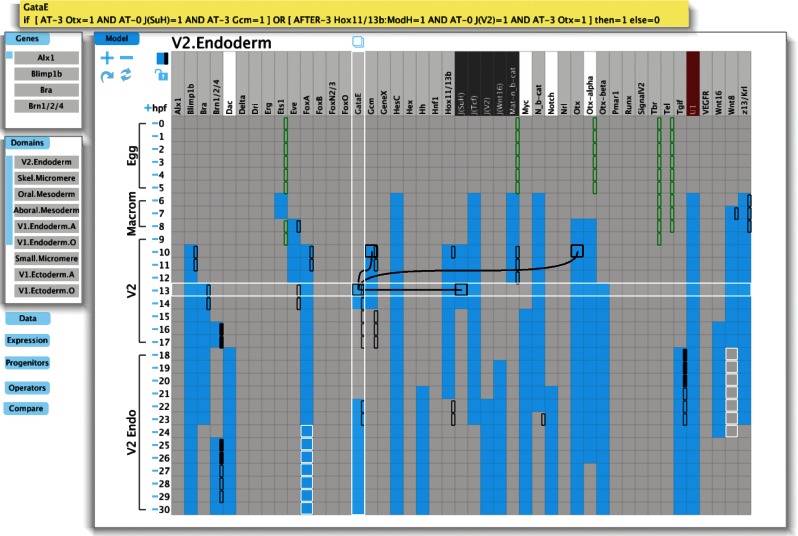
Table 1 is a transformation table that relates specific biological features of the developmental control process to the form of their representation within GeNeTool and summarizes the rationale behind each feature of the computational output of the program. Figure 1 is a screenshot of GeNeTool.
Table 1.
Representation of Biological Features in GeNeTool Software
| Biological feature | Formal statement | Explanation |
|---|---|---|
| Regulatory gene expression | Vector equation output, i.e., output of 1 or 0 at each time interval | Gene expression is modeled on the principle that the outcome of cis-regulatory interactions determines presence or absence of the regulatory gene product |
| Regulatory gene interactions (in the GRN) | Input variables in vector equations | Cis-regulatory inputs to given regulatory genes are outputs of other regulatory genes in the network |
| Cis-regulatory logic | Boolean logic operators upon variables in vector equations | Captures cis-regulatory logic operations given the inputs per module |
| Cis-regulatory modules | Vector equations for individual modules | Vector equation represents the invariant regulatory system encoded in the DNA of each gene |
| Module choice | Boolean logic operators that determine which modular vector equations are in effect | Captures module choice apparatus operating in genes expressed in multiple developmental conditions |
| Spatial domains and their changing geometry in development | Matrix of domain contiguity/noncontiguity relationships in developmental time | Captures morphological interrelations of regulatory state domains during embryogenesis |
| Signaling interactions | Janus function: target genes are ON in cells receiving signal, otherwise turned OFF | Converts signaling function into status of transcriptional regulatory response |
| Observed gene expression | Boolean matrix of observed gene expression in time/space | Represents expression status of each gene through time and space as experimentally determined |
| GRN output | Boolean matrix of computed gene expression in time/space | Represents expression status of each gene through time and space as computationally generated in dynamic Boolean model |
| Temporal dynamics of gene expression patterns | Step time dynamics used to relate gene interactions to real-time intervals | Computation incorporates real-time observations of interval from activation of an upstream gene to activation of its target gene (step time) |
FIG. 1.
A screenshot of the GeNeTool computation of regulatory gene expression in a particular domain of the sea urchin embryo. The functions illustrated include the Boolean computation of expression or silence of each gene at each 1-hr step in time for a 30-hr developmental process; comparison of computed result with experimental expression data; use of vector equations containing logic statements for each gene (e.g., see the Boolean logic statement for gatae at top). For symbolism and details, see the GeNeTool tutorial available online as Supplementary Material.
GeNeTool is constructed for maximum flexibility so that diverse geometrical or temporal relations, or any different kind of cis-regulatory control system or gene interaction, can be input, and the consequences studied in silico. As illustrated in the accompanying Supplementary Material (available online at www.liertonline.com/cmb), the interactive graphics used in GeNeTool make it particularly easy to change vector equations for any gene, with respect to the inputs into its cis-regulatory system, the cis-regulatory module it uses in given contexts, the logic operators that characterize its function, or its temporal behavior with respect to its inputs. Similarly, genes can be inserted or added to the system by the simplest operations, and gene interactions can be altered in any desired manner. Thus GeNeTool will have many uses for many different developmental systems.
Three major classes of application can be envisioned. First, GeNeTool can be used to provide a direct test of the completeness and explanatory power of an experimentally determined GRN, and indeed, though critical in importance, completeness and explanatory power of GRNs are otherwise elusive parameters. Thus, if the observed and computed expression patterns through developmental time are largely convergent, the GRN used for construction of the vector equations is causally sufficient, otherwise not, and furthermore what lacunae exist become explicitly evident. Second, GeNeTool can be used to explore in silico the expected effects of given perturbations. These can be perturbations of cis-regulatory function (site-specific regulatory mutations in silico); perturbations of gene interactions (morpholino treatment or genetic lesions in silico); or redesign of GRN architecture to be carried out by synthetic methods (Damle and Davidson, 2012). Thus, GeNeTool can be used for network redesign and synthetic reengineering of developmental systems. Both of these classes of application are illustrated in the study of the sea urchin GRN to which GeNeTool was originally applied (Peter et al., 2012).
In the future lies another related application, and that is the use of GeNeTool to analyze and reconstruct evolutionary change in body plans, as such changes are the surviving consequences of the network reengineering that have occurred naturally in deep time in every animal lineage (Davidson and Erwin, 2006; Erwin and Davidson, 2009).
Methods
The accompanying online material, available at http://whis.caltech.edu/GeNeTool/GeNeTool.html, consists of a complete primer and tutorial on the use of GeNeTool. Both an overview and detailed instructions and examples are provided for each of the following topics: (1) Downloading and opening GeNeTool, handling models and menus, and exporting results; (2) entering explicit and implicit data from the GRN model; (3) inserting temporal step dynamics into the model; (4) inserting spatial information about the developmental system to be modeled; (5) dealing with signaling interactions; (6) setting initial conditions and other manual inputs; (7) setting responses that are dependent on input levels; and (8) the repertoire of computational functions available in GeNeTool. For examples of application of all the features described in the accompanying material, see Peter et al. (2012).
Supplementary Material
Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Damle S.S. Davidson E.H. Synthetic in vivo validation of gene network circuitry. PNAS. 2012;109:1548–1553. doi: 10.1073/pnas.1119905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H. Erwin D.H. Gene regularoty networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Erwin D.H. Davidson E.H. The evolution of hierarchucal gene regulatory networks. Nat. Rev. Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- Peter I.S. Faure E. Davidson E.H. Predictive computation of genomic logic processing functions in embryonic development. PNAS. 2012;109:16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.