Abstract
The human gut microbiota has become the subject of extensive research in recent years and our knowledge of the resident species and their potential functional capacity is rapidly growing. Our gut harbours a complex community of over 100 trillion microbial cells which influence human physiology, metabolism, nutrition and immune function while disruption to the gut microbiota has been linked with gastrointestinal conditions such as inflammatory bowel disease and obesity. Here, we review the many significant recent studies that have centred on further enhancing our understanding of the complexity of intestinal communities as well as their genetic and metabolic potential. These have provided important information with respect to what constitutes a ‘healthy gut microbiota’ while furthering our understanding of the role of gut microbes in intestinal diseases. We also highlight recently developed genomic and other tools that are used to study the gut microbiome and, finally, we consider the manipulation of the gut microbiota as a potential therapeutic option to treat chronic gastrointestinal disease.
Keywords: gastrointestinal disease, gut health, microbial diversity, microbial manipulation
Introduction
The human intestinal tract harbours a diverse and complex microbial community which plays a central role in human health. It has been estimated that our gut contains in the range of 1000 bacterial species and 100-fold more genes than are found in the human genome [Ley et al. 2006a; Qin et al. 2010]. This community is commonly referred to as our hidden metabolic ‘organ’ due to their immense impact on human wellbeing, including host metabolism, physiology, nutrition and immune function. It is now apparent that our gut microbiome coevolves with us [Ley et al. 2008] and that changes to this population can have major consequences, both beneficial and harmful, for human health. Indeed, it has been suggested that disruption of the gut microbiota (or dysbiosis) can be significant with respect to pathological intestinal conditions such as obesity [Ley et al. 2006b; Zhang et al. 2009] and malnutrition [Kau et al. 2011], systematic diseases such as diabetes [Qin et al. 2012] and chronic inflammatory diseases such as inflammatory bowel disease (IBD), encompassing ulcerative colitis (UC) and Crohn’s disease (CD) [Frank et al. 2007].
The role of the gut microbiome in human health and disease is becoming clearer thanks to high throughput sequencing technologies (HTS) as well as parallel recent developments in nongenomic techniques. The purpose of this review is to summarize the very significant major developments that have occurred with respect to revealing the microbial diversity of the human gut and how this intestinal microbiota impacts on gastrointestinal (GI) disease. We also discuss the state-of-the-art tools that can be used to study the gut microbiome and look to future therapeutic options, such as the manipulation of the gut microbiota, to address GI conditions.
Tools for studying the gut microbiome
Understanding the composition and functional capacity of the gut microbiome represents a major challenge. However, research in this area is ever expanding and currently a number of different approaches are being used and developed to determine gut microbial composition, genetic content and function.
Traditionally, culture-based techniques were used to determine the composition of the gut microbiota. These approaches have generally focused on the ‘easy-to-culture’ microbes of the gut and have become less popular due to indications that just 10–50% of the gut bacteria are culturable [Eckburg et al. 2005]. Culturing-based methods certainly have their limitations and do not readily provide an overview of the gut microbial composition. It should be noted, however, that there have been some advances in this area through the increased availability of specialized media to cultivate more fastidious organisms [Goodman et al. 2011]. A recent study constitutes a further development in this area and has resulted in the coining of the term ‘microbial culturomics’ [Lagier et al. 2012]. Microbial culturomics introduces an array of new culturing techniques, coupled with matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MS), to identify a range of previously uncultivated microbiota from the gut. This strategy includes the elimination of the ‘easy-to-culture’, or more abundant, populations that are present in high numbers to facilitate the enrichment of the more difficult to culture organisms by methods such as diverse filtration or the use of antibiotics and phage cocktails, leading to the identification of 174 species not previously described in the human gut [Lagier et al. 2012].
Despite these recent successes, it is clear that culture-independent approaches are better suited to providing a more rapid insight into the gut microbiota. In particular, the development and application of fast and low-cost DNA sequencing methods has been revolutionary. HTS has been widely used to examine the complexity of the gut microbiome due to the speed, scale and precise information provided. For compositional analysis, the 16S rRNA gene has been most frequently targeted due to its presence in all prokaryotes and the existence of variable domains that allow different taxa to be distinguished. Although the majority of HTS studies to date have relied on the Roche 454 pyrosequencing platforms, other sequencing technologies, such as those provided by Illumina (San Diego, CA, USA), are becoming more popular [Caporaso et al. 2011b]. Other HTS technologies that can be applied include the SOLid system (Applied Biosystems, Foster City, CA, USA), the Ion platforms (Life Technologies, Carlsbad, CA, USA) and SMRT system (Pacific Biosystems, Menlo Park, CA, USA), while additional platforms, such as those that rely on nanopore technology, are in development [Clarke et al. 2009; Rosenstein et al. 2012; Schadt et al. 2010].
While 16S rRNA studies provide data in relation to the microbial composition of an ecosystem, these do not provide direct information regarding the microbial viability or the functional potential of the populations present. Metagenomic (or shotgun sequencing) studies go beyond the 16S rRNA gene to characterize the full genetic content of a community, thereby providing an insight into the potential functional capacity of the microbes present [Kurokawa et al. 2007; Qin et al. 2010; Turnbaugh et al. 2009b]. Regardless of the approach taken, it is important to note that these sequencing technologies require detailed bioinformatic analyses to deal with the large volumes of data generated (for a review, see Kuczynski and colleagues) [Kuczynski et al. 2012]. Indeed, increasingly, the major bottleneck has moved from being the generation of data to the storage of these data and the availability of scientists with the appropriate specialist bioinformatic skills. Furthermore, although these gene-centric approaches have provided much information regarding the content of the gut, we also need to understand the activity of these genes and the impact on the metabolic networks within the gut. To further determine specific microbial activity, it is necessary to analyse gene expression (metatranscriptomics), protein products (metaproteomics) and metabolic profiles (metabolomics). These techniques can be complex and, to different extents, are still somewhat in their infancy. To date, metatranscriptomics, based on large-scale sequencing of 16S rRNA transcripts, has been used to look at the composition of the active microbiota in healthy individuals and has revealed that the transcriptional profile across individuals is more similar than indicated by the associated taxonomic diversity [Gosalbes et al. 2011, 2012]. The faecal metaproteome of healthy adults was also recently investigated using liquid chromatography–tandem MS [Kolmeder et al. 2012]. Metaproteomics has an advantage over RNA-based studies as it analyses a more stable gene product. This study showed that the metaproteome retained considerable temporal stability over time and contained a proteome core that included metabolic enzymes, chaperones and stress proteins [Kolmeder et al. 2012]. The field of metabolomics has advanced dramatically and developments with respect to nuclear magnetic resonance (NMR) and MS make it possible to analyze 1000s of metabolites simultaneously [Nicholson et al. 2005]. NMR has been used to investigate metabolite compositions of the gut microbiota in very many instances [Marchesi et al. 2007; Mestdagh et al. 2012; Saric et al. 2008]. Although an extremely valuable tool, NMR can be limited by resolution and sensitivity. In some cases, ion cyclotron resonance–Fourier transform MS, which has an extremely high mass resolution and which can detect small variations between metabolite signals [Rossello-Mora et al. 2008], may merit consideration.
Large-scale studies of the gut microbiome
In recent years a number of large funding initiatives were undertaken with a view to understanding the complexity of the human microbiome, including the gut environment. The European Metagenomics of the human intestinal tract (MetaHIT) [Arumugam et al. 2011; Qin et al. 2010] and the US human Microbiome Project (HMP) [HMP Consortium, 2012a, 2012b] have both, through large-scale sequencing, worked towards establishing the baseline healthy gut microbiota and how this is altered in a disease state.
MetaHIT focused on investigating the correlation between the gut microbiome and intestinal pathologies, particularly obesity and IBD [Qin et al. 2010]. In one instance, this consortium sequenced faecal DNA from a cohort of 124 individuals, including healthy subjects and those with IBD or obesity, to establish a catalogue of nonredundant genes from the intestinal tract [Qin et al. 2010]. This project indicated that 40% of genes were shared among the majority of individuals and therefore represented a core metagenome. It was also found that 99.1% of genes were of bacterial origin, with the majority of the remaining genes belonging to the archeal kingdom, with a relatively small number of eukaryotic and viral genes also being detected [Qin et al. 2010].
The HMP assessed the diversity of the microbiota across multiple body sites in healthy subjects, including the GI tract, to determine the baseline composition of the healthy human microbiome [HMP Consortium, 2012a]. Large-scale sequencing for meta-analyses has produced 16S rRNA data from 690 samples from 300 subjects and across 15 body sites [Turnbaugh et al. 2007]. The HMP also generated a catalogue of microbial genomes from the human microbiome, which consists of approximately 800 reference genomes from multiple body sites to date [HMP Consortium, 2012b] (see also http://hmpdacc.org). Both consortia provided a hugely valuable microbial catalogue that highlights the substantial variation in microbial species and genes in the gut. In addition, together with others, this work helps our understanding of what constitutes a ‘healthy’ gut microbiota while revealing novel potential associations between the gut microbiota and GI diseases [Qin et al. 2010, Arumugam et al. 2011; HMP Consortium, 2012a, 2012b].
The ‘healthy’ gut microbiota
The intestinal microbiota of healthy individuals is known to confer a number of health benefits relating to, for example, pathogen protection, nutrition, host metabolism and immune modulation [O’Hara and Shanahan, 2006; Sekirov et al. 2010] (Figure 1). Historically, culture-based analysis has indicated that the gut of a healthy adult share bacterial species that are common among the majority of individuals. In contrast, however, the application of more recently developed technologies, which facilitate the culture-independent examination of the gut microbiota, have indicated that there is large interindividual microbial diversity, with only a small phylogenetic overlap between people [HMP Consortium, 2012a]. It should also be noted that the many HTS-based studies undertaken to describe the normal GI microbial community have differed with respect to the health, age, location and diet of the individuals included [Qin et al. 2010, 2012; Tap et al. 2009; Turnbaugh et al. 2009a], in the specific molecular methods used [Claesson et al. 2009; Hamady and Knight, 2009] and in how the data have been analysed [Wooley and Ye, 2009]. It has been established, however, that there is a high overall temporal stability of the microbial community within an individual, which suggests the existence of an individual core microbial population [Caporaso et al. 2011a; Costello et al. 2009; Jalanka-Tuovinen et al. 2011]. Even here, a number of factors including aging, diet, antibiotic use and environmental factors can cause changes.
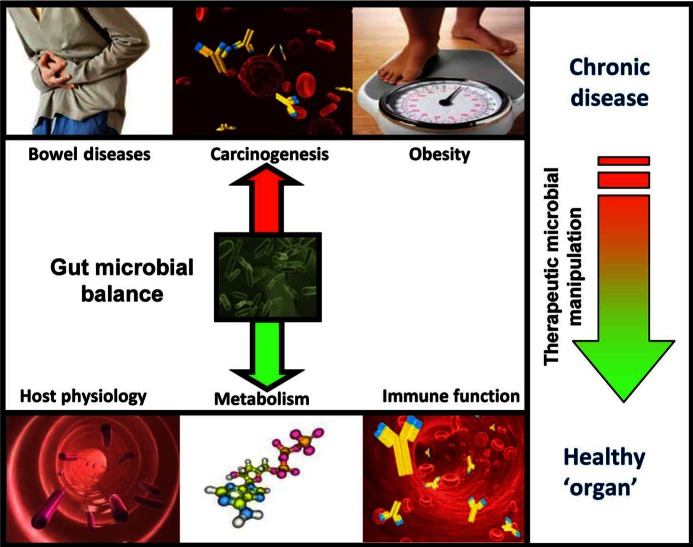
Figure 1.
The gut microbiota in health and intestinal disease. The gastrointestinal microbiota play a role in host physiology, metabolism and nutrition. An alteration in the gut microbial community is linked to a number of intestinal conditions, including cancer, obesity and a variety of bowel disorders. The contribution of beneficial components of the gut microbiome to host physiology, metabolism and immune function has become the focus of ever more attention, and will undoubtedly lead to new therapeutic approaches.
Infants are generally thought to be born with intestines that are sterile or that, at most, contain a very low level of microbes [Jimenez et al. 2008]. However, the infant GI tract is rapidly colonized following delivery. The composition of the infant gut can vary significantly based on a number of factors, including mode of delivery, feeding type, or due to antibiotic, prebiotic or probiotic use (for a review see Fouhy and colleagues) [Fouhy et al. 2012]. Despite this, the infant intestinal microbiota remains less complex than that of adults. Early colonizers include enterobacter and enterococci followed by anaerobic organisms such as bifidobacteria, clostridia, Bacteroides spp. and anaerobic streptococci [Adlerberth and Wold, 2009]. These populations continue to evolve and by age 2 the infant gut microbiota is thought to display a community structure similar to the adult gut [Palmer et al. 2007].
As noted above, interindividual variation within the adult gut microbiota is very large. Turnbaugh and colleagues established that the faecal microbiome of identical twins share less than 50% of species phylotypes [Turnbaugh et al. 2010]. However, based on HTS 16S rRNA-based, studies, it is apparent that in general the adult gut is dominated by two bacterial phyla, Firmicutes and Bacteroidetes, with other phyla including Actinobacteria, Proteobacteria, Verrucomicrobia and Fusobacteria being present in lower proportions [Eckburg et al. 2005; Tremaroli and Backhed, 2012]. Greater variations exist below the phylum level, although certain butyrate-producing bacteria, including Faecalibacterium prausnitzii, Roseburia intestinalis and Bacteroides uniformis, have been identified as key members of the adult gut microbiota [Qin et al. 2010]. Further knowledge relating to the species and functional composition of the gut was gleaned through the analysis of sequence data from 22 faecal metagenomes from individuals across four countries. This has led to a suggestion that the human gut microbiome consists of three enterotypes that vary with respect to the associated microbial species and their functional potential [Arumugam et al. 2011]. These clusters were named to reflect their dominant members, that is, Bacteroides (enterotype 1), Prevotella (enterotype 2) and Ruminococcus (enterotype 3). It was claimed that the most frequent of these was enterotype 3, which is enriched in Ruminococcus in addition to the co-occurring Akkermansia [Arumugam et al. 2011]. It has since been indicated, however, that the three enterotype divisions are not as distinct as first thought and in particular the Ruminococcus-dominant enterotype appears less evident than initially claimed [Jeffery et al. 2012a; Wu et al. 2011].
The elderly intestinal microbiota has also been the subject of a number of studies in recent years. This is particularly timely as an ageing population is now becoming a general feature of Western countries [Biagi et al. 2010; Claesson et al. 2011, 2012]. It has been noted that there are age-related physiological changes in the GI tract of older people that are characterized by a chronic low-grade inflammation (inflammageing) [Franceschi, 2007] which can lead to a microbial imbalance in the intestine [Guigoz et al. 2008]. HTS analysis has indicated that the composition of the gut microbiota of older people (>65 years) is distinct from that of younger adults and, although extremely variable between individuals, has a general dominance of the phylum Bacteroidetes [Claesson et al. 2011]. Claesson and colleagues further established a relationship between diet, the health status and the gut microbial population of older people [Claesson et al. 2012]. In summary, taxonomic assignments showed that the microbiota of people in a long-stay care environment had a high proportion of Bacteroidetes, whereas individuals living in the community had a high level of Firmicutes. Notably, the microbiota of individuals in long-stay care was significantly less diverse and a loss of the community-associated microbiota correlated with increased frailty [Claesson et al. 2012]. This and other work has strongly implied that the GI microbiota is extremely important to the health and in the progression of disease and frailty in older people [Claesson et al. 2012; Guigoz et al. 2008]. Regardless of age, the development of a clearer understanding of what constitutes a healthy microbiota allows one to establish what, if anything, is unusual within the microbiota of those with various diseases.
The gut microbiota and disease
As the volume of data relating to the composition and functional potential of the gut microbiota increases, the number of diseases that have been linked with alterations in our gut microbial community has also expanded. Indeed, the many instances of such potential associations are too great to summarize in this review and thus here the focus is on associations that have been the focus of greatest attention, that is, the possibility of a link between the gut microbiota and chronic GI diseases, including irritable bowel syndrome (IBS) and IBD, systemic diseases such as type 2 diabetes (T2D) and obesity, as well as the onset of colorectal cancer (CRC) (Table 1 and Figure 1).
Table 1.
Microbial associations with chronic intestinal diseases.
| Condition | Microbial association* | References$ |
|---|---|---|
| IBS | Increased: Firmicutes:Bacteroidetes ratio Ruminococcus Dorea Clostridium Gammaproteobacteria (pIBS) Haemophilus influenzae (pIBS) Decreased: Bifidobacterium Faecalibacterium Bacteroides |
Ghoshal et al. [2012]; Jeffery et al. [2012b]; Rajilic-Stojanovic et al. [2011]; Saulnier et al. [2011] |
| IBD (incl. CD and UC) | Increased: bacterial numbers in mucosa (CD) Gamma–proteobacteria Enterobacteraceae adherent invasive Escherichia coli (CD) Clostridium spp. Decreased: bacterial diversity Firmicutes Bacteroidetes Lachnospiracheae Clostridium leptum and coccoides group (Faecalibacterium prausnitzii) Roseburia Phascolarctobacterium |
Frank et al. [2007]; Garrett et al. [2010]; Li et al. [2012]; Manichanh et al. [2006]; Morgan et al. [2012] |
| CRC | Increased: Fusobacterium spp. E. coli (pks+) |
Arthur et al. [2012]; Castellarin et al. [2012]; Kostic et al. [2012]; McCoy et al. [2013] |
| Obesity | Increased: Firmicutes:Bacteroidetes ratio‡ Actinobacteria Bacteroides ‡ Prevotellaceae Decreased: bacterial diversity C. leptum group (Ruminococcus flavefaciens) Bifidobacterium Methanobrevibacter |
Clarke et al. [2012]; Duncan et al. [2007]; Ley et al. [2005]; Schwiertz et al. [2009]; Turnbaugh et al. [2006, 2009a]; Zhang et al. [2009] |
| T2D | Increased: Opportunistic pathogens (Clostridium spp., E. coli, Eggerthella lenta) Akkermansia muciniphilia Bacteroides spp. Decreased: Butyrate-producing organisms (Roseburia spp., Faecalibacterium spp., Eubacterium spp.) Firmicutes |
Qin et al. [2012]; Larsen et al. [2010] |
Examples of certain documented microbial changes associated with disease status.
See also reviews by Cho and Blaser [2012]; Clarke et al. [2012]; Shanahan [2012].
Varying results among studies.
CD, Crohn’s disease; CRC, colorectal cancer; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome; pIBS, paediatric IBS; pks+, polyketide synthase positive; T2D, type 2 diabetes; UC, ulcerative colitis.
Irritable bowel syndrome
Functional bowel disorders such as IBS are defined solely on symptom-based diagnostic criteria. IBS is characterized by abdominal pain or discomfort and altered bowel habits. Although the aetiology is multifactorial, recent understanding of the pathophysiology of IBS has revealed that variations in the normal gut microbiota may have a role to play in the low-grade intestinal inflammation associated with the syndrome [Brint et al. 2011; Ponnusamy et al. 2011]. Microbial dysbiosis in the gut is thought to be involved in IBS pathogenesis through facilitating adhesion of pathogens to the bowel wall (for a review, see Ghoshal and colleagues [Ghoshal et al. 2012]). Specifically, a study involving phylogenetic microarrays and quantitative polymerase chain reaction (qPCR) analysis revealed a clear separation between the GI microbiota of patients with IBS and that of the controls, that is, IBS was characterized by an increase in Firmicutes and, more specifically, in the numbers of Ruminococcus, Clostridium and Dorea, in addition to a marked reduction in Bifidobacterium and Faecalibacterium spp. [Rajilic-Stojanovic et al. 2011]. In a similar study of paediatric patients with the syndrome, an alteration in members of Firmicutes and Proteobacteria, also with a higher abundance of Dorea, Ruminococcus and Haemophilus parainfluenzae, was noted. Furthermore, members of the genus Bacteroides were found to be present at a lower level in paediatric patients with IBS than in the healthy controls and an increase in Alistipes was linked with a greater frequency of pain [Saulnier et al. 2011]. Other work by Jeffery and colleagues found subgroups among the patients with IBS with varying microbial signatures, however generally an increase in the Firmicutes to Bacteroidetes ratio was evident in patients with IBS who differed from normal populations [Jeffery et al. 2012b, 2012c]. These HTS studies suggest that a link between the gut microbiota and IBS may exist, which could, in time, lead to the design of therapeutic options.
Inflammatory bowel disease
IBD, encompassing both UC and CD, is characterized by a chronic and relapsing inflammation of the GI tract. UC and CD are generally described as chronic IBDs, although are distinct diseases that differ both in their symptoms and inflammation pattern. Specifically CD is a chronic, segmental inflammation of the GI tract [Loftus, 2004] and although the aetiology is not yet clear, it is defined as a complex trait that results from the interaction between the host genetics and the gut microbial population [Elson, 2002]. UC is generally characterized by inflammation and ulceration of the lining of the colon. The onset of both conditions is, in general, not thought to be due to a single causal organism but by a general microbial dysbiosis in the gut [Lepage et al. 2011; Martinez et al. 2008]. Nonetheless, this continues to be the subject of much debate. A role for gut microbes in the manifestation of IBD has been indicated by a number of studies and the gut microbiota are thought to be essential components in the development of mucosal lesions (for a review see Manichanh and colleagues) [Manichanh et al. 2012]. Intestinal inflammation is generally believed to be associated with a reduced bacterial diversity and, in particular, a lower abundance of, and a reduced complexity in, the Bacteroidetes and Firmicutes phyla with a specific reduction of abundance in the Clostridium leptum and Clostridium coccoides groups [Manichanh et al. 2006; Sokol et al. 2006]. It has also been indicated that while Firmicutes are reduced there is an increase in gammaproteobacteria in patients with CD [Li et al. 2012]. In contrast to the general microbial dysbiosis theory, some researchers have suggested the involvement of specific taxa, for example the Enterobacteriaceae have been associated with the microbiota of patients with UC [Garrett et al. 2010] and adherent invasive E. coli have been identified in the ileal mucosa of patients with CD [Darfeuille-Michaud et al. 2004]. There have been a number of studies that have also highlighted a lower abundance of F. prausnitzii (a member of the C. leptum group) in patients with CD and UC [Frank et al. 2007; Martinez-Medina et al. 2006; Sokol et al. 2009] and a role for this microorganism in combating bacterial dysbiosis in CD has been suggested [Sokol et al. 2008]. In addition, recent work analyzing intestinal biopsies and stool samples from patients with IBD and healthy subjects documented an association of the disease status of IBD with alterations in the abundances of Enterobacteriaceae, Ruminococcaceae and Leuconostocaceae, while at genus level, Clostridium levels increased whereas butyrate producer Roseburia and succinate producer Phascolarctobacterium were significantly reduced in both UC and CD conditions [Morgan et al. 2012]. Regardless of the microbial population or pathogen in question, and although specific causality has not yet been clarified, these and other studies have certainly outlined a link between the gut microbiota and IBD.
Colorectal cancer
A role for the gut microbiome in the pathogenesis of CRC has been suggested in a number of recent publications [Arthur et al. 2012; Kostic et al. 2012; Plottel and Blaser, 2011]. Although a single causative organism has not been identified, a number of studies have implicated an association for Fusobacterium members with CRC [Castellarin et al. 2012; Kostic et al. 2012; McCoy et al. 2013]. More specifically, a recent study using fluorescent in situ hybridization analysis indicated a link between Fusobacteria and CRC, with higher numbers identified in tumours compared with control samples [Kostic et al. 2012]. This observation was supported by 16S rDNA sequencing analysis of the colorectal microbiome that revealed members of the Fusobacterium genus, including Fusobacterium nucleatum, Fusobacterium mortiferum and Fusobacterium necrophorum sequences, were enriched in tumour tissue. These changes were found to be accompanied by broad phylum-level changes, including a significant reduction in Firmicutes and Bacteroidetes. This may suggest that Fusobacterium spp. contribute to tumourigenesis through an inflammatory mechanism [Kostic et al. 2012]. Chronic inflammation is an established risk factor for carcinogenesis [Balkwill and Mantovani, 2001] and a tumour-associated or ‘tumour-elicited’ inflammation can be a feature of CRCs [Grivennikov et al. 2010]. Notably, another study, which relied on the use of metagenomic sequence and qPCR data, confirmed the association between this genus and CRC, revealing an overabundance of Fusobacterium sequences in tumour tissue compared with normal contr-ols [Castellarin et al. 2012]. Members of Fusobacterium, interestingly, have also been associated with a number of other intestinal pathologies including IBD [Strauss et al. 2011] and acute appendicitis [Guinane et al. 2013; Swidsinski et al. 2011].
The link between microbially induced inflammation and CRC has also been highlighted in a number of other studies. Indeed it has been established that microbial products can enter barrier-defective colonic tumours, trigger inflammation through a host immune response and, in turn, increase tumour growth [Grivennikov et al. 2012]. HTS studies have also revealed a link between inflammation and the gut microbial composition in colitis-susceptible, interleukin-10 deficient mice [Arthur et al. 2012]. This study revealed that mice with colitis had a less diverse gut microbial composition, which was accompanied by an increase in Proteobacteria, and particularly in E. coli levels, in the presence of intestinal inflammation [Arthur et al. 2012]. Ultimately, the role of some E. coli in CRC was linked to a polyketide synthase (pks) pathogenicity island encoding a genotoxin (colibactin). This was supported by the observations that isogenic mutants lacking the pks island brought about decreased tumour growth and invasion in mice than their wild-type pks + counterparts [Arthur et al. 2012]. Although these studies suggest that a combination of host inflammation and specific microorganisms contribute to CRC tumourigenesis, it is evident that further research in this area is needed.
Obesity and type 2 diabetes
Obesity and related disorders, such as T2D and metabolic syndrome, have become increasingly common in recent decades. Obesity is a complex syndrome that develops from a prolonged imbalance of energy intake and energy expenditure. Although lifestyle factors, diet and exercise contribute largely to the modern epidemic, it has also been indicated by an ever-increasing body of work that the microbial communities within the human intestine play an important role in obesity [Ley, 2010; Ley et al. 2005; Tilg and Kaser, 2011; Turnbaugh et al. 2006]. Although it has been suggested that increased energy harvest due to the presence of specific microbial populations contributes to obesity [Ley et al. 2005; Turnbaugh et al. 2006], this has not always been found to be the case [Murphy et al. 2010] and, indeed, it is becoming increasingly apparent that there can be very many other ways in which the microbiota can influence weight gain and host metabolism (for a review see Clarke and colleagues) [Clarke et al. 2012]. The identity of the key populations/taxa that may be associated with weight gain has also been the subject of much debate. Although a number of studies of the microbiota of lean and obese mice have indicated that genetically (ob/ob) and diet-induced obese mice contain higher proportions of Firmicutes and lower levels of Bacteroidetes than their lean counterparts [Ley et al. 2005], the situation in humans is less clear despite the fact that there have been a number of studies that have focused on the gut microbiota of lean and obese individuals (for a review see Clarke and colleagues) [Clarke et al. 2012]. Indeed, Ley and colleagues, found a decrease in the Firmicutes to Bacteroidetes ratio following weight loss in human subjects [Ley et al. 2006b]. Further work by Turnbaugh and colleagues indicated a lower proportion of Bacteroidetes in obese individuals, an increased abundance of Actinobacteria while the levels of Firmicutes remained unaltered [Turnbaugh et al. 2009a]. The importance of the Firmicutes to Bacteroidetes ratio in obesity, however, is still not clear with some conflicting studies published to date in this area [Duncan et al. 2007; Schwiertz et al. 2009].
T2D has, in recent years, become a health issue worldwide. T2D is principally linked with obesity-related insulin resistance. However, several genetic and environmental factors are thought to influence the condition. Here again, alterations in the composition of the gut microbiota of adults with T2D, relative to that of healthy controls, has been noted. Although in many instances the question as to whether these changes represent a cause or an effect remains unresolved, it is anticipated that further research in this area will clarify this issue. Regardless, a considerable number of fascinating studies have recently appeared. Larsen and colleagues employed 16S rRNA compositional sequencing to reveal that the proportions of the Firmicutes, and specifically the Clostridia class, were reduced, while the Bacteroidetes and the class Betaproteobacteria were enriched in a group with T2D compared with controls [Larsen et al. 2010]. More recently, an impressive large metagenome-wide association study identified gut microbial markers which might be useful in classifying T2D [Qin et al. 2012]. Overall, this study found a moderate degree of gut dysbiosis in patients with T2D. Of the idenitifiable bacterial species in this study it was indicated that control samples were enriched in various butyrate-producing bacteria, while patients with T2D were characterized by an increase in certain opportunistic pathogens, such as a number of Clostridium spp. in addition to important gut microbes including Akkermansia muciniphilia, Bacteroides spp. and Desulfovibrio spp. [Qin et al. 2012]. The identification of these gut microbial markers may be important in classifying T2D or perhaps other obesity or metabolic-related diseases.
Strategies to manipulate the gut microbiota
As shown in the above, there is growing evidence that the gut microbiota plays a central role in human GI health and disease. It is therefore logical that modulating the gut microbiota should be considered as a therapeutic strategy to treat chronic disease. The approaches investigated include the use of prebiotics, supplementation with probiotics, reconstitution of bacterial populations by faecal transplantation or by employing antimicrobials to eliminate pathogens or manipulate the gut microbiota in a way that will benefit host health.
Prebiotics and probiotics are becoming increasingly popular (for a review see Vyas and Ranganathan) [Vyas and Ranganathan, 2012]. Prebiotics are nutritional compounds used to promote the growth of beneficial commensals and thus have the potential to improve GI health. Use of oral probiotic cultures to restore the gut microbiota has led to promising results in the treatment of intestinal disorders such as UC and obesity [Andreasen et al. 2010; Bibiloni et al. 2005; Kadooka et al. 2010]. While it can be argued, however, that oral probiotic doses do not provide sufficient microbial numbers to fully influence the populations of the colon, it may be that these microbes exert their influence through complex means, such as the production of an antimicrobial or a modulation of the immune system. Faecal microbial transplantation (FMT) is becoming a more commonly used approach to replenishing the GI microbiota (for reviews see Borody and Khoruts, and Floch) [Borody and Khoruts, 2011; Floch, 2012]. The aim of FMT is to reintroduce a stable community of GI microbes from a healthy donor to replace the disrupted populations in a diseased individual. In particular, FMT has been used in the treatment of recurrent Clostridium difficile infection when standard treatment has failed. FMT has been found to be successful in C. difficile treatment, with disease remission reported in up to 92% of cases [Gough et al. 2011].
In addition to being a viable therapeutic option, antibiotics can have potentially damaging effects through the perturbation of the gut microbiota. In particular, broad spectrum antibiotics can inflict significant ‘collateral damage’, as has been revealed recently by HTS technologies (for a review see Cotter and colleagues) [Cotter et al. 2012]. As a consequence, a number of investigations have focused on antimicrobials other than classical antibiotics. It is thus particularly notable that the ability to produce bacteriocins is a common feature among gut microbes. Bacteriocins are ribosomally synthesized small antimicrobial peptides produced by bacteria with either a broad or narrow spectrum and to which the producing bacterium is immune [Cotter et al. 2005]. Bacteriocins with a narrow spectrum of activity against a target microorganism can offer a therapeutic alternative to traditional antibiotics. Gut-associated bacteriocin producers also have the advantage of producing the antimicrobial in situ, and therefore, in these situations, the antimicrobial peptide is not affected by proteolysis during gastric transit or does not need to be encapsulated. Bacteriocins have been shown to be useful in controlling a number of GI pathogens in vivo, including Listeria monocytogenes [Corr et al. 2007], Salmonella spp. [Casey et al. 2004], Campylobacter jejuni [Stern et al. 2006] and C. difficile [Rea et al. 2011].
In addition to employing antimicrobials with a view to controlling pathogens in the GI tract, it has also been suggested that antimicrobials could be employed to manipulate the microbiota to treat other GI disorders, such as obesity [Murphy et al. 2013a, 2013b]. In one instance, Murphy and colleagues explored the concept of targeting the gut microbiota diet-induced obese mice through two different antimicrobial strategies with a view to, in turn, assessing the impact on obesity-associated metabolic abnormalities [Murphy et al. 2013a]. The two interventions employed involved oral administration of the antibiotic vancomycin and the Abp 118 bacteriocin-producing probiotic Lactobacillus salivarius UCC 118 respectively. Both strategies altered the gut populations in distinct ways. For example, vancomycin administration resulted in a dramatic increase in Proteobacteria levels accompanied with a decrease in the Firmicutes and Bacteroidetes phyla, but only vancomycin resulted in an improvement in the metabolic abnormalities associated with obesity. These results further highlighted the link between the gut microbiota and health, and indicate the potential benefits of using gut microbiota-manipulating strategies to improve health [Murphy et al. 2013a, 2013b].
Concluding remarks
Our gut microbiota evolves with us and plays a pivotal role in human health and disease. We now know that the resident microbiota influence host metabolism, physiology and immune system development while perturbation of the microbial community can result in chronic GI disease. While the revolution in molecular technologies has provided us with the tools necessary to more accurately study the gut microbiota, we now need to more accurately elucidate the relationships between the gut microbiota and several intestinal pathologies. Understanding the part that microbial populations play in GI disease is fundamental to the ultimate development of appropriate therapeutic approaches. The concept of altering our gut community by microbial intervention in an effort to improve GI health is currently a topic that is receiving considerable interest. The targeting of specific components of the gut microbiome will potentially allow the removal of the harmful organisms and enrich the beneficial microbes that contribute to our health.
Footnotes
Funding: Research in P.D.C. lab is supported by Science Foundation of Ireland (SFI) funded Centre for Science, Engineering and Technology, the Alimentary Pharmabiotic Centre (APC) and P.D.C. is also supported by a SFI PI award “Obesibiotics” (11/PI/1137).
Conflict of interest statement: The authors declare no conflicts of interest in preparing this article.
Contributor Information
Caitriona M. Guinane, Teagasc Food Research Centre, Moorepark, Fermoy, Co. Cork, Ireland
Paul D. Cotter, Teagasc Food Research Centre, Moorepark, Fermoy, Co. Cork, Ireland The Alimentary Pharmabiotic Centre, University College Cork, Cork, Ireland
References
- Adlerberth I., Wold A. (2009) Establishment of the gut microbiota in Western infants. Acta Paediatr 98: 229–238 [DOI] [PubMed] [Google Scholar]
- Andreasen A., Larsen N., Pedersen-Skovsgaard T., Berg R., Moller K., Svendsen K., et al. (2010) Effects of Lactobacillus acidophilus NCFM on insulin sensitivity and the systemic inflammatory response in human subjects. Br J Nutr 104: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Arthur J., Perez-Chanona E., Muhlbauer M., Tomkovich S., Uronis J., Fan T., et al. (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338: 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D., et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F., Mantovani A. (2001) Inflammation and cancer: back to Virchow? Lancet 357: 539–545 [DOI] [PubMed] [Google Scholar]
- Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., et al. (2010) Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5: e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibiloni R., Fedorak R., Tannock G., Madsen K., Gionchetti P., Campieri M., et al. (2005) VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol 100: 1539–1546 [DOI] [PubMed] [Google Scholar]
- Borody T., Khoruts A. (2011) Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol 9: 88–96 [DOI] [PubMed] [Google Scholar]
- Brint E., MacSharry J., Fanning A., Shanahan F., Quigley E. (2011) Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol 106: 329–336 [DOI] [PubMed] [Google Scholar]
- Caporaso J., Lauber C., Costello E., Berg-Lyons D., Gonzalez A., Stombaugh J., et al. (2011a) Moving pictures of the human microbiome. Genome Biol 12: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J., Lauber C., Walters W., Berg-Lyons D., Lozupone C., Turnbaugh P., et al. (2011b) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108(Suppl. 1): 4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey P., Casey G., Gardiner G., Tangney M., Stanton C., Ross R., et al. (2004) Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett Appl Microbiol 39: 431–438 [DOI] [PubMed] [Google Scholar]
- Castellarin M., Warren R., Freeman J., Dreolini L., Krzywinski M., Strauss J., et al. (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I., Blaser M. (2012) The human microbiome: at the interface of health and disease. Nat Rev Genet 13: 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M., Cusack S., O’Sullivan O., Greene-Diniz R., de Weerd H., Flannery E., et al. (2011) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 108(Suppl. 1): 4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M., Jeffery I., Conde S., Power S., O’Connor E., Cusack S., et al. (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184 [DOI] [PubMed] [Google Scholar]
- Claesson M., O’Sullivan O., Wang Q., Nikkila J., Marchesi J., Smidt H., et al. (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4: e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J., Wu H., Jayasinghe L., Patel A., Reid S., Bayley H. (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270 [DOI] [PubMed] [Google Scholar]
- Clarke S., Murphy E., Nilaweera K., Ross P., Shanahan F., O’Toole P., et al. (2012) The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes 3: 186–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corr S., Li Y., Riedel C., O’Toole P., Hill C., Gahan C. (2007) Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA 104: 7617–7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E., Lauber C., Hamady M., Fierer N., Gordon J., Knight R. (2009) Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P., Hill C., Ross R. (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3: 777–788 [DOI] [PubMed] [Google Scholar]
- Cotter P., Stanton C., Ross R., Hill C. (2012) The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov Med 13: 193–199 [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A., Barnich N., et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127: 412–421 [DOI] [PubMed] [Google Scholar]
- Duncan S., Belenguer A., Holtrop G., Johnstone A., Flint H., Lobley G. (2007) Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73: 1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P., Bik E., Bernstein C., Purdom E., Dethlefsen L., Sargent M., et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson C. (2002) Genes, microbes, and T cells – new therapeutic targets in Crohn’s disease. N Engl J Med 346: 614–616 [DOI] [PubMed] [Google Scholar]
- Floch M. (2012) The power of poop: probiotics and fecal microbial transplant. J Clin Gastroenterol 46: 625–626 [DOI] [PubMed] [Google Scholar]
- Fouhy F., Ross R., Fitzgerald G., Stanton C., Cotter P. (2012) Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes 3: 203–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C. (2007) Inflammaging as a major characteristic of old people: can it be prevented or cured? Nutr Rev 65: S173–S176 [DOI] [PubMed] [Google Scholar]
- Frank D., St Amand A., Feldman R., Boedeker E., Harpaz N., Pace N. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett W., Gallini C., Yatsunenko T., Michaud M., DuBois A., Delaney M., et al. (2010) Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8: 292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal U., Shukla R., Ghoshal U., Gwee K., Ng S., Quigley E. (2012) The gut microbiota and irritable bowel syndrome: friend or foe? Int J Inflam 2012: 151085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A., Kallstrom G., Faith J., Reyes A., Moore A., Dantas G., et al. (2011) Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA 108: 6252–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes M., Abellan J., Durban A., Perez-Cobas A., Latorre A., Moya A. (2012) Metagenomics of human microbiome: beyond 16s rDNA. Clin Microbiol Infect 18(Suppl. 4): 47–49 [DOI] [PubMed] [Google Scholar]
- Gosalbes M., Durban A., Pignatelli M., Abellan J., Jimenez-Hernandez N., Perez-Cobas A., et al. (2011) Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6: e17447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough E., Shaikh H., Manges A. (2011) Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53: 994–1002 [DOI] [PubMed] [Google Scholar]
- Grivennikov S., Greten F., Karin M. (2010) Immunity, inflammation, and cancer. Cell 140: 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S., Wang K., Mucida D., Stewart C., Schnabl B., Jauch D., et al. (2012) Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491: 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigoz Y., Dore J., Schiffrin E. (2008) The inflammatory status of old age can be nurtured from the intestinal environment. Curr Opin Clin Nutr Metab Care 11: 13–20 [DOI] [PubMed] [Google Scholar]
- Guinane C., Tadrous A., Fouhy F., Ryan C., Dempsey E., Murphy B., et al. (2013) Microbial composition of human appendices from patients following appendectomy. MBio 4:pii: e00366–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Knight R. (2009) Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project (HMP) Consortium (2012a) Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project (HMP) Consortium (2012b) A framework for the human microbiome research. Nature 486: 215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J., Salonen A., Nikkila J., Immonen O., Kekkonen R., Lahti L., et al. (2011) Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One 6(7): e23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery I., Claesson M., O’Toole P., Shanahan F. (2012a) Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol 10: 591–592 [DOI] [PubMed] [Google Scholar]
- Jeffery I., O’Toole P., Ohman L., Claesson M., Deane J., Quigley E., et al. (2012b) An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 61: 997–1006 [DOI] [PubMed] [Google Scholar]
- Jeffery I., Quigley E., Ohman L., Simren M., O’Toole P. (2012c) The microbiota link to irritable bowel syndrome: An emerging story. Gut Microbes 3: 572–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez E., Marin M., Martin R., Odriozola J., Olivares M., Xaus J., et al. (2008) Is meconium from healthy newborns actually sterile? Res Microbiol 159: 187–193 [DOI] [PubMed] [Google Scholar]
- Kadooka Y., Sato M., Imaizumi K., Ogawa A., Ikuyama K., Akai Y., et al. (2010) Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur J Clin Nutr 64: 636–643 [DOI] [PubMed] [Google Scholar]
- Kau A., Ahern P., Griffin N., Goodman A., Gordon J. (2011) Human nutrition, the gut microbiome and the immune system. Nature 474: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmeder C., de Been M., Nikkila J., Ritamo I., Matto J., Valmu L., et al. (2012) Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One 7: e29913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A., Gevers D., Pedamallu C., Michaud M., Duke F., Earl A., et al. (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22: 292–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J., Lauber C., Walters W., Parfrey L., Clemente J., Gevers D., et al. (2012) Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Itoh T., Kuwahara T., Oshima K., Toh H., Toyoda A., et al. (2007) Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14: 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J., Armougom F., Million M., Hugon P., Pagnier I., Robert C., et al. (2012) Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Larsen N., Vogensen F., van den Berg F., Nielsen D., Andreasen A., Pedersen B., et al. (2010) Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5: e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P., Hasler R., Spehlmann M., Rehman A., Zvirbliene A., Begun A., et al. (2011) Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141: 227–236 [DOI] [PubMed] [Google Scholar]
- Ley R. (2010) Obesity and the human microbiome. Curr Opin Gastroenterol 26: 5–11 [DOI] [PubMed] [Google Scholar]
- Ley R., Backhed F., Turnbaugh P., Lozupone C., Knight R., Gordon J. (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102: 11070–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R., Hamady M., Lozupone C., Turnbaugh P., Ramey R., Bircher J., et al. (2008) Evolution of mammals and their gut microbes. Science 320: 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R., Peterson D., Gordon J. (2006a) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124: 837–848 [DOI] [PubMed] [Google Scholar]
- Ley R., Turnbaugh P., Klein S., Gordon J. (2006b) Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- Li Q., Wang C., Tang C., Li N., Li J. (2012) Molecular-phylogenetic characterization of the microbiota in ulcerated and non-ulcerated regions in the patients with Crohn’s disease. PLoS One 7: e34939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus E., Jr (2004) Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 126: 1504–1517 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Borruel N., Casellas F., Guarner F. (2012) The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 9: 599–608 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., et al. (2006) Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J., Holmes E., Khan F., Kochhar S., Scanlan P., Shanahan F., et al. (2007) Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res 6: 546–551 [DOI] [PubMed] [Google Scholar]
- Martinez C., Antolin M., Santos J., Torrejon A., Casellas F., Borruel N., et al. (2008) Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol 103: 643–648 [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M., Aldeguer X., Gonzalez-Huix F., Acero D., Garcia-Gil L. (2006) Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis 12: 1136–1145 [DOI] [PubMed] [Google Scholar]
- McCoy A., Araujo-Perez F., Azcarate-Peril A., Yeh J., Sandler R., Keku T. (2013) Fusobacterium is associated with colorectal adenomas. PLoS One 8: e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh R., Dumas M., Rezzi S., Kochhar S., Holmes E., Claus S., et al. (2012) Gut microbiota modulate the metabolism of brown adipose tissue in mice. J Proteome Res 11: 620–630 [DOI] [PubMed] [Google Scholar]
- Morgan X., Tickle T., Sokol H., Gevers D., Devaney K., Ward D., et al. (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Clarke S., Marques T., Hill C., Stanton C., Ross R., et al. (2013b) Antimicrobials: strategies for targeting obesity and metabolic health? Gut Microbes 4: 48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Cotter P., Healy S., Marques T., O’Sullivan O., Fouhy F., et al. (2010) Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Murphy E., Cotter P., Hogan A., O’Sullivan O., Joyce A., Fouhy F., et al. (2013a) Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut 62: 220–226 [DOI] [PubMed] [Google Scholar]
- Nicholson J., Holmes E., Wilson I. (2005) Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol 3: 431–438 [DOI] [PubMed] [Google Scholar]
- O’Hara A., Shanahan F. (2006) The gut flora as a forgotten organ. EMBO Rep 7: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C., Bik E., DiGiulio D., Relman D., Brown P. (2007) Development of the human infant intestinal microbiota. PLoS Biol 5: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plottel C., Blaser M. (2011) Microbiome and malignancy. Cell Host Microbe 10: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy K., Choi J., Kim J., Lee S., Lee C. (2011) Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol 60: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K., Manichanh C., et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: 55–60 [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Biagi E., Heilig H., Kajander K., Kekkonen R., Tims S., et al. (2011) Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141: 1792–1801 [DOI] [PubMed] [Google Scholar]
- Rea M., Dobson A., O’Sullivan O., Crispie F., Fouhy F., Cotter P., et al. (2011) Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci USA 108(Suppl. 1): 4639–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein J., Wanunu M., Merchant C., Drndic M., Shepard K. (2012) Integrated nanopore sensing platform with sub-microsecond temporal resolution. Nat Methods 9: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossello-Mora R., Lucio M., Pena A., Brito-Echeverria J., Lopez-Lopez A., Valens-Vadell M., et al. (2008) Metabolic evidence for biogeographic isolation of the extremophilic bacterium Salinibacter ruber. ISME J 2: 242–253 [DOI] [PubMed] [Google Scholar]
- Saric J., Wang Y., Li J., Coen M., Utzinger J., Marchesi J., et al. (2008) Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J Proteome Res 7: 352–360 [DOI] [PubMed] [Google Scholar]
- Saulnier D., Riehle K., Mistretta T., Diaz M., Mandal D., Raza S., et al. (2011) Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141: 1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E., Turner S., Kasarskis A. (2010) A window into third-generation sequencing. Hum Mol Genet 19: R227–R240 [DOI] [PubMed] [Google Scholar]
- Schwiertz A., Taras D., Schafer K., Beijer S., Bos N., Donus C., et al. (2009) Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18: 190–195 [DOI] [PubMed] [Google Scholar]
- Sekirov I., Russell S., Antunes L., Finlay B. (2010) Gut microbiota in health and disease. Physiol Rev 90: 859–904 [DOI] [PubMed] [Google Scholar]
- Shanahan F. (2012) The microbiota in inflammatory bowel disease: friend, bystander, and sometime-villain. Nutr Rev 70(Suppl. 1): S31–S37 [DOI] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L., Gratadoux J., et al. (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Seksik P., Furet J., Firmesse O., Nion-Larmurier I., Beaugerie L., et al. (2009) Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189 [DOI] [PubMed] [Google Scholar]
- Sokol H., Seksik P., Rigottier-Gois L., Lay C., Lepage P., Podglajen I., et al. (2006) Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis 12: 106–111 [DOI] [PubMed] [Google Scholar]
- Stern N., Svetoch E., Eruslanov B., Perelygin V., Mitsevich E., Mitsevich I., et al. (2006) Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob Agents Chemother 50: 3111–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J., Kaplan G., Beck P., Rioux K., Panaccione R., Devinney R., et al. (2011) Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 17: 1971–1978 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Dorffel Y., Loening-Baucke V., Theissig F., Ruckert J., Ismail M., et al. (2011) Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60: 34–40 [DOI] [PubMed] [Google Scholar]
- Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J., et al. (2009) Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11: 2574–2584 [DOI] [PubMed] [Google Scholar]
- Tilg H., Kaser A. (2011) Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest 121: 2126–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V., Backhed F. (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489: 242–249 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P., Hamady M., Yatsunenko T., Cantarel B., Duncan A., Ley R., et al. (2009a) A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P., Ley R., Hamady M., Fraser-Liggett C., Knight R., Gordon J. (2007) The human microbiome project. Nature 449: 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P., Ley R., Mahowald M., Magrini V., Mardis E., Gordon J. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P., Quince C., Faith J., McHardy A., Yatsunenko T., Niazi F., et al. (2010) Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA 107: 7503–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P., Ridaura V., Faith J., Rey F., Knight R., Gordon J. (2009b) The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas U., Ranganathan N. (2012) Probiotics, prebiotics, and synbiotics: gut and beyond. Gastroenterol Res Pract 2012: 872716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley J., Ye Y. (2009) Metagenomics: facts and artifacts, and computational challenges. J Comput Sci Technol 25: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Chen J., Hoffmann C., Bittinger K., Chen Y., Keilbaugh S., et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., DiBaise J., Zuccolo A., Kudrna D., Braidotti M., Yu Y., et al. (2009) Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 106: 2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]